Abstract
RNA editing in plants alters specific nucleotides from C to U in mRNAs in plastids and in mitochondria. I here characterize the nuclear gene MITOCHONDRIAL EDITING FACTOR9 (MEF9) that is required for RNA editing of the site nad7-200 in the nad7 mitochondrial mRNA in Arabidopsis (Arabidopsis thaliana). The MEF9 protein belongs to the E subfamily of pentatricopeptide repeat proteins and unlike the three previously identified mitochondrial editing factors MEF1 and MEF11 in Arabidopsis and OGR1 in rice (Oryza sativa) does not contain a DYW C-terminal domain. In addition, the E domain is incomplete, but seems to be functionally required, since one of the two independent EMS mutants encodes a MEF9 protein truncated by a stop codon at the beginning of the E domain. In both mutant plants premature stop codons in MEF9 inactivate RNA editing at site nad7-200. The homozygous mutant plants are viable and develop rather normally. The lack of RNA editing at site nad7-200 thus seems to be tolerated although this editing event is conserved in most plant species or the genomic sequence already codes for a T at this position, resulting in a generally conserved amino acid codon.
RNA editing in flowering plants has been documented in mitochondria where about 450 and in plastids where 30 to 40 selected cytosines are changed to uridines in mRNAs and in some tRNAs (Giegé and Brennicke, 1999; Handa, 2003; Sasaki et al., 2006; Shikanai, 2006; Takenaka et al., 2008). So far the enzyme(s) involved in the biochemical reaction have not yet been identified, but several nuclear-encoded proteins required for RNA editing at specific sites have been characterized. All of these specific factors are pentatricopeptide repeat (PPR) proteins, a family of about 450 proteins composed of a variable number of repeats of about 35 amino acids and additional domains that latter group these proteins by structure but not by function (Small and Peeters, 2000; Schmitz-Linneweber and Small, 2008). Most of these PPR proteins are predicted to be targeted to mitochondria and/or plastids (Lurin et al., 2004).
In plastids, several such nuclear factors for specific RNA editing events have been identified, some of which are members of the class of DYW-PPR proteins, others are E-class PPR proteins that do not contain the DYW C terminus (Kotera et al., 2005; Shikanai, 2006; Okuda et al., 2007, 2008; Chateigner-Boutin et al., 2008; Zhou et al., 2008; Robbins et al., 2009; Yu et al., 2009). In plant mitochondria, so far three such factors have been described, all with DYW extensions (Kim et al., 2009; Verbitskiy et al., 2009; Zehrmann et al., 2009). The DYW extension has been found to contain a signature characteristic of zinc-containing cytidine deaminases and has consequently been proposed to be potentially involved in the enzymatic reaction of the C to U RNA editing in the two plant organelles (Salone et al., 2007). An in vitro investigation of a DYW-PPR protein (At2g02980), however, revealed an endonuclease activity rather than the catalysis of a deamination reaction (Nakamura and Sugita, 2008). The in vivo function of this protein (At2g02980) is, however, not known so far and the endonuclease activity may be specific to this moiety. Furthermore, in the CRR22 and CRR28 plastid factors the DYW domains are not needed for RNA editing in vivo, since deletion of these regions does not detract from their function (Okuda et al., 2009).
Since several PPR proteins required for specific editing events in plastids are classified as containing E extensions but no DYW similarity, additional factors are being postulated for the enzymatic activity. In this scenario, a PPR protein recognizes a specific RNA sequence motif, binds there, and recruits through protein-protein interactions one or more additional proteins including one with the enzymatic activity for the deamination step (Kotera et al., 2005; Okuda et al., 2007).
Analysis of the RNA-binding properties of the CRR4 protein showed that this polypeptide specifically recognizes an RNA region extending about 20 to 30 nucleotides upstream and 10 nucleotides downstream of its cognate editing site in the translation initiation codon of the ndhD gene (Okuda et al., 2006). The size and extent of this region is in the range of the specific cis-regions delineated by in vitro assays for several plastid editing sites (Chaudhuri and Maliga, 1996; Hirose and Sugiura, 2001; Miyamoto et al., 2002; Sasaki et al., 2006). Recognition of mitochondrial editing sites seems to occur through similar cis-element motifs that in the last few years have been characterized by in organello and in vitro analyses of several such sites (Farré et al., 2001; Neuwirt et al., 2005; van der Merwe et al., 2006; Takenaka et al., 2008).
The number of specific sites addressed by the trans-acting PPR proteins varies. Several of the plastid RNA editing factors presently identified address single sites, others of these PPR proteins seem to be required for editing of two sites in different plastid mRNAs and only one is needed at three sites in this organelle (Kotera et al., 2005; Okuda et al., 2007; Chateigner-Boutin et al., 2008). Tentative connections between editing sites in plastids had been postulated from transgenic and in vitro assays, in which introduction of an additional editing site into the plastid system lowered RNA editing at other sites in trans (Kobayashi et al., 2007; Heller et al., 2008).
