Abstract
Cell division depends on the correct localization of the cyclin-dependent kinases that are regulated by phosphorylation, cyclin proteolysis, and protein-protein interactions. Although immunological assays can define cell cycle protein abundance and localization, they are not suitable for detecting the dynamic rearrangements of molecular components during cell division. Here, we applied an in vivo approach to trace the subcellular localization of 60 Arabidopsis (Arabidopsis thaliana) core cell cycle proteins fused to green fluorescent proteins during cell division in tobacco (Nicotiana tabacum) and Arabidopsis. Several cell cycle proteins showed a dynamic association with mitotic structures, such as condensed chromosomes and the preprophase band in both species, suggesting a strong conservation of targeting mechanisms. Furthermore, colocalized proteins were shown to bind in vivo, strengthening their localization-function connection. Thus, we identified unknown spatiotemporal territories where functional cell cycle protein interactions are most likely to occur.
Eukaryotic cells are highly compartmentalized, and a correct localization of proteins is required for their functions (Pines, 1999). Localization data support or exclude putative protein interactions and must be taken into consideration when studying the proteome. Identification of spatiotemporal protein patterns is particularly significant for molecules involved in the cell division process, because they behave dynamically and their action is often restricted to a certain time window. Much effort has been made to unravel mechanisms governing the cell cycle progression. As a result, many key components of this machinery have been discovered. Diverse cyclins, cyclin-dependent kinases (CDKs), CDK inhibitors (CKIs), homologs of the retinoblastoma protein, and E2F/DP transcription factors have been identified in mammals, yeast, and plants (Schafer, 1998; Vandepoele et al., 2002; Bähler, 2005). The cell cycle protein families of plants are larger than those of mammals and yeast, most probably due to genome duplication events (Criqui and Genschik, 2002; Vandepoele et al., 2002; Menges et al., 2005; Sterck et al., 2007). Although the basic functional characteristics of the cell cycle machinery are conserved in all eukaryotes (Nasmyth, 1996; Novak et al., 1998), plants have acquired unique regulatory mechanisms to accommodate specialized cell divisions (Sheen and Key, 2004) and have developed unique cytoskeletal structures, the preprophase band (PPB) and the phragmoplast, to position the cell division plane and to construct the cell plate (Mineyuki, 1999; Van Damme and Geelen, 2008). Thus, the occurrence of plant-specific structures implies that proteins conserved between plants and animals might have adapted different functions and localizations. Until now, only a few studies have addressed the localization of core cell cycle regulatory proteins in plants (Supplemental Table S1).
The passage through successive cell cycle phases is controlled by CDKs (CDKA;1 in Arabidopsis [Arabidopsis thaliana]; Nigg, 1995) that are activated mainly upon binding of regulatory proteins (such as cyclins) and phosphorylation (Jeffrey et al., 1995; Russo et al., 1996). The localization of CDKs probably restricts the timing and level of their activity with respect to the specific cellular compartments and structures. The Medicago species and rice (Oryza sativa) CDKA proteins are associated with chromatin in the interphase and are located in the microtubule arrays, including the PPB, the spindle, the spindle midzone, and the phragmoplast (Stals et al., 1997; Weingartner et al., 2001). The B-type CDKs that are unique for the plant kingdom (Joubès et al., 2000) were also localized to the PPB, spindle, and phragmoplast in Medicago and rice (Mészáros et al., 2000; Lee et al., 2003).
Plant cyclins have been classified based on their sequence homology with their animal counterparts (Vandepoele et al., 2002), and in general, they execute similar functions (Breyne and Zabeau, 2001; Potuschak and Doerner, 2001). In tobacco (Nicotiana tabacum), Medicago, and Arabidopsis, the A2-type and A3-type cyclins localized to the interphase nucleus (Criqui et al., 2001; Yu et al., 2003; Imai et al., 2006). In contrast, the cytoplasmic in interphase B1-type cyclins localized to the nucleus as cells entered mitosis, associated with the chromosomes during prophase, and were destroyed after metaphase (Criqui et al., 2001). Plant A-type cyclins, with the exception of the maize (Zea mays) CYCA1;1 (Mews et al., 1997), are unstable proteins that are degraded at early stages of mitosis, prior to degradation of the B-type cyclins (Hunt et al., 1992; Mews et al., 1997; den Elzen and Pines, 2001; Geley et al., 2001).
The CDK subunit (CKS) proteins are scaffold proteins of CDKs that, in contrast to cyclins, are not required for kinase activation but are indispensable for appropriate phosphorylation activity (Tang and Reed, 1993; Bourne et al., 1996; Patra and Dunphy, 1998; Patra et al., 1999), because they serve as adaptors for targeting CDKs to mitotic substrates (Hayles et al., 1986; Tang and Reed, 1993). Animal CKS proteins function both in mitotic and meiotic progression (McKim and Hawley, 1995; Spruck et al., 2003; Pearson et al., 2005), in ubiquitin-mediated proteolysis of CKIs (Ganoth et al., 2001; Spruck et al., 2001), and in transcriptional activation (Morris et al., 2003; Yu et al., 2005). Although the function and localization of plant CKS proteins still await thorough examination, the Arabidopsis CKS proteins have been implicated in cell division control and meristem maintenance (De Veylder et al., 2001a).
CKIs and Kip-related proteins (KRPs) bind and inhibit CDK activity (Harper et al., 1993). Plant KRPs localize to the nucleus in interphase, with KRP1, KRP3, KRP4, and KRP5 displaying a subnuclear punctate pattern of distribution (Jasinski et al., 2002; Zhou et al., 2003; Weinl et al., 2005; Bird et al., 2007).
Although high-throughput fluorescent tagging localization studies of plant proteins have been initiated (Tian et al., 2004; Koroleva et al., 2005; Li et al., 2009), dynamic information about the subcellular localization of the core cell cycle players and their interacting partners is still limited. Therefore, we analyzed the subcellular localization of a set of core cell cycle proteins throughout cell division in tobacco cv Bright Yellow 2 (BY2) and Arabidopsis cells. A-type and B-type cyclins, KRPs, and CSK proteins dynamically associated with the condensed chromosomes during mitosis. The chromosome-associated KRPs directly bound the A-type cyclins, indicating a role for the KRPs in regulating mitosis besides their previously described role in G1-to-S transition. Furthermore, we show that KRP1 and KRP4 associated with CDKA;1 and that the complex localized to the chromosomes during cell division. Thus, KRPs can either act as scaffolds in depleting active CDKA;1/cyclin/CKS complexes from the cytoplasm or regulate CDK activity in the chromosomes. Additionally, five cell cycle proteins associated with the microtubule arrays, such as the PPB, allowed us to identify a putative CDK/cyclin complex and its regulatory subunits.
