Abstract
Traditionally, phenotype-driven forward genetic plant mutant studies have been among the most successful approaches to revealing the roles of genes and their products and elucidating biochemical, developmental, and signaling pathways. A limitation is that it is time consuming, and sometimes technically challenging, to discover the gene responsible for a phenotype by map-based cloning or discovery of the insertion element. Reverse genetics is also an excellent way to associate genes with phenotypes, although an absence of detectable phenotypes often results when screening a small number of mutants with a limited range of phenotypic assays. The Arabidopsis Chloroplast 2010 Project (www.plastid.msu.edu) seeks synergy between forward and reverse genetics by screening thousands of sequence-indexed Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutants for a diverse set of phenotypes. Results from this project are discussed that highlight the strengths and limitations of the approach. We describe the discovery of altered fatty acid desaturation phenotypes associated with mutants of At1g10310, previously described as a pterin aldehyde reductase in folate metabolism. Data are presented to show that growth, fatty acid, and chlorophyll fluorescence defects previously associated with antisense inhibition of synthesis of the family of acyl carrier proteins can be attributed to a single gene insertion in Acyl Carrier Protein4 (At4g25050). A variety of cautionary examples associated with the use of sequence-indexed T-DNA mutants are described, including the need to genotype all lines chosen for analysis (even when they number in the thousands) and the presence of tagged and untagged secondary mutations that can lead to the observed phenotypes.
Decoding of the Arabidopsis (Arabidopsis thaliana) genome sequence earlier this decade (Arabidopsis Genome Initiative, 2000) provided the opportunity to determine the functions of approximately 27,000 protein-coding genes. One or more functions of a small percentage of genes are currently experimentally determined, typically from mutant or transgenic analysis or through biochemistry. However, roles for the vast majority of plant genes are either more or less accurately predicted by DNA sequence homology or unpredictable based upon DNA sequence (Arabidopsis Genome Initiative, 2000; Cho and Walbot, 2001; Rhee et al., 2008; for recent specific examples, see Gao et al., 2009; Schilmiller et al., 2009). Because of the uncertainty associated with homology-based function assessment, high-throughput approaches to gene function identification are needed to expand the universe of genes with experimental annotation.
In contrast to organisms amenable to targeted gene replacement, such as bacteria, yeast, and mouse (Wendland, 2003; Wu et al., 2007; Adams and van der Weyden, 2008), obtaining a gene knockout is not as efficient in flowering plants. In Arabidopsis, the conventional way of creating a gene knockout is by insertional mutagenesis via Agrobacterium tumefaciens-mediated transformation (Krysan et al., 1999). Using this technique, a large piece of T-DNA is inserted into the genome in an untargeted manner (Alonso et al., 2003). If it lands within a coding or regulatory region, the T-DNA can influence the expression of the corresponding gene. While the probability of any single insertion element causing a mutation in a gene of interest is low, sequencing of hundreds of thousands of independent insertion sites has led to a collection of mutants in the majority of genes (http://signal.SALK.edu/tabout.html; Alonso et al., 2003).
T-DNA mutants can be a valuable tool for forward genetics, in which hundreds or thousands of mutants are subjected to phenotypic assays (Feldmann, 1991; Kuromori et al., 2006), but reverse genetics is the most common way in which these mutant collections are utilized. Typically, a small number of candidate genes are tested for a role in a particular biological process by reducing or increasing gene expression and assaying one or more phenotypes (for review, see Page and Grossniklaus, 2002; Alonso and Ecker, 2006). The availability of a gene-indexed T-DNA mutant collection allows researchers to rapidly obtain mutant lines for their genes of interest (http://signal.SALK.edu/cgi-bin/tdnaexpress). The availability of a large collection of indexed mutant or RNA interference lines in other model organisms has facilitated large-scale reverse genetics studies (Piano et al., 2000; Giaever et al., 2002; Ho et al., 2009).
In the course of a large reverse genetics project (The Chloroplast 2010 Project; http://www.plastid.msu.edu/), more than 3,500 T-DNA lines harboring insertions in nuclear genes, most of which were computationally predicted to encode chloroplast-targeted proteins, were subjected to a diverse set of phenotypic screens (Lu et al., 2008). In total, 85 phenotypic observations ranging from quantitative metabolite measurements to qualitative phenotypic observations are collected for each mutant line, and the data are stored in a relational database (http://bioinfo.bch.msu.edu/2010_LIMS). This approach seeks to take advantage of the best features of forward and reverse genetics by screening a large number of lines with mutations in known genes. Unlike conventional genetics screens, where plants are assayed for one or a small number of traits, this project surveys varied phenotypes.