In mitochondria, the three PPR proteins identified to be required for specific RNA editing events, MITOCHONDRIAL EDITING FACTOR1 (MEF1), MEF11, and OGR1, all appear to recognize and bind to several RNA motifs present at least at three different editing sites (Kim et al., 2009; Verbitskiy et al., 2009; Zehrmann et al., 2009). In mutants of the OGR1 factor, seven editing events are affected (Kim et al., 2009). The different degrees of residual editing at two of these sites suggest that these are indirectly influenced by the ogr1-induced loss of an upstream event, which would leave five sites as primary targets including the one upstream of the other two sites.
Judging by the sheer numbers, it seems to be necessary that at least in mitochondria a given PPR protein will have to address several editing events. Even the large group of PPR proteins with 450 members in plants would not be able to provide one specific protein dedicated to each of the 400 to 500 editing events, particularly since several of the PPR proteins have been shown to be active also in other posttranscriptional processes in mitochondria as well as in plastids. Some of the PPR proteins are required for specific intron splicing events, some are required for processing of multicistronic pre-mRNAs, for stabilization of pre-tRNAs, and yet others have been implicated in translation of specific genes in plastids (Beick et al., 2008; Williams-Carrier et al., 2008; for review, see Delannoy et al., 2007; Schmitz-Linneweber and Small, 2008). In mitochondria, a PPR protein has been found to be involved in an intron splicing event (de Longevialle et al., 2007). Taking into account these multiple functions of PPR proteins in various RNA maturation processes, the 450 genes for PPR proteins can only accommodate addressing the more than 450 editing sites in mitochondria plus the more than 30 sites in plastids in addition to the various processing and splicing events required in both organelles if at least in RNA editing multiple sites are addressed by a given protein. This problem is further exaggerated in plant species where more than 1,000 editing events are required for functional open reading frames such as in the quillwort Isoetes engelmanii (Grewe et al., 2009).
I here identify the trans-factor MEF9 for RNA editing in plant mitochondria that is required for at least one editing event in the nad7 mRNA. This PPR protein does not contain a DYW domain and only a partial E domain. Accordingly this protein may act as a specificity factor and may require one or more further proteins for functional RNA editing at the target site.
RESULTS
Identification of Mutant Plants with No Detectable RNA Editing at Site nad7-200
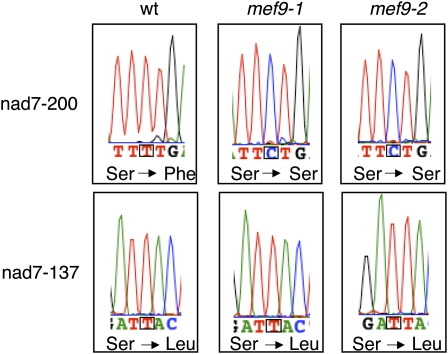
To find genes involved in RNA editing at specific sites in plant mitochondria, I developed a procedure to identify mutants defective in RNA editing at individual sites. With this method I select such mutants by a forwards genetic screen in a collection of chemically mutagenized Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col) plants. This editing-site-targeted search is performed with the recently developed multiplexed single nucleotide extension protocol (Takenaka and Brennicke, 2009) and complements the RNA editing gene identification through ecotype-specific variations (Zehrmann et al., 2008, 2009). The screen for mutants deficient in editing at the nad7-200 site yielded two mutant plants, mef9-1 and mef9-2, which have lost detectable editing at this site (Fig. 1, top section). Another site in the same transcript, nad7-137, is edited normally in both mutants (Fig. 1, bottom section).
Figure 1.
RNA editing is not detectable at the nad7-200 editing site in mitochondria of mef9-1 and mef9-2 mutant plants. Comparison of the cDNA sequence analysis of two RNA editing sites (boxed) in the mitochondrial nad7 mRNA between wild-type Arabidopsis (wt) and the mef9-1 and mef9-2 mutant plants shows that both mutants have lost the ability of C to U editing at site nad7-200, but not at another site in the same mRNA, site nad7-137. This latter site is correctly edited in wild type and in both mutant plants changing Ser to Leu codons. The absence of RNA editing event nad7-200 results in incorporation of the genomically encoded Ser rather than the Phe specified by the edited codon. In the cDNA strand analyzed, the detected T nucleotide (red trace) corresponds to the edited U, the observed C (blue trace) is derived from an unedited C.
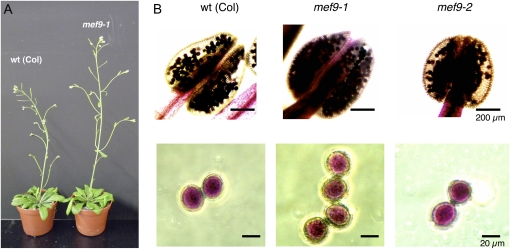
The mutant line mef9-1 shows a normal growth phenotype under standard growth chamber conditions comparable to the wild-type plant (Fig. 2). Mutant plant mef9-2 displays a somewhat slower growth with bolting and flower set delayed by about 2 weeks. Other phenotypic parameters like sizes and shapes of leaves, their numbers at the time of bolting, flower shapes, and numbers as well as seed set are comparable between both mutants and the wild type. However, the number of homozygous mutant plants in the F2 generation obtained by selfing the heterozygous F1 of the cross between mutant line mef9-1 and wild-type Landsberg erecta (Ler) plants is with 1:5 lower than the expected 1:3 ratio to the sum of the homozygous wild-type and the heterozygous plants. One possible cause for this difference could be a lower viability of the mutant pollen.