RESULTS
Systematic GFP Tagging of Core Cell Cycle Proteins
Open reading frames of 60 core cell cycle proteins (Supplemental Table S2) were C-terminally fused to GFP with the Gateway technology and stably expressed in tobacco BY2 cells. The selected proteins included 57 previously annotated core cell cycle proteins (Vandepoele et al., 2002) and three proteins recently implicated in cell cycle control, namely CCS52A2, CCS52B, and CDKG;2 (Fülöp et al., 2005; Menges et al., 2005). C-terminal tagging was chosen to avoid the expression of truncated protein fusions. CCS52A2 and CYCB1;2, were N-terminally tagged because the C-terminal constructs did not yield fluorescent signals (Supplemental Table S2). The functionality of C-terminal fusions of CDKA;1 and CYCA2;3 had been confirmed previously by mutant complementation analysis (Weingartner et al., 2001; Imai et al., 2006). The N-terminal fusion of CCS52A2 rescued the ccs52a2 mutant phenotype (data not shown) similarly to the C-terminal construct under the control of the endogenous promoter (Vanstraelen et al., 2009).
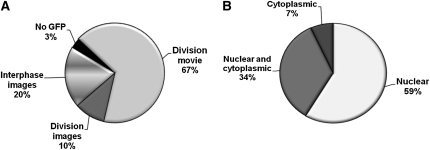
Single and time-course images were analyzed for at least two independent transformation events to ensure consistency of the observed localization patterns. Our results provide localization data for 58 (97%) of the 60 analyzed proteins. The remaining two proteins (DEL1 and DEL2; 3%) were not imaged due to the absence of fluorescent signal (Fig. 1A; Supplemental Table S2). Of the transformed constructs, 67% were followed during division by time-lapse imaging; for 10%, only single images of cell division were recorded, whereas for the remaining 20%, only interphase cells were imaged. The images and movies showing tagged proteins in interphase as well as during division were gathered in a publicly available online database (http://www.psb.ugent.be/split-gfp/localisome.html).
Figure 1.
The localization patterns of the Arabidopsis cell cycle proteins in interphase cells. A, Pie chart representing 60 analyzed proteins according to their imaging status. B, Interphase localization distribution of analyzed cell cycle proteins.
Subcellular Localization in Interphase BY2 Cells
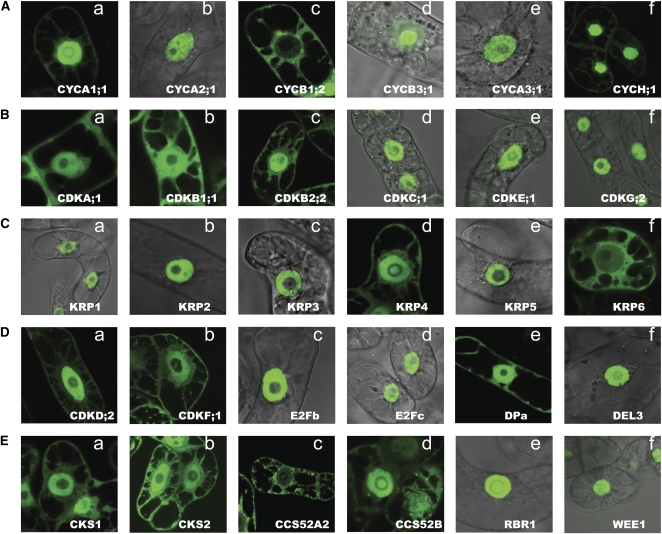
BY2 cells are very amenable to rapid cell biological analysis of cell division-related processes (Geelen and Inzé, 2001). We took advantage of these assets and initially analyzed the cell cycle fusion constructs in transformed BY2 callus cultures. The interphase BY2 cells were marked by an intact nuclear envelope and visible nucleolus in the differential interference contrast channel. The majority of the core cell cycle proteins localized exclusively to the nucleus or to both the nucleus and cytoplasm, namely 34 (59%) and 20 (34%) proteins, respectively (Fig. 1B), and four (7%) proteins localized to the cytoplasm only. As expected, cyclins showed the most variable localization patterns in interphase cells (Supplemental Fig. 1, A and B), of which 18 cyclins, including types B2, A2, and A3, were nuclear (Fig. 2, Ab, Ad, and Ae), four (CYCB1;1, CYCB1;2, CYCB1;3, and CYCB1;4) were exclusively cytoplasmic (Fig. 2Ac), and four (CYCA1;1, CYCD4;1, CYCD4;2, and CYCH;1) were present in both compartments (Fig. 2, Aa and Af; Supplemental Fig. 1B). Interestingly, besides the nuclear localization, a nucleolar localization was observed in cells expressing CYCB3;1 fusion protein (Fig. 2Ad). In spite of being driven by the 35S promoter, A-type and B-type cyclins were expressed at relatively low levels or were unstable in the interphase cells, as inferred by the weak GFP signals (data not shown).
Figure 2.
Interphase subcellular localization of selected GFP-tagged proteins representing patterns of localization in BY2. Subcellular localization is shown for cyclin proteins (A), CDKs (B), KRPs (C), CDK-activating kinases and transcription factors (D), and CKS proteins and two CCS52s, RBR1 and WEE1 (E).
The A-type and B-type CDKs were distributed in both the nucleus and the cytoplasm (Fig. 2, Ba–Bc; Supplemental Fig. 1A), in contrast to some members of the CDK families of type C, E, and G, which were predominantly nuclear (Fig. 2, Bd–Bf). The CDK-cyclin complex inhibitors ICK1/KRP1, ICK2/KRP2, ICK6/KRP3, ICK3/KRP5, and ICK5/KRP7 were exclusively nuclear (Fig. 2, Ca–Cc and Ce; Supplemental Fig. 1A), whereas ICK7/KRP4 and ICK4/KRP6 were also detected in the cytoplasm (Fig. 2, Cd and Cf). Of all nuclear KRPs, KRP1, KRP3, and KRP5 showed a more complex subcellular patterning, which included the association with distinct punctate structures inside the nucleus (Fig. 2, Ca, Cc, and Ce).
The CDK-activating kinases CDKD;2 and CDKF;1 localized to the nucleus and cytoplasm (Fig. 2, Da and Db; Supplemental Fig. 1A), whereas CDKD;1 and CDKD;3 were retained solely inside the nucleus (see images in the online database). Different localization patterns were observed for the transcription factors: the two DPs and E2Fa were nuclear and cytoplasmic (Fig. 2De), while DEL3, E2Fb, and E2Fc were exclusively nuclear (Fig. 2, Dd and Df; Supplemental Fig. 1A).
Both CDK subunits, CKS1 and CKS2, were highly and homogenously expressed in the nucleus and the cytoplasm (Fig. 2, Ea and Eb; Supplemental Fig. 1A), while the two components of the anaphase-promoting complex, CCS52A2 and CCS52B, localized to the nucleus and cytoplasm (Fig. 2, Ec and Ed), where CCS52A2, but not CCS52B associated with multiple punctae. RBR1 and WEE1 proteins localized exclusively to the nucleus (Fig. 2, Ee and Ef).