In this study, a variety of phenotypic variants were analyzed. In some cases, independent mutants of the same gene were found to have similar phenotypes, revealing new information about those genes. In other examples, a single homozygous mutant allele was found to have a detectable phenotype. These run the gamut from cases where secondary mutations are strongly implicated in causing the phenotype, to an example where an analogous maize (Zea mays) mutant is known to have a similar phenotype, to other instances where the causative mutation is yet to be identified. In several examples of secondary mutations, the phenotype was not due to a T-DNA insertion, reinforcing the idea that these untagged alleles are a cause for concern in conducting large-scale reverse genetics screens (Vitha et al., 2003; Adham et al., 2005; Zolman et al., 2008), while providing opportunities for gene function discovery by map-based cloning or whole genome sequence analysis.
RESULTS
Examples of Confirmed Mutants from the Chloroplast 2010 Project
New Phenotypes for a Previously Characterized Mutant: Short-Chain Dehydrogenase At1g10310
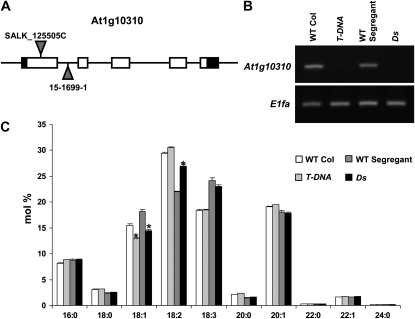
The Chloroplast 2010 Project identified a variety of T-DNA lines with phenotypes different from the parental ecotype Columbia plants, including changes not previously described in the Arabidopsis literature. One such example was discovered in the assay for changes in leaf fatty acid methylesters (FAMEs): SALK_125505C leaf samples have an approximately 10% decrease in mol % (1.34% ± 0.05% for wild-type samples and 1.16% ± 0.11% for SALK_125505C; P < 0.01) of 18:1Δ9 FAMEs (18:1, number of carbons:number of double bonds; Δ9, double bond between carbon 9 and 10 counting from the carboxyl end), as determined by gas chromatography-flame ionization detection (GC-FID) of FAMEs. SALK_125505C carries a T-DNA insertion in the first exon of At1g10310 (Fig. 1A), an oxidoreductase that belongs to the short-chain dehydrogenase/reductase (SDR) gene family. Surprisingly, this gene is annotated as encoding a pterin aldehyde reductase based upon the biochemical work of Noiriel et al. (2007). This annotation was based upon in vitro NADPH-dependent pterin aldehyde reductase activity of recombinant protein produced in Escherichia coli. When seed fatty acid composition of the mutant was compared with that in the wild type, a 15% reduction of 18:1Δ9 FAME was observed (Fig. 1C). This phenotype is consistent with data in AtGenExpress (Schmid et al., 2005) and Noiriel et al. (2007), indicating that At1g10310 mRNA accumulation is highest in mature seed. A second allele, a transposon-tagged line (RIKEN line 15-1699-1; Ito et al., 2002) in the Nossen ecotype carrying a Ds element in the second intron of the SDR gene, was analyzed to test whether the lesion in At1g10310 was actually responsible for the observed decrease in 18:1Δ9. Indeed, the transposon-tagged line also possessed a similar decrease in seed 18:1Δ9 methylester (approximately 20%) accompanied by a substantial increase in 18:2 (approximately 23%) that was not observed for the first allele (Fig. 1C). Because triacylglycerols account for the majority of lipids in the seed, it is not unexpected that these FAME differences were confirmed in triacylglycerols extracted from seeds (Supplemental Fig. S1). Consistent with the hypothesis that these are loss-of-function alleles, reverse transcription-PCR analysis of the two mutant lines confirmed that both T-DNA lines have dramatically reduced levels of At1g10310 mRNA (Fig. 1B).
Figure 1.