Figure 2.
Phenotype and pollen analysis of the mef9-1 and mef9-2 mutant plants. A, The growth habitus of the mef9-1 mutant plants is indistinguishable from the wild-type (wt) Col plant and shows flowers and beginning seed set after 6 weeks. B, Anther and pollen staining shows that the mef9-1 and mef9-2 mutant plants are not male sterile and differentiate viable pollen comparable to the wild-type plants. Staining was done according to Alexander (1969). [See online article for color version of this figure.]
One of the first tissues affected by mitochondrial dysfunctions in plants is the tapetum layer and the development of functional pollen, which in severe cases leads to cytoplasmic male sterility. For example, in a mitochondrial mutant of tobacco (Nicotiana tabacum) the nad7 gene has become nonfunctional by genomic recombination events, resulting in early abortion of the developing pollen and male sterility (Gutierres et al., 1997). To investigate whether pollen development is affected I compared the structure and composition of anthers and pollen in wild type and the two mef9 mutant plant lines (Fig. 2B). Pollen numbers, shape, and contents look indistinguishable between the three plants and the Alexander (Alexander, 1969) staining showed that nearly all pollen grains are viable.
Since the delayed growth of mef9-2 is only observed in one of the mutants, this phenotypic defect is probably due to another EMS-induced mutational event and not directly caused by the mutation in MEF9. On the other hand a differential effect of the distinct mutations in mef9-1 and mef9-2 cannot be excluded at present. Extensive backcrossing would be required to determine if the mef9-2 mutation can be separated from the slower growth habitus. The virtually normal growth pattern of mef9-1 is surprising when considering that this editing event changes an amino acid (S > F) in the NAD7 protein subunit. Detailed biochemical analyses will have to be done to determine whether the function of the respiratory chain complex I, the NADH-dehydrogenase complex is affected by this amino acid alteration in subunit 7.
Mapping of the MEF9 Gene
To locate the gene locus affected by the mutation in MEF9, the mutant line mef9-1 in the Arabidopsis Col ecotype was crossed with wild-type plants of ecotype Ler. All F1 plants are competent for editing at site nad7-200, suggesting a recessive nature of mef9-1. Of the F2 generation, 167 plants were screened for individuals with the phenotype of defect editing at the nad7-200 site. The 25 plants identified by this criterion are presumably homozygous in the mutant mef9-1 allele. These mutant plants were then mapped for crossover events with Ler and Col specific markers, which delineated a window of 1.7 Mb on chromosome 1 for the mutant allele.
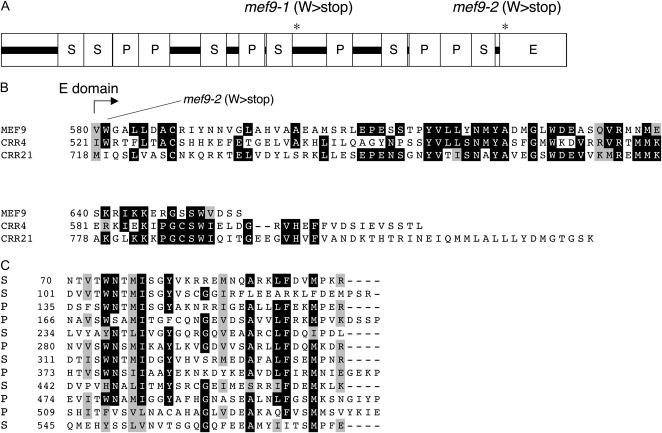
In this region 262 genes are annotated, including three genes coding for PPR proteins, the likely candidates for editing factors. These three genes, At1g60770, At1g61870, and At1g62260 were sequenced in the mutant and compared to the respective wild-type sequences. The first two genes encode P-class PPRs and show no differences in their sequence compared to the wild-type genes. The E-class PPR protein encoded by At1g62260 shows one altered nucleotide, a C to T transition typical for an EMS-induced mutation, which is different from the polymorphisms (http://polymorph.weigelworld.org) between Ler and Col, suggesting this open reading frame as a likely candidate gene. The mutation in this line mef9-1 alters a Trp codon to a termination signal (W346stop) in the center of the open reading frame and thus effectively truncates the translation product by half (Fig. 3).
Figure 3.
Schematic structure of the MEF9 PPR protein encoded by locus At1g62260. A, The mef9-1 and mef9-2 mutant plants each have a stop codon truncating the MEF9 protein in different positions (denoted by stars). Both are C to T transitions in CGA codons resulting in TGA translational stops. The amino acid sequences in the boxes marked by black rectangles show little structural similarity and are not predicted to form further S or P repeats. The MEF9 protein contains an E domain at the C terminus, but is lacking a DYW region. B, Amino acid alignment of the C-terminal regions of the mitochondrial editing factor MEF9 with two chloroplast editing factors of the E class, CRR4 and CRR21. MEF9 is the shortest PPR protein identified so far to be involved in specific RNA editing events in either mitochondria or plastids. In the mef9-2 mutant, the premature stop codon removes the entire E domain. Amino acids in inverse shading are identical between at least two of three aligned sequences, similar amino acids are underlayered with gray. C, The amino acid alignment of the S and P domains in MEF9 reveals the varying lengths of the so-called 35er repeats and shows that only few amino acids are actually shared by most of these repeat regions. Amino acids in inverse shading are identical between at least eight of the 12 aligned sequences, those highlighted against a gray background are similar in eight or more of the 12 repeats.