Subcellular Localization during Cell Division in BY2 Cells
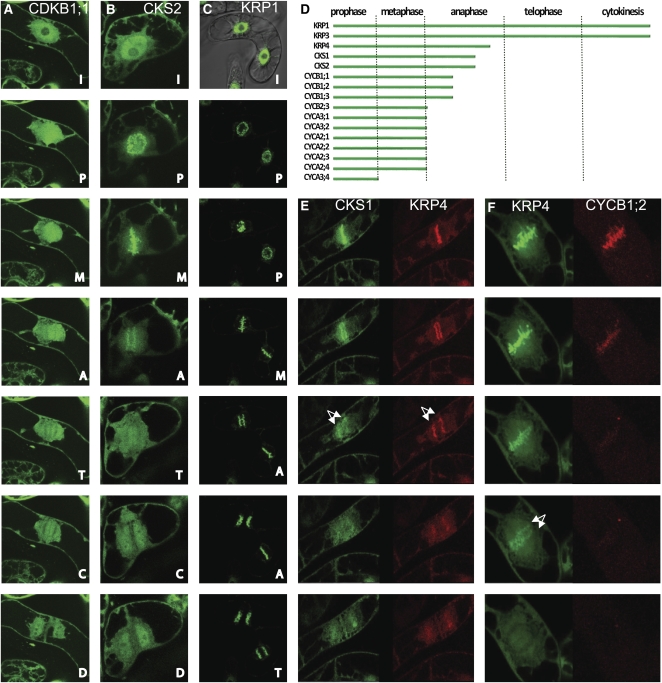
To build a more comprehensive map of the localization patterns of core cell cycle proteins, time-lapse recordings were made throughout mitosis. After nuclear envelope breakdown in late prophase, the majority of the fluorescent cell cycle proteins diffused into the cytoplasm and did not associate with any particular structures (Supplemental Table S2). After completion of cytokinesis and formation of the daughter nuclei, the localization remained as in the interphase cells (Fig. 3A). During cell division, the CDKB1;1-GFP signal occurred in the cytoplasm at the spindle position, whereas afterward the labeling diffused to the phragmoplast and was absent from the cell plate.
Figure 3.
A to C, Time-lapse analysis of subcellular localization in BY2 cells: CDKB1;1 (A), CKS2 (B), and KRP1 (C). Labels are as follows from top: I, interphase; P, prophase; M, metaphase; A, anaphase; T, telophase; C, cytokinesis; D, daughter cells after division. D, Schematic representation of the timing of protein association with chromosomes and disappearance of the GFP signal from these structures. E and F, Colocalization analysis of chromosomally associated proteins in BY cells: CKS1-GFP and KRP4-RFP (E) and KRP4-GFP and cyclin B1;2-RFP (F). Arrows indicate chromosomes in anaphase.
In contrast, a set of 16 proteins, consisting of CYCA2;1, CYCA2;2, CYCA2;3, CYCA2;4, CYCA3;1, CYCA3;2, CYCA3;4, CYCB1;1, CYCB1;2, CYCB1;3, CYCB2;3, CKS1, CKS2, KRP1, KRP3, and KRP4, localized to chromosomes in prophase, metaphase, or anaphase (Supplemental Table S2). Although these proteins localized to similar mitotic structures, different dynamic patterns were observed. For example, the fluorescent signal of the CKS2-GFP fusion (Fig. 3B) associated with the condensing chromosomes from prophase, persisted at the equatorial plane when the chromosome arms were fully condensed, and diffused during anaphase, when sister chromatids separated, whereas KRP1 (Fig. 3C) and KRP3 (see the online database) did not diffuse away from the chromosome arms after metaphase, but their chromosomal signal persisted throughout telophase and cytokinesis and remained associated with the nucleus when chromatin decondensed. Analysis of the timing of the chromosomal association and fluorescence disappearance (Fig. 3D; Supplemental Table S2) revealed that A-type and B2-type cyclins disappeared before the chromosome separation. In contrast, the fluorescent signals of B1-type cyclins, CKS proteins, and KRP4 persisted only until anaphase, and some weak fluorescence was observed in the region of the spindle during metaphase and anaphase (Fig. 3, D and E). Interestingly, CYCA3;4 was observed on condensing prophase chromatin, but this protein no longer associated with the aligned metaphase chromosomes (see online database).
To more accurately time the dynamics of the fluorescence patterning, GFP-tagged and red fluorescent protein (RFP)-tagged versions of the cell cycle proteins were coexpressed and simultaneously monitored. For example, the colocalization of KRP4 with either CKS1 or CYCB1;2 was analyzed (Fig. 3, D and E). Both pairs of fluorescent proteins associated with the condensing chromosomes; however, while the KRP4 and CKS1 fluorescence was visible until mid anaphase, the fluorescent signal of CYCB1;2 disappeared in early anaphase.
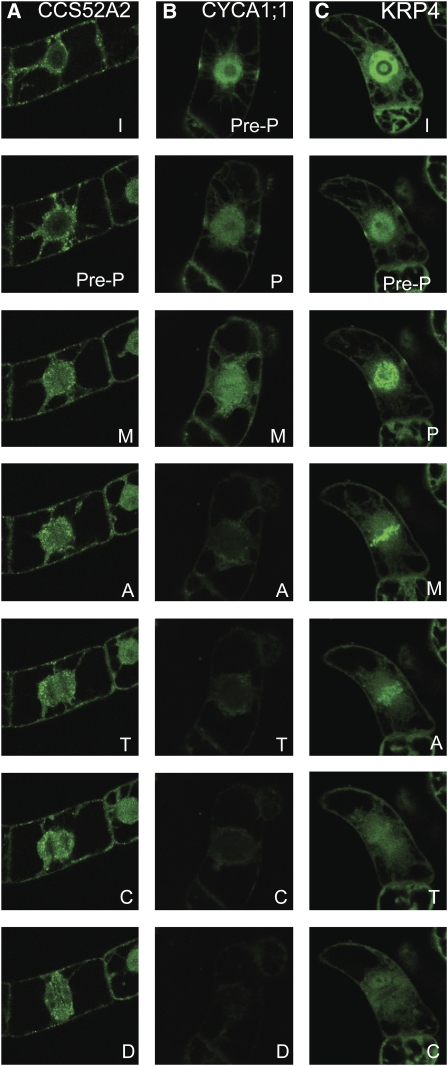
A mitosis-specific protein association, distinct from that mentioned above, was visible for CCS52A2 but not for its close homolog CCS52B (Fig. 4A). At the onset of mitosis, the fluorescent CCS52A2 fusion localized to the nucleus and to cytoplasmic punctae. In metaphase, the GFP-labeled punctae moved from the cell cortex toward the division plane and the GFP-CCS52A2 protein accumulated in the zone around and between the condensed chromosomes in metaphase. Interestingly, the CCS52A2 protein transiently accumulated at the PPB (Fig. 4A). Besides CCS52A2, several other proteins localized to the PPB, namely CYCA1;1 (Fig. 4B), KRP4 (Fig. 4C), and CKS2. All of them transiently marked the PPB zone, and while CKS2 and KRP4 later associated with the chromosomes (Figs. 3B and 4C), CYCA1;1 was excluded from the metaphase plate but was present at the spindle and disappeared before anaphase (Fig. 4B).
Figure 4.
Subcellular localization of proteins associating with the PPB: CCS52A2 (A), CYCA1;1 (B), and KRP4 (C). Labels are as follows from top: I, interphase; Pre-P, preprophase; P, prophase; M, metaphase; A, anaphase; T, telophase; C, cytokinesis; D, daughter cells after division.