Mutation of gene At1g10310 causes decreased seed 18:1Δ9 fatty acids. A, Schematic representation of the At1g10310 gene model and two insertion alleles. White rectangles represent exons, black rectangles represent 5′ and 3′ untranslated regions, solid lines represent introns and intergenic regions, and gray triangles represent a T-DNA insertion for SALK_125505C and a Ds element for RIKEN line 15-1699-1. B, At1g10310 steady-state mRNA levels in wild-type Columbia (WT Col), a line derived from a wild-type segregant plant (WT Segregant), SALK_125505C (T-DNA), and RIKEN mutant line 15-1699-1 (Ds). C, Seed fatty acid composition of a line derived from a wild-type segregant plant (WT Segregant), the Columbia ecotype (WT Col), and the two mutant lines, T-DNA and Ds, as determined by GC-FID of FAMEs. Error bars represent sd values for five biological replicates (each replicate consisted of 30 seeds). Differences between the mutant and the wild type of greater than 15% that are statistically significant (Student's t test, P < 0.01) compared with the wild type are marked by asterisks.
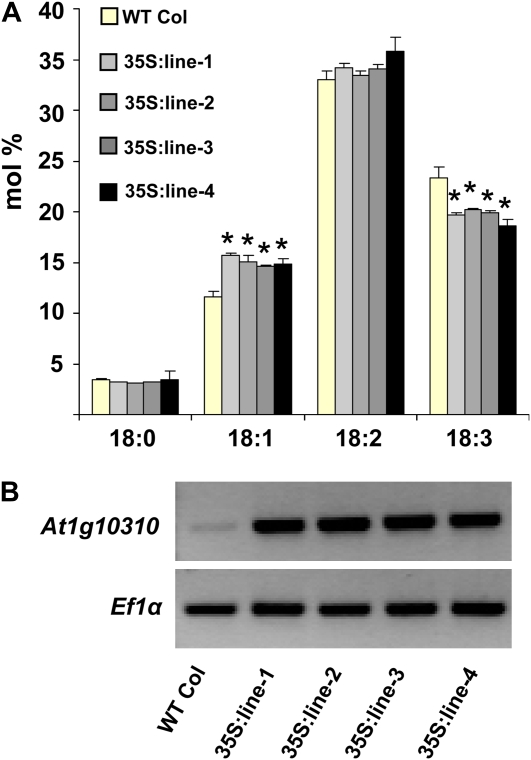
To further investigate the influence of this gene in modulating fatty acid composition, transgenic lines were generated by expression of the wild-type At1g10310 cDNA sequence under control of the 35S-cauliflower mosaic virus promoter. Reproducible changes were seen in seed fatty acid composition in T3 overexpresser lines, with an approximately 25% increase in 18:1Δ9 methylester and concomitant approximately 15% decrease in 18:3 (Fig. 2). No differences in patterns were observed for leaf FAMEs in the mRNA-overexpressing lines, suggesting that this gene product is not normally limiting for 18:1Δ9 in leaves. While Noiriel et al. (2007) demonstrated that this protein is active toward a number of different aldehydes, including two- to nine-carbon aliphatic aldehydes, aromatic aldehydes, and pterin aldehydes, they did not detect a pterin aldehyde-associated phenotype in the Ds mutant 15-1699-1. Taken together, knockout lines and ectopic overexpression of At1g10310 implicate this cytosolic SDR (Supplemental Fig. S2) in 18-carbon seed fatty acid metabolism. These results reinforce the value of large-scale reverse genetics mutant screening in discovering novel functions for genes, although understanding the mechanism by which this gene affects 18-carbon metabolism requires further investigation.
Figure 2.
35S:At1g10310 mRNA overexpresser lines have more 18:1Δ9 and less 18:3 seed fatty acids than the wild type. A, Eighteen-carbon seed fatty acid composition for the wild type (WT Col) and four transgenic lines (35S:line-1 to -4) transformed with a 35S:At1g10310 construct. Each replicate consisted of 30 seeds, and the error bars represent sd values for three biological replicates. Considerable changes (>15%) relative to the wild type that are statistically significant (Student's t test, P < 0.01) are marked by asterisks. B, Semiquantitative PCR shows that the four lines are high expressers of At1g10310 mRNA. Plants were grown under 16-h-light/8-h-dark conditions. [See online article for color version of this figure.]