To corroborate that this locus indeed codes for a protein required for the editing event at nad7-200, the status of this gene locus was analyzed in the second mutant plant line mef9-2. In this line mef9-2, the reading frame encoded by At1g62260 is also prematurely terminated by a single nucleotide alteration, albeit in a location further downstream at the beginning of the E domain (W581stop; Fig. 3). This result confirms that the two mutant lines mef9-1 and mef9-2 arose by independent mutation events, each involving distinct nucleotide changes. The correlation between the phenotype of loss of RNA editing at the nad7-200 site and two independent mutations in this nuclear genomic locus identifies the At1g62260 gene as the MEF9 gene. A T-DNA insertion line (SALK 107370) annotated to contain an insertion at the very beginning of the reading frame in the first S-PPR repeat could not be verified by PCR analysis.
The Wild-Type MEF9 Gene Restores RNA Editing in the Mutants
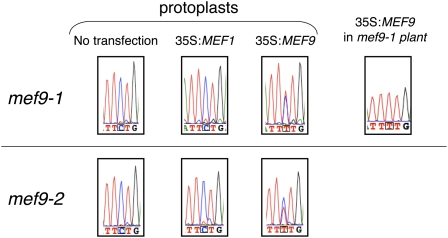
The connection between the MEF9 gene and RNA editing at the nad7-200 site was further assayed by complementation experiments with protoplasts from the two mutant plant lines. The Col wild-type MEF9 gene was transfected into the mef9-1 and mef9-2 mutant protoplasts, respectively, and was transiently expressed under the control of a 35S-cauliflower mosaic virus promoter. When this construct was brought into the mutant protoplasts, the ability for RNA editing at the nad7-200 site was restored (Fig. 4; 35S:MEF9).
Figure 4.
The Col MEF9 gene sequence restores the ability for RNA editing in protoplasts of the EMS mutant lines mef9-1 and mef9-2. The cDNA sequence tracings compare the effects of the protoplast complementation assays of the mef9-1 and mef9-2 mutants with the Col MEF9 gene (35S:MEF9) on RNA editing at site nad7-200 (boxed nucleotide) at 48 h after transfection. The regained ability to edit this site is detected by the increased T-signal trace (red). For control, transfection with the MEF1 gene is assayed. This gene is involved in editing at other mitochondrial editing sites (Zehrmann et al., 2009) and does not complement the mef9-1 or mef9-2 mutant protoplasts. Untransfected protoplasts do not recover any editing at the site monitored. On the very right-hand side the cDNA analysis of a mef9-1 mutant stably transformed with the wild-type MEF9 gene shows that RNA editing at site nad7-200 is fully recovered in vivo and that no trace of an unedited C is detectable.
Independent repetitions of these complementation assays yielded varying rates of the recovery of RNA editing. In some assays, about 50% successful editing is observed already after 24 h incubation and increases only little after incubation for 48 h. In other experiments, recovery is only observed after prolonged incubation of the transfected protoplasts. With the previously identified editing factors MEF1 and MEF11 editing can usually be recovered already after 24 h (Verbitskiy et al., 2009; Zehrmann et al., 2009). The reason for this occasional delay of editing recovery with MEF9 in comparison to analogous factors is unclear at present.
The undetectable editing in untransfected protoplasts of mef9-1 and mef9-2 and in control transfections with the unrelated editing factor MEF1 gene indicates that RNA editing at this site nad7-200 is specifically recovered by the introduced MEF9 gene (Fig. 4). To further corroborate this specificity, the wild-type MEF9 gene was stably transformed into the mef9-1 mutant plants. Analysis of the transgenic plants revealed a fully edited nad7-200 site with no detectable background of the unedited precursor (Fig. 4, right section labeled 35S:MEF9 in mef9-1 plant). These results show that a functional MEF9 gene is essential for RNA editing at the nad7-200 site.