In Vivo Protein Localization in Arabidopsis
To determine the localization of the core cell cycle proteins in a tissue context, all GFP fusion constructs were stably transformed in Arabidopsis (Supplemental Table S2) and homozygous lines exhibiting no macroscopic phenotypes were selected for further analysis. Several endogenous promoter open reading frame-GFP fusions were created to confirm the localization patterns observed with the 35S promoter. Interphase and dividing cells from roots and cotyledons of 2- to 4-d-old seedlings were imaged. As observed in BY2, CKS1 and CKS2 displayed identical localization patterns (Fig. 5, A and B) and accumulated in both the nucleus and the cytoplasm (Fig. 5, Aa and Ba) in interphase cells. In metaphase, the fluorescent signal associated with the chromosomes (Fig. 5, Ab and Bc). The CKS2 localization to the PPB was more pronounced in Arabidopsis than in BY2 cells (Fig. 5Bb). The specific association of CKS1 with the chromosomes was also confirmed with the endogenous promoter in Arabidopsis (Supplemental Fig. 2A). Notably, the expression level of the endogenous promoter-driven CKS1 transcript was higher than that under the control of the 35S promoter (Supplemental Fig. 2D).
Figure 5.
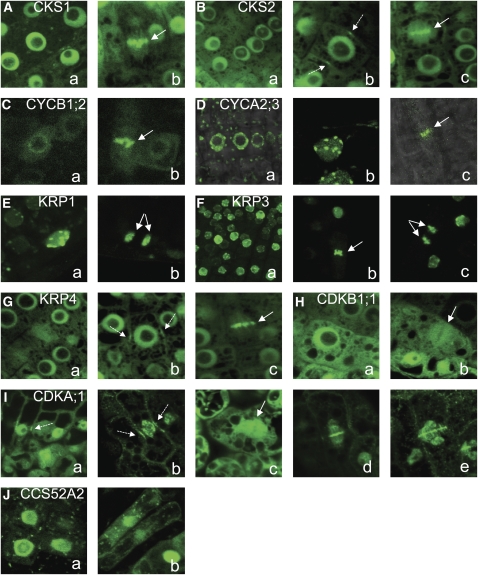
Subcellular localization of GFP-tagged cell cycle proteins driven by the 35S promoter in Arabidopsis plants. A, CKS1. B, CKS2. C, CYCB1;2. D, CYCA2;3. E, KRP1. F, KRP3. G, KRP4. H, CDKB1;1. I, CDKA;1. J, CCS52A2. Solid arrows indicate metaphase and anaphase chromosomes, and dashed arrows point to the preprophase band signal.
Similar to the observation in BY2 cells, CYCB1;2 was cytoplasmic in interphase cells (Fig. 5Ca) and associated with chromosomes during cell division (Fig. 5Cb). Interestingly, in both BY2 and Arabidopsis cells, a ring of brighter fluorescence was observed outside the nucleus prior to the division event, corresponding to either the nuclear envelope or the prophase spindle (Figs. 2Ac and 5C; see also online database).
The localization of the CYCA2;3-GFP fusion protein was studied in Arabidopsis plants upon overnight estradiol induction (Zuo et al., 2000), because plants transformed with the fusion under the control of the 35S promoter yielded a very low fluorescence. As previously reported (Imai et al., 2006), the CYCA2;3 fluorescent signal was detected exclusively in the nuclei of interphase cells, where it associated with multiple nuclear speckles (Fig. 5, Da and Db). During mitosis, CYCA2;3-GFP associated with metaphase chromosomes, and later, the signal disappeared from the separating chromatids (Fig. 5Dc).
In Arabidopsis, KRP1 and KRP3 localized to the nucleus and displayed a dotted-like pattern inside the nuclei (Fig. 5, Ea and Fa, respectively), whereas KRP4 was uniformly nuclear, cytoplasmic, and marked the PPB (Fig. 5, Ga and Gb), in agreement with the BY2 localization data. The chromosomal association of KRP1 and KRP3 persisted in anaphase and the following stages (Fig. 5, Eb and Fb), whereas the KRP4 signal was detected on metaphase chromosomes (Fig. 5Gc). The chromosomal association of KRP4 was confirmed in an Arabidopsis line expressing the KRP4-GFP fusion under the control of the endogenous promoter (Supplemental Fig. 2B) and displaying a significantly lower KRP4 transcript level when compared with the KRP4 overexpression line (Supplemental Fig. 2D).
The localization pattern of CDKB1;1 was nuclear and cytoplasmic in interphase cells (Fig. 5Ha) and diffused to the cytoplasm upon nuclear envelope breakdown. Later, it weakly associated with the phragmoplast, confirming the localization pattern in BY2 cells (Fig. 5Hb).
In interphase cells of young cotyledons, the CDKA;1-GFP signal was nuclear and cytoplasmic (Fig. 5Ia). In dividing cells, the CDKA;1 signal associated with the PPB (Fig. 5Ia), and in some cells, it persisted through later stages of mitosis (Fig. 5Ib). In prophase, CDKA;1 associated with condensing chromatin in the nucleus before the nuclear envelope broke down (Fig. 5Ib). Afterward, the protein localized to the mitotic spindle apparatus, was excluded from the metaphase plate (Fig. 5Ic), and associated with the midzone of the anaphase spindle (Fig. 5Id). Subsequently, the protein clearly marked the early and expanding phragmoplast (Fig. 5Ie). Identical localization patterns (Supplemental Fig. 1B) were observed when CDKA;1 fused to YFP was expressed in Arabidopsis under the control of its endogenous promoter (Dissmeyer et al., 2007).
In Arabidopsis, the CCS52A2 protein localized to the nucleus and the cytoplasm (Fig. 5Ja). The punctate localization pattern, although present, was less pronounced than in BY2 cells (Fig. 5Jb). Overall, the protein localization in Arabidopsis plants confirmed the patterns observed in BY2 cells.
Protein-Protein Interaction Analysis
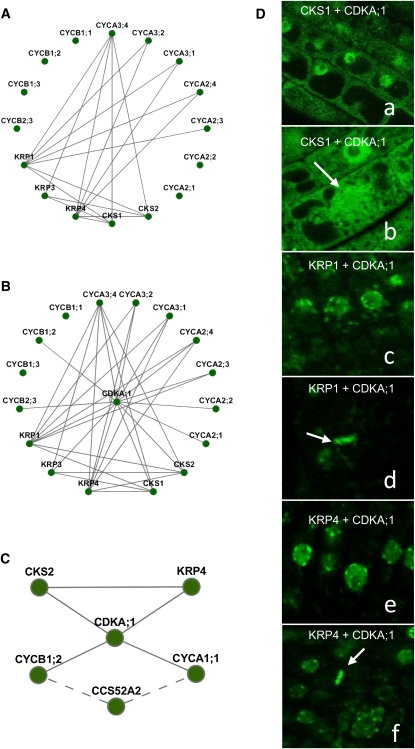
Because several core cell cycle proteins displayed overlapping localization patterns, we hypothesized that they might form functional complexes. Thus, all proteins associating with chromosomes and the PPB were tested for direct binary interactions among each other using the bimolecular fluorescence complementation (BiFC) assay in tobacco epidermal cells (Supplemental Table S3). The protein-protein interaction networks (Fig. 6, A and C) indicated 18 binary interactions among the chromosome-associated proteins and seven binary interactions among the PPB-localized proteins. Although BiFC failed to reveal a direct interaction between CCS52A2 and any of the PPB-localized proteins, CCS52A2 was included in the network because it had been detected previously in a complex with CYCA1;1 and CYCB1;2 by immunoprecipitation experiments (Fig. 6C; Fülöp et al., 2005).