Mutants for Genes with Bona Fide Functions: Acyl Carrier Protein4
High-throughput reverse genetics can also be used to confirm and refine our current knowledge. For example, characterization of Arabidopsis Acyl Carrier Protein4 (ACP4) antisense transgenic plants revealed that down-regulation of multiple ACPs results in altered fatty acid composition, compromised photosynthetic efficiency, and a bleached appearance (Branen et al., 2003). The observed phenotypes are consistent with known ACP function, as these small polypeptides play critical roles in de novo fatty acid biosynthesis and lipid metabolism. In Arabidopsis, there are five known ACP genes (Beisson et al., 2003), which are differentially expressed (Hlousek-Radojcic et al., 1992). ACP4 is the most abundant leaf isoform (Bonaventure and Ohlrogge, 2002), but the influence that each isoform has on leaf fatty acid biosynthesis and photosynthesis is not clear, because ACP2 and ACP3 isoforms were also reduced by 25% to 50% in the reported antisense lines (Branen et al., 2003). Insertional mutant analysis can help determine whether the ACP4 isoform has the most important function in leaf tissue or whether their roles overlap.
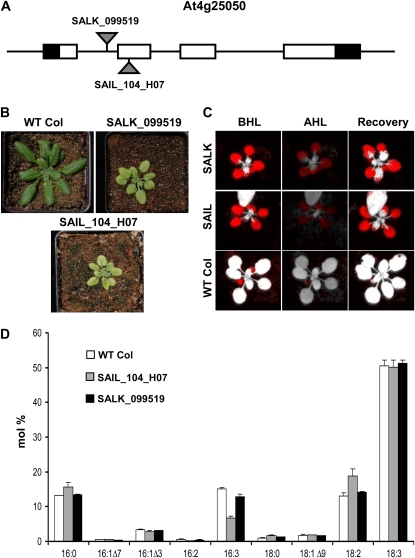
The Chloroplast 2010 Project obtained phenotypic results for two independent T-DNA lines, SALK_099519 and SAIL_104_H07, confirmed by our project to harbor insertions in intron 1 and exon 2 of ACP4, respectively (Fig. 3A; Supplemental Table S1). Both T-DNA mutants are slow growing, have small rosettes, and are chlorotic in appearance (Fig. 3B). Rosette leaves of SALK_099519 are uniformly light green, whereas chlorosis in SAIL_104_H07 is more severe, revealing dark green veins and leaf margins that are often necrotic. Measurements of maximum quantum yield of PSII (Fv/Fm) showed that both lines are impaired in photosynthetic electron transport, as portrayed by false-color images in Figure 3C. The FAME profiles of both mutants also differed from that of the wild type (Fig. 3D), especially the more severely affected mutant SAIL_104_H07. The prominent decrease in 16:3 methylester was also observed for the ACP4 antisense line (Branen et al., 2003). Taken together, these data indicate that ablation of expression of the ACP4 isoform alone is sufficient to cause a chlorotic phenotype, reduce photosynthetic competence, and alter fatty acid composition. The SALK_099519 allele was renamed acp4-1 and the SAIL_104_H07 mutation is now acp4-2.
Figure 3.
Molecular basis and associated phenotypes for two ACP4 (At4g25050) T-DNA alleles. A, SALK_099519 contains an insertion in intron 1 of At4g25050, and SAIL_104_H07 contains an insertion in exon 2 of At4g25050. Schematic representation of the gene model and alleles is as described in the Figure 1 legend. B, Small size and pale green appearance of SALK_099519 and SAIL_104_H07 compared with a Columbia wild-type plant (WT Col). C, Fv/Fm values were calculated before high light (BHL), after plants were exposed to 3 h of high light (1,500−1,700 μE m−2 s−1; AHL), and after a 2-d recovery period (Recovery) using the MAXI version of the IMAGING-PAM M-series chlorophyll fluorescence system (Heinz-Walz Instruments). Wild-type control plants were used to assign cutoff values for Fv/Fm under each condition. Red coloring in the false-color images indicates tissues that were below the wild-type cutoff value for Fv/Fm. D, Altered leaf fatty acid composition of SALK_099519 and SAIL_104_H07 compared with wild-type plants. Error bars represent sd values for two biological replicates.