Transcript Patterns of nad7
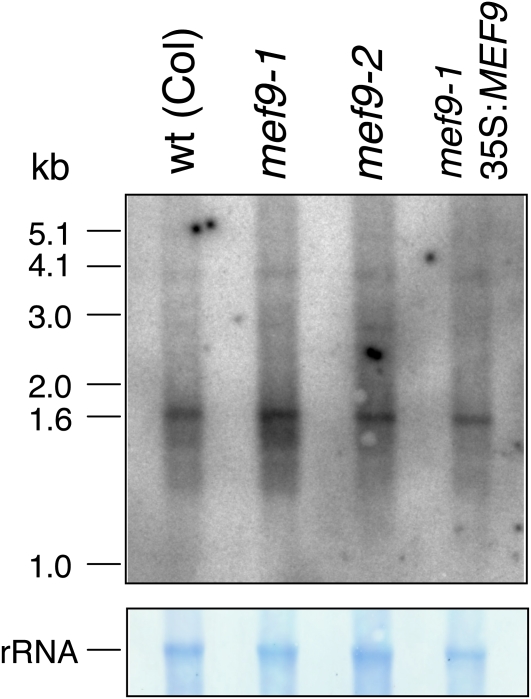
The absence of the RNA editing event at site nad7-200 could be caused by indirect effects, for example disturbed transcription or processing of the nad7 mRNA rather than a direct interference of the MEF9 protein at the editing site. To investigate this possibility I compared the patterns of the nad7 transcripts between the Col wild-type plants, the two mutant lines mef9-1 and mef9-2, and the stable transformant of mef9-1 in which the wild-type MEF9 gene is expressed from a 35S promoter (Fig. 5). In this northern analysis, the four transcript patterns look comparable with the mature nad7 transcript of 1,612 nucleotides being the prominent mRNA species and much less abundant precursors detectable as very weak signals. This result suggests that the mutations in the MEF9 gene do not disturb other processing events and specifically and directly affect RNA editing of this site nad7-200 in the nad7 mRNA.
Figure 5.
Northern analysis of the nad7 transcript pattern in the mef9-1 and mef9-2 mutant plants. The top section shows the nad7 transcripts visualized by autoradiography of the radioactive nad7 probe in wild-type (wt) plants, in the mef9-1 and mef9-2 mutant plants, and in the mef9-1 mutant stably complemented with a wild-type MEF9 gene. Besides the mature mRNA of 1,612 nucleotides low-abundant precursor transcripts can be detected. The bottom section shows the large rRNA in a methylene blue stain of the membrane to confirm the intactness of the RNA in these preparations. Five micrograms of total RNA were loaded into each slot, the blot was exposed overnight. [See online article for color version of this figure.]
Are Further Editing Sites Affected by the MEF9 Locus?
To investigate whether other RNA editing sites are targeted by the MEF9 gene, I analyzed 289 of the about 440 editing sites annotated in Arabidopsis (Giegé and Brennicke, 1999) by the SNaPshot assay in mutant plants homozygous for the mef9-1 allele. All RNA editing sites appear to be normally edited and consistently show T nucleotides. This result suggests that the MEF9 gene may be necessary for only the RNA editing event at the nad7-200 site, and is not strictly required for other sites. Since only 90% of the about 440 RNA editing events were directly analyzed, it is still possible that one of the sites not tested is another target of the MEF9 gene. Furthermore, other editing sites within or outside the number of sites investigated may be only partially affected by the truncated MEF9 proteins in mef9-1 and mef9-2 and thus escape direct experimental identification. Alternatively, at other MEF9 target sites other editing factors may compensate for the mutations in the MEF9 proteins in mef9-1 and mef9-2.
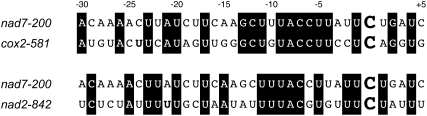
To investigate also the editing sites not directly tested, I searched for likely targets among all annotated editing sites by screening for sequence similarities in the region generally identified as the cis-specificity determinant in the RNA. Within this region of 35 nucleotides covering positions −30 to +5 relative to the edited C, I found the two sites cox2-581 and nad2-842 to share 18 and 17 nucleotides with site nad7-200, respectively (Fig. 6). However, both these sites are not affected by the mutations in mef9-1 or mef9-2 and are edited normally in the two mutant plant lines. The feasibility of MEF target site identification by this approach of similarity scans was recently documented by the identification of a bona fide target site for MEF11 through such an in silico screen for shared nucleotides (Verbitskiy et al., 2009). These results suggest that the MEF9 specificity factor possibly is required as an essential, unreplaceable element only for the editing of site nad7-200 in the mitochondrial transcriptome.
Figure 6.
Comparison of nucleotide identities in the cis-recognition sequence around the nad7-200 editing site with other editing sites. The presumed recognition sequence between nucleotides −30 and +5 of the MEF9 target site nad7-200 was used to scan all other RNA editing sites in the mitochondrial transcriptome of Arabidopsis for sites with shared nucleotide identities. Sites cox2-581 and nad2-842 share with 18 and 17 nucleotides, respectively (plus the edited C), the most nucleotides. Nevertheless, both of these sites are edited normally in the mef9-1 or mef9-2 mutants.
MEF9 Does Not Affect RNA Editing in Plastids
Most of the PPR proteins found in silico are predicted to be targeted to either or both, mitochondria and plastids (Lurin et al., 2004; Andrés et al., 2007). The here identified PPR protein MEF9 is indicated by one of the target programs (Predotar) to be located in mitochondria, while another program (TargetP) cannot make a clear distinction between the endoplasmic reticulum (0.476) and mitochondria (0.397). To investigate a potential influence of MEF9 on RNA editing in plastids, I analyzed the status of the editing sites in plastids in the mutant mef9-1. All of the 34 annotated editing sites in the plastid are edited normally, confirming that MEF9 is involved in RNA editing in mitochondria, but not in chloroplasts.