Figure 6.
Binary protein-protein interaction analysis by the BiFC assay. A and B, Interaction networks of chromosomally associated proteins (A) and the PPI network of chromosomally associated proteins with CDKA;1 (B). C, Interaction network of PPB-associated proteins. Solid lines indicate the interactions detected in this study with the BiFC assay, and dashed lines depict previously published interactions (with the coimmunoprecipitation assay). D, Confocal images of Arabidopsis cells coexpressing binding protein complexes tested in the BiFC assay. Subcellular localization of complexes is as follows: CKS1 and CDKA;1 (a and b), KRP1 and CDKA;1 (c and d), KRP4 and CDKA;1 (e and f), and KRP3 and CDKA;1 (g). Arrows indicate cells in metaphase.
Because of its chromatin association (Stals et al., 1997; Weingartner et al., 2001), CDKA;1 was tested for interaction with all chromosome-associated proteins in BiFC (Supplemental Table S3). Interestingly, the incorporation of CDKA;1 into the network increased the number of edges to 33 (Fig. 6B), indirectly supporting the presence of CDKA;1 on condensed chromosomes and suggesting that the localization of the CDK component in mitosis might depend on its interacting partners. To verify this hypothesis, we analyzed Arabidopsis plants stably expressing selected binary CDK complexes (Fig. 6D). Arabidopsis plants expressing the C-terminal fragment of GFP (cGFP) fused to CDKA;1 were stably cotransformed with individual constructs containing CKS1, KRP1, or KRP4 proteins tagged with the N-terminal fragment of GFP (nGFP). CKS1 formed a complex with CDKA;1 in the nucleus and in the cytoplasm (Fig. 6D). In metaphase cells, the complex diffused to the cytoplasm and the spindle zone but did not associate with chromosomes (Fig. 6Db). KRP1 and KRP4 interacted with CDKA;1 exclusively in the nucleus and showed characteristics of the KRP-dotted subnuclear localization pattern (Fig. 6, Dc and De). Interestingly, in dividing root meristematic cells, the KRP1/CDKA;1 and KRP4/CDKA;1 complexes associated with metaphase chromosomes (Fig. 6, Dd and Df), suggesting that the localization of CDKA;1 depended on the interacting partners.
DISCUSSION
Dynamic Localization of the Arabidopsis Core Cell Cycle Proteins
Cell cycle progression is controlled by assembly and disassembly of regulatory protein complexes that influence gene transcription, posttranslational modifications, and subcellular localization. Despite the increasing knowledge on the plant cell cycle components, little is known about the spatiotemporal occurrence of different cell cycle proteins during cell division and their functions. Here, we analyzed the localization patterns of the Arabidopsis core cell cycle proteins during cell division. We used tobacco BY2 cell suspension culture (Nagata and Kumagai, 1999) as a convenient system to follow the division of single cells expressing GFP-tagged cell cycle proteins. BY2 cells have certain advantages over Arabidopsis cell culture cells, especially with regard to amenability for microscopic analysis (Geelen and Inzé, 2001). The use of strongly constitutive promoters ensured the high expression that was required for the detection and subcellular localization of the fusion proteins. Such an overexpression approach had also been used in the field of the animal cell cycle (Nguyen et al., 2002; Clay-Farrace et al., 2003). In our experiment, 97% of the transformation events resulted in BY2 calli expressing the fusion protein at a level allowing imaging. For a few constructs, a low fluorescence signal was observed, probably due to protein instability, counterselection against strongly expressing lines, or silencing. Although the localization of most of the cell cycle proteins in dividing cells of BY2 and Arabidopsis was similar, additional targeting and accumulation assays will be required to confirm their location (Millar et al., 2009).
Chromosome-Associated Arabidopsis Core Cell Cycle Proteins
Our localization analysis showed that the Arabidopsis cyclins of types B1, B2, A2, and A3 associated with a common structure, the chromosomes. In all cases, the B-type cyclins persisted longer during cell division than did the A-type cyclins. Similarly, animal A-type and B-type cyclins are recruited to chromatin at specific times in the cell cycle to mediate different transitions (Pines and Hunter, 1991; Cardoso et al., 1993; Kim and Kaelin, 2001; Frouin et al., 2002). In addition, mammalian B-type cyclins might specifically function in chromosome condensation, as supported by the fact that the cyclinB/Cdk complex from frog (Xenopus laevis) induced this process (Shimada et al., 1998). Although the function of the mammalian cyclinB/Cdk complexes cannot simply be extrapolated to plants, the chromosomal localization of some Arabidopsis A-type and B-type cyclins and their interaction with CDKA;1 can suggest that in plants similar complexes exhibit chromosome-associated functions. Both animal and plant A-type cyclins are expressed and degraded earlier than the B-type cyclins (Geley et al., 2001; Menges et al., 2005) to exert their prometaphase role (Lehner and O'Farrell, 1990; Guadagno and Newport, 1996; Gong et al., 2007). It is proposed that human cyclin A functions as a part of the machinery that activates the CYCB/CDC2 (Fung et al., 2007). In plants, it remains to be clarified whether and which A-type cyclin acts upstream of the B-type cyclin complex at the onset of mitosis and whether the functions of the A-type and B-type cyclin family members are redundant in their functions.
The two Arabidopsis CKS proteins displayed identical localization patterns in both interphase and dividing cells. However, while Arabidopsis CKS2 is G2/M specific, the transcript of CKS1 is not cell cycle regulated (Menges et al., 2005), suggesting different nonredundant functions. Similarly, the two animal Cks proteins differ in function, as Cks1 facilitated the ubiquitin-mediated proteolysis of CKI (Ganoth et al., 2001; Spruck et al., 2001), while its close homolog Cks2 regulates spindle morphogenesis and chromosome alignment (McKim and Hawley, 1995; Spruck et al., 2003; Pearson et al., 2005). The colocalization of Arabidopsis CKS proteins with A-type and B-type cyclins at the chromosomes in prophase and prometaphase might hint at a functional interaction at the onset of mitosis. Although we did not detect a direct binding between these proteins in the BiFC assay, the interaction might require a CDK component, in agreement with previously published work (De Veylder et al., 1997). Alternatively, CKS might be involved in the KRP-mediated inhibition of the cyclin/CDK complexes, as observed in animals, where phosphorylation of the APC components and activation of the degradation machinery depend on the Cks binding to the cyclinB/CDK complexes (Patra and Dunphy, 1998; Shteinberg et al., 1999; Rudner and Murray, 2000). Because CKS proteins directly bind KRPs, it is not excluded that CKS proteins directly activate KRP degradation in plants, but this remains to be proven experimentally.