Phenotypes That Appear to Make Sense Sometimes Do Not
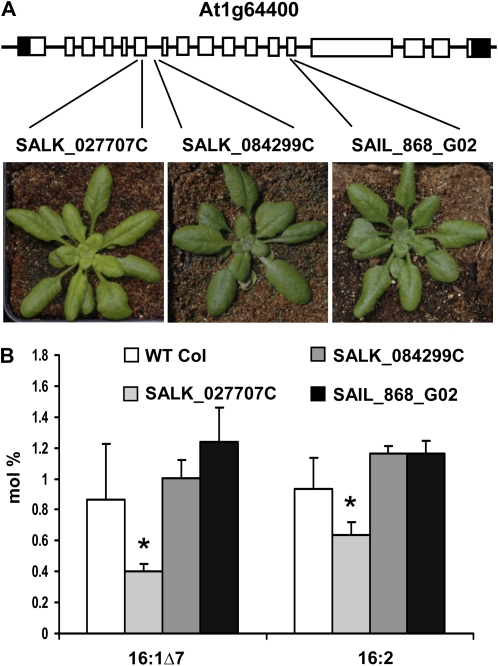
SALK_027707C stands out in the Chloroplast 2010 phenotypic pipeline due to altered whole plant morphology and leaf FAMEs. This line has lighter green leaves than the wild type, but the rosette size and leaf shape are normal (Fig. 4A). Levels of chloroplastic 16:1Δ7 and 16:2 FAMEs are approximately 50% and 20% lower than in the wild type, respectively. This mutant is annotated as having a T-DNA insertion in exon 6 of a long-chain acyl-CoA synthetase (ACS3; At1g64400; Fig. 4A). Acyl-CoA synthetases play central roles in fatty acid metabolism and transport because they activate free fatty acids to fatty acyl-CoAs by an ATP-dependent thioesterification (Schnurr et al., 2002; Shockey et al., 2002). Considering the biochemical function of these enzymes in fatty acid biology, it appeared reasonable to hypothesize that lower levels of 16:1Δ7 and 16:2 are due to a perturbation in fatty acid metabolism, suggesting that this mutation defines a role for this specific gene. To test whether mutation of ACS3 caused the phenotypes, two other independent homozygous mutant alleles were tested. Contrary to expectation, SALK_084299C and SAIL_868_G02 possessed wild-type leaf color and fatty acid composition (Fig. 4). Therefore, the phenotypes observed for SALK_027707C are presumably caused by one or more mutations outside of the ACS3 gene. This underscores the importance of having multiples lines of evidence to associate mutation of a gene with an altered phenotype.
Figure 4.
Inconsistent phenotypes of three At1g64400 T-DNA insertion lines. A, Insertion sites and photographs of representative plants for each mutant line. Schematic representation of the gene model and alleles is as described in the Figure 1 legend. B, 16:1Δ7 and 16:2 FAME levels of wild-type (WT Col), SALK_027707C, SALK_084299, and SAIL_868_G02 lines as measured by GC-FID. Statistically significant differences between each mutant and the wild type (Student's t test, P < 0.01) are depicted by asterisks. Error bars represent sd values of four biological replicates for SALK_027707C and SAIL_868_G02 and three biological replicates for the wild type and SALK_084299.
Confirming Genotype Is Essential
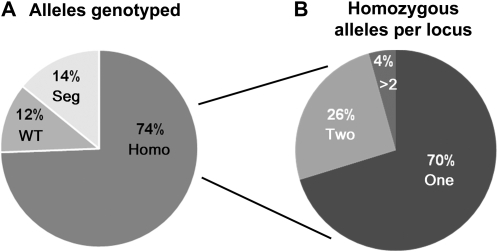
The previous example illustrates that the value of data generated by reverse genetics projects is critically dependent on the number and quality of available homozygous mutants. The collection of publicly available Arabidopsis insertion lines is an unprecedented resource for plant functional genomics. However, as our project sought to confirm the phenotype of lines selected randomly or chosen based upon their phenotypes, some were found not to contain a detectable insertion at the annotated locus. Because these lines complicate the interpretation of data generated by the project, a relatively high-throughput genotyping assay was used to test all individuals entering the pipeline (see “Materials and Methods”). Out of 3,673 independent lines tested, 74% genotyped as homozygous for the proposed insertion, 14% had individuals with a combination of any of the three genotypes but were not all homozygous (called segregating lines), and 12% yielded results consistent with all tested lines being wild type for the proposed insertion (Fig. 5A; Supplemental Table S1). Phenotypic data from plants that did not yield genotypic data consistent with being homozygous for the target mutation are filtered from the project database to increase the robustness of the phenotypic data.
Figure 5.