DISCUSSION
An E-Class PPR Protein Is Involved in RNA Editing in Mitochondria
In flowering plants about 450 different PPR proteins are encoded, more than in any other group of organisms. Of the PPR proteins identified to be required for distinct RNA editing events in plastid mRNAs some contain C-terminal E and adjacent DYW regions, while others terminate after the E domain (Kotera et al., 2005; Okuda et al., 2006, 2007; Chateigner-Boutin et al., 2008). The DYW domain adjacent to the E region has been suggested from in silico analyses to harbor a deaminating activity for this type of RNA editing (Salone et al., 2007; Rüdinger et al., 2008). PPR proteins containing only the E domain without the adjacent DYW region have been proposed to interact with other proteins, including the as yet hypothetical editing enzyme (Kotera et al., 2005; Okuda et al., 2006). Similar to these plastid editing factors, the here identified mitochondrial MEF9 PPR protein terminates before the DYW extension, while the three previously identified PPRs required for specific editing sites in mitochondria contain highly conserved DYW regions (Kim et al., 2009; Verbitskiy et al., 2009; Zehrmann et al., 2009).
Notably, the E domain in MEF9 is the shortest observed in any of the so far characterized editing factors (Fig. 3B). Both the C-terminal part of the E region and the conserved 15-amino acid stretch at this border of the E domain are not present in MEF9.
The observation of an analogous requirement of E-type or DYW-type PPR proteins for individual RNA editing events in mitochondria as well as in plastids further supports the similarity between the RNA editing processes in the two organelles. Since either the E- or the DYW-PPR proteins appear to act as specificity determinants, the enzymatic moiety for the deamination reaction can be expected to be very similar if not identical in both organelles. This elusive RNA editing enzyme may even be encoded by a single gene and may be dually targeted to both organelles. The requirement of this activity for many editing events could still be met, since for example the dually targeted aminoacyl synthetases likewise have to be continuously active at many tRNA molecules (Duchêne et al., 2005). Alternatively, the plastid and mitochondrial enzymes may be encoded by a small gene family similar to the organellar RNA polymerases in plants (Hedtke et al., 2000).
18 or 17 of 35 Nucleotides Shared with Other RNA Editing Sites Are Not Sufficient for Recognition by MEF9
When comparing the nad7-200 RNA editing site addressed by the MEF9 protein to the most similar sequence motifs around other editing sites in the presumed specific recognition regions, 18 and 17 nucleotide identities out of 35 are found to be shared with sites cox2-581 and nad2-842, respectively (Fig. 6). These two sites are edited normally in the mutants mef9-1 and mef9-2, suggesting that the nucleotide positions deviating at these two sites from the nad7-200 RNA editing site are not tolerated by MEF9. Alternatively, as mentioned above, these sites may be genuine MEF9 target sites, but can also be recognized by other editing factors. These then compensate for the disabled MEF9 proteins in the two mutant plant lines and successfully edit these nucleotide positions.
Functional Consequences of the RNA Editing Event at Site nad7-200
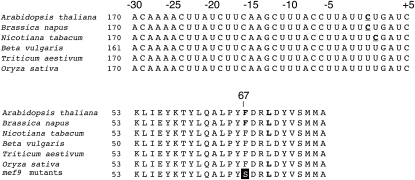
The amino acid sequence in the NAD7 protein surrounding amino acid 67, which is altered by the nad7-200 RNA editing event, is identical between various flowering plants, including monocots and dicots (Fig. 7). Since such a high degree of conservation usually reflects a functional importance, it seems the more surprising that there is little detectable phenotype in the mef9-1 and mef9-2 mutant plants (Fig. 2). On the other hand, the NAD7 protein is only altered in one amino acid position in the mef9-1 and mef9-2 mutant plants, which may be tolerated and compensated under standard greenhouse conditions. This inference may be supported by the previous observations that in spite of the high conservation of the NAD7 protein sequence, the complete absence of NAD7 is detrimental but not lethal in mitochondrial mutants of tobacco, in which the nad7 gene has been interrupted by genomic recombination events (Gutierres et al., 1997). Detailed biochemical characterization of complex I at least in the mef9-1 mutant plants may clarify if the single amino acid alteration by the absence of this RNA editing event does alter the properties of this respiratory chain complex and if this change shows biochemical or physiological effects under some conditions.
Figure 7.
Nucleotide and amino acid sequences at and around the nad7-200 editing site are conserved in flowering plants. The top part shows the nucleotide sequence alignment around the nad7-200 editing site in several plant species. The editing site here at nucleotide number 0 is conserved only between Arabidopsis and Brassica napus. The other plants compared encode a genomic T at this position. In tobacco, a new C residue one nucleotide downstream is now edited to the U present in the other monocot and dicot plants. In the bottom part, amino acids of the NAD7 proteins around the site of editing are aligned from several plant species. The nad7-200 editing event alters amino acid 67 in the protein, another editing event changes the codon, but not the identity of amino acid 70. Nucleotides and amino acids at RNA editing sites are highlighted in bold. The Ser residue encoded in the unedited mef9 mutant mRNAs is inversely shaded.