In interphase cells, all C-terminally GFP-tagged KRPs localized to the nucleus. As described before, KRP1, KRP3, and KRP5 localize to subnuclear speckles (Jasinski et al., 2002; Zhou et al., 2003; Weinl et al., 2005; Bird et al., 2007). Unexpectedly, KRP1, KRP3, and KRP4 localized to metaphase and anaphase chromosomes. In dividing cells, the KRP fluorescence persisted during mitosis, with the exception of KRP4, of which the fluorescent signal disappeared from separating chromosomes during anaphase. The seven Arabidopsis KRPs have diverse structures, with regard to the CDK consensus phosphorylation site, nuclear localization signals, and PEST domains (De Veylder et al., 2001b); therefore, their different localization patterns are plausible. We do not exclude that the position of the GFP tag influenced the localization. For example, the C-terminal GFP fusions of KRP4 and KRP6 displayed an additional cytoplasmic localization, in contrast to data obtained with N-terminal fusions (Bird et al., 2007). The results of our BiFC experiment showed that all KRPs, when tagged C terminally, directly bound CKS proteins and A-type cyclins, indicating a role for these KRPs in regulating mitosis, in addition to their previously described role in the G1-to-S transition. Furthermore, we show that KRP1 and KRP4 mediated the association of CDKA;1 with the chromosomes during mitosis. We hypothesize that KRPs either bind to the CDK/cyclin/CKS complex to modulate its activity in a concentration-dependent fashion or act as scaffolds to deplete active CDKA;1/cyclin/CKS complexes from the cytoplasm. Interestingly, in fruit fly (Drosophila melanogaster), a specific Cdk/cyc inhibitor, Roughex, can stimulate Cdk/cyc activity when present at low concentrations (Foley et al., 1999; Foley and Sprenger, 2001). We cannot exclude that the three chromosome-localized KRPs display specific functions, as they are expressed in diverse tissues (Wang et al., 1998, 2008; De Veylder et al., 2001b; Jasinski et al., 2002; Ormenese et al., 2004). Interestingly, misexpression of the KRP1 gene in Arabidopsis trichomes induced cell death (Schnittger et al., 2003), in the same manner as the cytoplasmic Cip/Kip proteins mediated the apoptotic process after DNA damage in animals (Gorospe et al., 1996, 1997; Bunz et al., 1998).
The PPB-Associated Arabidopsis Core Cell Cycle Proteins
The PPB is a transient microtubule array that demarcates the future cortical division zone, where the cell will separate into two daughter cells (Van Damme and Geelen, 2008; Müller et al., 2009). It is dismantled in prophase but leaves a molecular landmark behind, which later guides the phragmoplast and a forming cell plate (Smith, 2001). Some nuclear factors are thought to be involved in the regulation of PPB formation, because the PPB encircles the nucleus and only in this configuration can it be degraded as cells progress through prophase (Mineyuki et al., 1991). Under normal conditions, the PPB of BY2 and Arabidopsis is short lived and occurs during early prophase to disappear prior to nuclear envelope breakdown. Maize CDKA and CYCB1;2 have been immunodetected at narrow PPB structures, suggesting that PPB associates just before its disintegration. Hence, it was speculated that CDKA;1 regulates the PPB microtubule stability (John et al., 2001). Although the CDKA;1/CYCB kinase complex causes rapid disassembly of the PPB in hair cells of spider lily (Tradescantia virginiana; Hush et al., 1996), other cyclins and different kinases might also regulate the PPB dynamics. We found that several cell cycle proteins associate with the PPB: CYCA1;1, CCS52A2, CKS2, and KRP4. CYCA1;1 interacted with CDKA;1 and with CCS52A2 (Fülöp et al., 2005), and both CKS2 and KRP4 formed a complex with CDKA;1 (our results). A possible scenario could be that the PPB-localized KRP4 keeps the CDKA;1/CYCA1;1 and/or CDKA;1/CYCB1;2 complexes inactive until required, whereas CKS2 is responsible for promoting the kinase activity on preprophase substrates. The presence of CCS52A2 in this structure implies a kinase regulation mechanism, dependent on the local cyclin degradation. The CDKA;1/CYCA1;1 complex could regulate the microtubule dynamics by phosphorylating microtubule-associated proteins (MAPs). For instance, the Xenopus microtubule-stabilizing factor XMAP215/MOR1 protein associated with Cdc2/cyclinB and lost its ability to promote tubulin polymerization when phosphorylated, leading to microtubule rearrangements (Charrasse et al., 1998; Tournebize et al., 2000). In plants, no MOR1-interacting proteins have been found so far (Kawamura et al., 2006). Similarly, MAP4 mediates the interaction of Cdc2/cyclinB binding to microtubules in starfish (Asterina pectinifera) oocytes, but this protein has not been isolated in plants (Ookata et al., 1995). We observed CYCB1;2 and CDKA;1 on the spindle, where they could play a role as a complex. Medicago CDKA;2 and tubulin copurify with microtubules from mitotic cell extracts, suggesting that plant CDKA proteins might regulate microtubule organization throughout mitosis (Weingartner et al., 2001). However, the plant microtubule-targeting mechanisms for the CDKA;1/cyclin complex and the cell cycle-cytoskeleton link in preprophase as well remain unknown.
MATERIALS AND METHODS
Fusion Constructs
The set of 60 cell cycle genes was cloned into the Gateway entry vector pDONR221 (Karimi et al., 2007). The recombination reactions were done according to the manufacturer's instructions (Invitrogen). The GFP fusion constructs carried a kanamycin resistance gene, whereas those with RFP were selectable on hygromycin. The p35S-driven GFP fusion constructs were generated in the destination vectors pK7FWG2 and pK7WGF2 for C-terminal and N-terminal fusions, respectively. RFP fusions were constructed similarly (in the destination vector pH7RWG2 for C-terminal fusions), except that GFP was substituted with RFP in the destination vector. The split GFP fusions were generated as described (Boudolf et al., 2009). Binary vectors were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation.
Plant Growth Conditions and Transformations
Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were grown under long-day conditions (16 h of light, 8 h of darkness) at 22°C on half-strength Murashige and Skoog germination medium. Plants were transformed by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on kanamycin-containing half-strength Murashige and Skoog solid medium. The split GFP lines with expression pairs of interacting proteins were created by supertransformation of (kanamycin-resistant) CDKA;1-cGFP or CDKB1;1-cGFP lines with (hygromycin-resistant) nGFP-tagged interactor constructs.
Stable tobacco (Nicotiana tabacum ‘Bright Yellow 2’) transformation was carried out as described (Geelen and Inzé, 2001). Several independently transformed calli were imaged for each construct.
For colocalization studies, a few calli of chosen constructs were selected. BY2 callus harboring the GFP-tagged protein fusion was resuspended in the liquid medium and then supertransformed with other RFP-tagged proteins. Positive double transformants were selected on a medium enriched for hygromycin.
Expression Analysis
Fresh Arabidopsis seedlings were ground. The RNA was extracted with the RNeasy kit (Qiagen). The RNA concentration was adjusted, and 1 μg of total RNA was used for cDNA amplification (SuperScript RNA polymerase III; Invitrogen). Quantitative PCR was done with the following primer pair for GFP amplification: 5′-GACAAGCAGAAGAACGGCATCAAGGTGA-3′ and 5′-CTTGTACAGCTCGTCCATGCCGAGAGTG-3′.
Confocal Imaging
Fluorescence was analyzed with a confocal microscope (LSM 510 version 3.2 [Zeiss] or FluoView FV1000 [Olympus]), both equipped with a 63× water-corrected objective (numerical aperture of 1.2) to scan cells. Dual GFP and RFP fluorescence was sequentially imaged in a multichannel setting with 488- and 543-nm light for GFP and RFP excitation, respectively. Emission fluorescence was captured in the frame-scanning mode, alternating GFP and RFP fluorescence via a 500- to 550-nm bandpass emission filter and a 560-nm cutoff filter, respectively. Images were recorded at 2× to 8× digital zoom. When no GFP fluorescence could be detected, instability of the fusion protein or lack of expression due to silencing or counterselection were assumed (Joubès et al., 2004). GFP-positive calli and Arabidopsis seedlings were further analyzed.
For live-cell recordings, samples were applied to a chambered cover glass (Lab-Tek) and immobilized in a thin layer of 200 μL of BY2 medium containing vitamins and 0.8% low-melting-point agar (Invitrogen). Cells were monitored over the course of several hours and imaged every 3 min (approximately 200 images).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of the localization patterns among the Arabidopsis core cell cycle proteins in interphase.
Supplemental Figure S2. Subcellular localization patterns of selected proteins expressed under the control of their endogenous promoters.
Supplemental Table S1. Overview of the Arabidopsis core cell cycle proteins and their subcellular localization.
Supplemental Table S2. Literature search on the localization of plant core cell cycle proteins.
Supplemental Table S3. Protein-protein interaction screen of the Arabidopsis core cell cycle proteins by the BiFC assay in tobacco epidermal cells.
Supplementary Material
Acknowledgments
We thank Raimundo Villarroel and Jan Zethof for help at the initial stages of this project, Lieven De Veylder for constructive suggestions and critical reading of the manuscript, and Martine De Cock for help in preparing it.
This work was supported by the European Union-Human Resources and Mobility for an Early Stage Training (grant no. MEST–CT–2004–514632 to J.B.) and the Research Foundation-Flanders (postdoctoral fellowship to D.V.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eugenia Russinova (eugenia.russinova@psb.vib-ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bähler J (2005) Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet 39 69–94 [DOI] [PubMed] [Google Scholar]
- Bird DA, Buruiana MM, Zhou Y, Fowke LC, Wang H (2007) Arabidopsis cyclin-dependent kinase inhibitors are nuclear-localized and show different localization patterns within the nucleoplasm. Plant Cell Rep 26 861–872 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E, et al (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, Watson MH, Hickey MJ, Holmes W, Rocque W, Reed SI, Tainer JA (1996) Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell 84 863–874 [DOI] [PubMed] [Google Scholar]
- Breyne P, Zabeau M (2001) Genome-wide expression analysis of plant cell cycle modulated genes. Curr Opin Plant Biol 4 136–142 [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282 1497–1501 [DOI] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H, Nadal-Ginard B (1993) Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74 979–992 [DOI] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C (1998) The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci 111 1371–1383 [DOI] [PubMed] [Google Scholar]
- Clay-Farrace L, Pelizon C, Santamaria D, Pines J, Laskey RA (2003) Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J 22 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Genschik P (2002) Mitosis in plants: how far we have come at the molecular level? Curr Opin Plant Biol 5 487–493 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Weingartner M, Capron A, Parmentier Y, Shen WH, Heberle-Bors E, Bögre L, Genschik P (2001) Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J 28 569–581 [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J (2001) Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol 153 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001. a) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beemster GTS, Beeckman T, Inzé D (2001. b) CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J 25 617–626 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Segers G, Glab N, Casteels P, Van Montagu M, Inzé D (1997) The Arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett 412 446–452 [DOI] [PubMed] [Google Scholar]
- Dissmeyer N, Nowack MK, Pusch S, Stals H, Inzé D, Grini PE, Schnittger A (2007) T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O'Farrell PH, Sprenger F (1999) Rux is a cyclin-dependent kinase inhibitor (CKI) specific for mitotic cyclin-Cdk complexes. Curr Biol 9 1392–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, Sprenger F (2001) The cyclin-dependent kinase inhibitor Roughex is involved in mitotic exit in Drosophila. Curr Biol 11 151–160 [DOI] [PubMed] [Google Scholar]
- Frouin I, Montecucco A, Biamonti G, Hübscher U, Spadari S, Maga G (2002) Cell cycle-dependent dynamic association of cyclin/Cdk complexes with human DNA replication proteins. EMBO J 21 2485–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülöp K, Tarayre S, Kelemen Z, Horváth G, Kevei Z, Nikovics K, Bakó L, Brown S, Kondorosi A, Kondorosi E (2005) Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4 1084–1092 [PubMed] [Google Scholar]
- Fung TK, Ma HT, Poon RYC (2007) Specialized roles of the two mitotic cyclins in somatic cells: cyclin A as an activator of M phase-promoting factor. Mol Biol Cell 18 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A (2001) The cell-cycle regulatory protein Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nat Cell Biol 3 321–324 [DOI] [PubMed] [Google Scholar]
- Geelen DNV, Inzé DG (2001) A bright future for the Bright Yellow-2 cell culture. Plant Physiol 127 1375–1379 [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T (2001) Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol 153 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Pomerening JR, Myers JW, Gustavsson C, Jones JT, Hahn AT, Meyer T, Ferrell JE Jr (2007) Cyclin A2 regulates nuclear-envelope breakdown and the nuclear accumulation of cyclin B1. Curr Biol 17 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi MC, Holbrook NJ (1997) p21Waf1/Cip1 protects against p53-mediated apoptosis of human melanoma cells. Oncogene 14 929–935 [DOI] [PubMed] [Google Scholar]
- Gorospe M, Wang X, Guyton KZ, Holbrook NJ (1996) Protective role of p21Waf1/Cip1 against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol Cell Biol 16 6654–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadagno TM, Newport JW (1996) Cdk2 kinase is required for entry into mitosis as a positive regulator of Cdc2-cyclin B kinase activity. Cell 84 73–82 [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75 805–816 [DOI] [PubMed] [Google Scholar]
- Hayles J, Aves S, Nurse P (1986) suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. EMBO J 5 3373–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Luca FC, Ruderman JV (1992) The requirements for protein synthesis and degradation, and the control of destruction of cyclins A and B in the meiotic and mitotic cell cycles of the clam embryo. J Cell Biol 116 707–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hush J, Wu L, John PCL, Hepler LH, Hepler PK (1996) Plant mitosis promoting factor disassembles the microtubule preprophase band and accelerates prophase progression in Tradescantia. Cell Biol Int 20 275–287 [DOI] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18 382–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Perennes C, Bergounioux C, Glab N (2002) Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiol 130 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP (1995) Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376 313–320 [DOI] [PubMed] [Google Scholar]
- John PCL, Mews M, Moore R (2001) Cyclin/cdk complexes: their involvement in cell cycle progression and mitotic division. Protoplasma 216 119–142 [DOI] [PubMed] [Google Scholar]
- Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudin JP (2000) CDK-related protein kinases in plants. Plant Mol Biol 43 607–620 [DOI] [PubMed] [Google Scholar]
- Joubès J, De Schutter K, Verkest A, Inzé D, De Veylder L (2004) Conditional, recombinase-mediated expression of genes in plant cell cultures. Plant J 37 889–896 [DOI] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E, Himmelspach R, Rashbrooke MC, Whittington AT, Gale KR, Collings DA, Wasteneys GO (2006) MICROTUBULE ORGANIZATION1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol 140 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TY, Kaelin WG Jr (2001) Differential control of transcription by DNA-bound cyclins. Mol Biol Cell 12 2207–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH (2005) High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J 41 162–174 [DOI] [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, Hashimoto J, Tsutsumi N, Uchimiya H, Umeda M (2003) Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J 34 417–425 [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH (1990) The roles of Drosophila cyclins A and B in mitotic control. Cell 61 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Park E, von Arnim AG, Nebenführ A (2009) The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Hawley RS (1995) Chromosomal control of meiotic cell division. Science 270 1595–1601 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41 546–566 [DOI] [PubMed] [Google Scholar]
- Mészáros T, Miskolczi P, Ayaydin F, Pettkó-Szandtner A, Peres A, Magyar Z, Horváth GV, Bakó L, Fehér A, Dudits D (2000) Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells. Plant Mol Biol 43 595–605 [DOI] [PubMed] [Google Scholar]
- Mews M, Sek FJ, Moore R, Volkmann D, Gunning BES, John PCL (1997) Mitotic cyclin distribution during maize cell division: implications for the sequence diversity and function of cyclins in plants. Protoplasma 200 128–145 [Google Scholar]
- Millar AH, Carrie C, Pogson B, Whelan J (2009) Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineyuki Y (1999) The preprophase band of microtubules: its function as a cytokinetic apparatus in higher plants. Int Rev Cytol 187 1–50 [Google Scholar]
- Mineyuki Y, Marc J, Palevitz BA (1991) Relationship between the preprophase band, nucleus and spindle in dividing Allium cotyledon cells. J Plant Physiol 138 640–649 [Google Scholar]
- Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI (2003) Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423 1009–1013 [DOI] [PubMed] [Google Scholar]
- Müller S, Wright AJ, Smith LG (2009) Division plane control in plants: new players in the band. Trends Cell Biol 19 180–188 [DOI] [PubMed] [Google Scholar]
- Nagata T, Kumagai F (1999) Plant cell biology through the window of the highly synchronized tobacco BY-2 cell line. Methods Cell Sci 21 123–127 [DOI] [PubMed] [Google Scholar]
- Nasmyth K (1996) At the heart of the budding yeast cell cycle. Trends Genet 12 405–412 [DOI] [PubMed] [Google Scholar]
- Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik-Rogers J, Pines J, Wolgemuth DJ, Koff A (2002) Characterization and expression of mammalian cyclin B3, a prepachytene meiotic cyclin. J Biol Chem 277 41960–41969 [DOI] [PubMed] [Google Scholar]
- Nigg EA (1995) Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays 17 471–480 [DOI] [PubMed] [Google Scholar]
- Novak B, Csikasz-Nagy A, Gyorffy B, Nasmyth K, Tyson JJ (1998) Model scenarios for evolution of the eukaryotic cell cycle. Philos Trans R Soc Lond B Biol Sci 353 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T (1995) Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J Cell Biol 128 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormenese S, de Almeida Engler J, De Groodt R, De Veylder L, Inzé D, Jacqmard A (2004) Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann Bot (Lond) 93 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D, Dunphy WG (1998) Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes Dev 12 2549–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra D, Wang SX, Kumagai A, Dunphy WG (1999) The Xenopus Suc1/Cks protein promotes the phosphorylation of G2/M regulators. J Biol Chem 274 36839–36842 [DOI] [PubMed] [Google Scholar]
- Pearson NJ, Cullen CF, Dzhindzhev NS, Ohkura H (2005) A pre-anaphase role for a Cks/Suc1 in acentrosomal spindle formation of Drosophila female meiosis. EMBO Rep 6 1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J (1999) Four-dimensional control of the cell cycle. Nat Cell Biol 1 E73–E79 [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T (1991) Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol 115 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4 501–506 [DOI] [PubMed] [Google Scholar]
- Rudner AD, Murray AW (2000) Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol 149 1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Pavletich NP (1996) Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol 3 696–700 [DOI] [PubMed] [Google Scholar]
- Schafer KA (1998) The cell cycle: a review. Vet Pathol 35 461–478 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schöbinger U, Hülskamp M (2003) Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Key S (2004) Cell signalling and gene regulation: exploring new functions and actions of regulatory molecules. Curr Opin Plant Biol 7 487–490 [Google Scholar]
- Shimada A, Ohsumi K, Kishimoto T (1998) An indirect role for cyclin B-Cdc2 in inducing chromosome condensation in Xenopus egg extracts. Biol Cell 90 519–530 [PubMed] [Google Scholar]
- Shteinberg M, Protopopov Y, Listovsky T, Brandeis M, Hershko A (1999) Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem Biophys Res Commun 260 193–198 [DOI] [PubMed] [Google Scholar]
- Smith LG (2001) Plant cell division: building walls in the right places. Nat Rev Mol Cell Biol 2 33–39 [DOI] [PubMed] [Google Scholar]
- Spruck C, Strohmaier H, Watson M, Smith APL, Ryan A, Krek W, Reed SI (2001) A CDK-independent function of mammalian Cks1: targeting of SCFSkp2 to the CDK inhibitor p27Kip1. Mol Cell 7 639–650 [DOI] [PubMed] [Google Scholar]
- Spruck CH, de Miguel MP, Smith APL, Ryan A, Stein P, Schultz RM, Lincoln AJ, Donovan PJ, Reed SI (2003) Requirement of Cks2 for the first metaphase/anaphase transition of mammalian meiosis. Science 300 647–650 [DOI] [PubMed] [Google Scholar]
- Stals H, Bauwens S, Traas J, Van Montagu M, Engler G, Inzé D (1997) Plant CDC2 is not only targeted to the pre-prophase band, but also co-localizes with the spindle, phragmoplast, and chromosomes. FEBS Lett 418 229–234 [DOI] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Vandepoele K, Rouzé P, Van de Peer Y (2007) How many genes are there in plants (…and why are they there)? Curr Opin Plant Biol 10 199–203 [DOI] [PubMed] [Google Scholar]
- Tang Y, Reed SI (1993) The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev 7 822–832 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol 2 13–19 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Geelen D (2008) Demarcation of the cortical division zone in dividing plant cells. Cell Biol Int 32 178–187 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E (2009) APC/CCCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA 106 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15 501–510 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Bird DA, Fowke LC (2008) Functions, regulation and cellular localization of plant cyclin-dependent kinase inhibitors. J Microsc 231 234–246 [DOI] [PubMed] [Google Scholar]
- Weingartner M, Binarova P, Drykova D, Schweighofer A, David JP, Heberle-Bors E, Doonan J, Bögre L (2001) Dynamic recruitment of cdc2 to specific microtubule structures during mitosis. Plant Cell 13 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJH, Nowack MK, Jakoby MJ, Hülskamp M, Schnittger A (2005) Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17 1704–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VPCC, Baskerville C, Grünenfelder B, Reed SI (2005) A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol Cell 17 145–151 [DOI] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li G, Brandizzi F, Fowke LC, Wang H (2003) The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant J 35 476–489 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.