While the nad7-200 RNA editing event may be tolerated in the NAD7 protein despite its conservation in many plants, amino acid changes caused by other editing events in the nad7 mRNA may be more detrimental. The initial screen for normally growing plants in the EMS mutant population would have selected against the inclusion of such mutants with stronger effects on growth patterns.
Are There Enough PPR Proteins for All RNA Processing Events in Organelles?
The observation that possibly only one RNA editing site is targeted by the MEF9 PPR protein raises the question whether the about 450 PPR proteins are indeed sufficient to address the 450 or more RNA editing sites in a given flowering plant mitochondrial transcriptome, the about 35 editing sites in the plastid, and the various other RNA processing events in which the PPR proteins are involved (Schmitz-Linneweber et al., 2005; de Longevialle et al., 2007; Beick et al., 2008; Williams-Carrier et al., 2008). Of the different PPR subclasses, the E-only and the E-DYW groups together number about 150 proteins (Lurin et al., 2004). The editing factors presently identified in plastids and in mitochondrial protein exclusively belong to these subgroups, which would not be sufficient to supply all of the specific editing factors if only single sites would be addressed by one protein as found for several plastid editing factors and possibly here for MEF9. However, the one PPR-one site ratio appears to be the exception, since most of the other PPR proteins identified as editing factors at least in mitochondria recognize several editing sites, the rice (Oryza sativa) OGR1 protein is even involved in seven editing events, although other factors may contribute here (Kim et al., 2009). The specificity of the MEF9 protein for one site thus appears to be an exception at least for mitochondria and may be corrected to include more sites upon further specific investigation.
MATERIALS AND METHODS
Plant Material and Preparation of Nucleic Acids
Arabidopsis (Arabidopsis thaliana) seeds for the Ler ecotype were kind gifts of J. Forner and S. Binder (Universität Ulm). The EMS (for ethylmethanesulfonate) mutant population of Arabidopsis ecotype Col was obtained commercially (Lehle Seeds). Growth of the Arabidopsis plants and preparation of DNA or RNA from the leaves were as described (Takenaka and Brennicke, 2007). Anthers and pollen were collected from mature flowers. The pollen kernels were stained for viability by the Alexander method (Alexander, 1969).
SNaPshot Assays and Mutant Analysis
The EMS mutant library was screened by multiplexed single base extension (Takenaka and Brennicke, 2009) for plants with altered RNA editing at specific sites. Plants were first analyzed in pools of 10 from which the deviant plants were recovered. In the identified individual plants, the compromised RNA editing phenotype was verified by cDNA sequence analysis for the status of the respective investigated editing site. In the mef9-1 mutant plants, the three candidate PPR genes in the 1.7 Mb window on chromosome 1 were investigated by direct sequence analysis of the relevant PCR products. Most sequences were obtained commercially from 4base lab, Reutlingen, Germany, or from Macrogen, Seoul, Korea.
Screening Assays of RNA Editing Sites with C or T Specific Reverse Transcription PCR
Specific cDNA fragments were generated by reverse transcription (RT)-PCR amplification by established protocols (Takenaka and Brennicke, 2007). The cDNA sequences were compared for C to T differences resulting from RNA editing. The 167 F2 generation plants obtained from the cross between the mef9-1 mutant plant line in a Col background and Ler wild-type plants were screened for homozygous mutants by RT-PCR. The downstream primers were designed with a single nucleotide mismatch and either a C or T specific 3′-terminal nucleotide (Verbitskiy et al., 2009). The mismatch yielded optimal effects when positioned three nucleotides upstream of the 3′ terminus. Products generated after PCR on a gene-specific cDNA strand yielded predominantly reciprocal products depending on the RNA editing status of this site. Generally the yes or no decision of RNA editing was possible even in the presence of cross-contaminating background.
Northern-Blot Analysis
Total cellular RNA was prepared from young rosette leaves with the Spektrum plant total RNA kit (Sigma). RNA was size fractionated on a 1.5% agarose gel in MOPS buffer and blotted to Hybond N membranes as previously described (Zehrmann et al., 2009). The membrane was stained with methylene blue and hybridized with an nad7 probe covering the entire reading frame labeled by 32P-alpha-dCTP as described (Zehrmann et al., 2009).
Protoplast Complementation Assays
Protoplasts were prepared from 3- to 4-week-old plantlets by the method of Yoo et al. (2007). Transfected genes, including GFP and the wild-type Col MEF1 as controls or wild-type Col MEF9 reading frames, were expressed from the 35S promoter in the cloning site of vector pSMGFP4. Efficiency of the transfection was monitored by the signals from separately introduced or cotransfected GFP genes in the cytoplasm. Typically the GFP fluorescence was detected in more than 80% of the transfected protoplasts. Total RNA was isolated after 24 or 48 h incubation at room temperature. Sequences of cDNAs were determined after RT-PCR with the respective specific primers. RNA editing levels were estimated by the relative heights of the respective nucleotide peaks in the sequence analyses (Zehrmann et al., 2009).
Acknowledgments
I thank Dagmar Pruchner (Molekulare Botanik, Universität Ulm) for excellent experimental support and Axel Brennicke (Molekulare Botanik, Universität Ulm) for help with the manuscript. I am very grateful to Stefan Binder, Joachim Forner, Angela Hölzle, and Christian Jonietz (Molekulare Botanik, Universität Ulm) for their gifts of seeds, markers, and other materials, and to Walther Vogel, Christiane Maier, and Bärbel Weber in the Institut für Humangenetik, Universität Ulm, for the use of the automated sequencing equipment and materials and their very constructive and helpful discussions throughout.
This work was supported by grants from the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mizuki Takenaka (mizuki.takenaka@uni-ulm.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44 117–122 [DOI] [PubMed] [Google Scholar]
- Andrés C, Lurin C, Small ID (2007) The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol Plant 129 14–22 [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small ID, et al (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56 590–602 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Maliga P (1996) Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J 15 5958–5964 [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID (2007) Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem Soc Trans 35 1643–1647 [DOI] [PubMed] [Google Scholar]
- Duchêne AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, Zaepfel M, Maréchal-Drouard L, Small ID (2005) Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA 102 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré JC, Leon G, Jordana X, Araya A (2001) Cis recognition elements in plant mitochondrion RNA editing. Mol Cell Biol 21 6731–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P, Brennicke A (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA 96 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Viehoever P, Weisshaar B, Knoop V (2009) A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res 37 5093–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (2000) One RNA polymerase serving two genomes. EMBO Rep 1 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller WP, Hayes ML, Hanson MR (2008) Cross-competition in editing of chloroplast RNA transcripts in vitro implicates sharing of trans-factors between different C-targets. J Biol Chem 283 7314–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugiura M (2001) Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast RNA: development of an in vitro RNA editing system. EMBO J 20 1144–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Yang JI, Moon S, Ryu CH, An K, Yim J, An G (2009) Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in plant mitochondria. Plant J 59 738–749 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Matsuo M, Sakamoto K, Wakasugi T, Yamada K, Obokata J (2007) Two RNA editing sites with cis-acting elements of moderate sequence identity are recognized by an identical site-recognition protein in tobacco chloroplasts. Nucleic Acids Res 36 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto JM, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Obokata J, Sugiura M (2002) Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol Cell Biol 22 6726–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Sugita M (2008) A conserved DYW domain of the pentatricopeptide protein possesses a novel endoribonuclease activity. FEBS Lett 582 4163–4168 [DOI] [PubMed] [Google Scholar]
- Neuwirt J, Takenaka M, van der Merwe JA, Brennicke A (2005) An in vitro RNA editing system from cauliflower mitochondria: editing site recognition parameters can vary in different plant species. RNA 11 1563–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga R, Motohashi K, Shinozaki K, Small I, Shikanai T (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Habata Y, Kobayashi Y, Shikanai T (2008) Amino acid sequence variations in Nicotiana CRR4 orthologs determine the species-specific efficiency of RNA editing in plastids. Nucleic Acids Res 36 6155–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Myouga R, Motohashi K, Shinozaki K, Shikanai T (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA 104 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T (2006) A pentatricopeptide repeat protein is a site-recognition factor in chloroplast RNA editing. J Biol Chem 281 37661–37667 [DOI] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger M, Polsakiewicz M, Knoop V (2008) Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in Jungermaniid but not in Marchantiid liverworts. Mol Biol Evol 25 1405–1414 [DOI] [PubMed] [Google Scholar]
- Salone V, Rüdinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small ID, Knoop V, Lurin C (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett 581 4132–4138 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yukawa Y, Wakasugi T, Yamada K, Sugiura M (2006) A simple in vitro RNA editing assay for chloroplast transcripts using fluorescent dideoxynucleotides: distinct types of sequence elements required for editing of ndh transcripts. Plant J 47 802–810 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small ID (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Barkan A (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ regions of mRNAs whose translation it activates. Plant Cell 17 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T (2006) RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol Life Sci 63 689–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25 46–47 [DOI] [PubMed] [Google Scholar]
- Takenaka M, Brennicke A (2007) RNA editing in plant mitochondria: assays and biochemical approaches. Methods Enzymol 424 439–458 [DOI] [PubMed] [Google Scholar]
- Takenaka M, Brennicke A (2009) Multiplex single base extension typing to identify nuclear genes required for RNA editing in plant organelles. Nucleic Acids Res 37 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8 35–46 [DOI] [PubMed] [Google Scholar]
- van der Merwe JA, Takenaka M, Neuwirt J, Verbitskiy D, Brennicke A (2006) RNA editing sites in plant mitochondria can share cis-elements. FEBS Lett 580 268–272 [DOI] [PubMed] [Google Scholar]
- Verbitskiy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M (2009) The PPR-protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J (in press) [DOI] [PubMed]
- Williams-Carrier R, Kroeger T, Barkan A (2008) Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59 1011–1023 [DOI] [PubMed] [Google Scholar]
- Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M (2008) Ecotype-specific variations in the extent of RNA editing in plant mitochondria. Mitochondrion 8 319–327 [DOI] [PubMed] [Google Scholar]
- Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M (2009) A DYW-domain containing PPR-protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammami K, Small ID, Huang J (2008) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58 82–96 [DOI] [PubMed] [Google Scholar]