Summary genotyping information for T-DNA lines genotyped by the Chloroplast 2010 pipeline. A, A total of 3,673 independent T-DNA lines annotated as homozygous for specific alleles were genotyped; 74% were homozygous (Homo), 12% were wild type (WT), and 14% had individuals with a combination of any of the three genotypes but were not all homozygous (Seg) for the proposed insertion. These could include seed stocks contaminated with wild-type seed or with seeds of other mutants. B, Out of the 2,733 homozygous SALK lines, 70% of loci have a single allele (One), 26% of genes have two independent homozygous mutants (Two), and 4% of genes have more than two homozygous mutants (>2).
Secondary Mutations That Cause the Detected Phenotypes
In theory, the use of sequence-indexed T-DNA mutant collections in large-scale phenotypic screens accelerates the rate of gene discovery, because identification of the affected locus by genetic mapping should not be necessary. However, occasionally, the observed phenotype is caused by a secondary mutation and not by the T-DNA-tagged gene associated with the mutant line. Thus, confirmation of an observed phenotype with a second line of evidence is a requirement if accurate gene-phenotype relationships are to be established. In the case of a high-throughput phenotyping platform such as the Arabidopsis Chloroplast 2010 Project, the most straightforward approach is to assay two or more T-DNA lines for every gene tested. Three years into the project, we have encountered a number of examples where a strong phenotype was caused by a known secondary mutation and not by the locus associated with the T-DNA-tagged line. The following are examples of cases where the locus responsible for the phenotype was identified and verified by analysis of the candidate locus.
SALK_006881C: a Mutant Lacking the Main 16-Carbon Unsaturated Fatty Acids
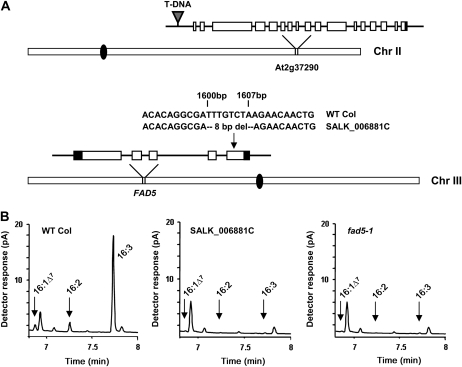
SALK_006881C contains a T-DNA insertion approximately 1,000 bp from the proposed start codon of At2g37290 (Fig. 6A), a protein that shows similarity to Rab GTPase activators. Rab proteins are a family of small GTPases that are involved in the regulation of vesicular membrane trafficking. Leaf FAMEs of SALK_006881C lack any detectable amounts of the unsaturated 16-carbon fatty acids 16:1Δ7, 16:2, and 16:3 (Fig. 6B). The mutant leaves also have increased levels of 16:0, suggesting that a block in 16:0Δ7 desaturation activity catalyzed by FAD5 (At3g15850) was the biochemical cause of the observed fatty acid phenotype (Heilmann et al., 2004a, 2004b). At the time, a second allele for At2g37290 was not available, and the FAD5 gene was sequenced from SALK_006881C. Indeed, the last exon of FAD5 in SALK_006881C was missing 8 bp (1,600–1,607 bp starting from the translational start site; Fig. 6A). This out-of-frame deletion is predicted to cause truncation of FAD5 protein, followed by insertion of 22 new amino acids. Therefore, we attribute the altered fatty acid phenotype in SALK_006881C to a lesion within the FAD5 locus and not to At2g37290. Based upon these results, the allele causing the FAME defect of SALK_006881C is designated as fad5-2, and the T-DNA insertion allele is named rgc1-1.
Figure 6.
Molecular basis of the altered FAME phenotype of mutant SALK_006881C. A, Locations and details of two mutations identified in SALK_006881C on chromosomes (Chr) II and III (centromeres are depicted by black ovals; gene models and mutation sites are schematically represented as described in the Fig. 1 legend). Based on gene model At3g15850.1, the 8-bp deletion (8 bp del; indicated with an arrow) occurs 1,600 bp downstream of the FAD5 translation start site. B, GC-FID chromatogram traces of total leaf FAME extracts of wild-type (WT Col), SALK_006881C, and fad5-1 plants (Heilmann et al., 2004a). Peaks for 16:1Δ7, 16:2, and 16:3 are indicated by arrows. Based upon these results, the FAD5 allele found in SALK_006881C is now designated fad5-2. The insertional mutation in At2g37290 has been renamed rgc1-1.
SALK_133788C: a Mutant with Low Afternoon Starch
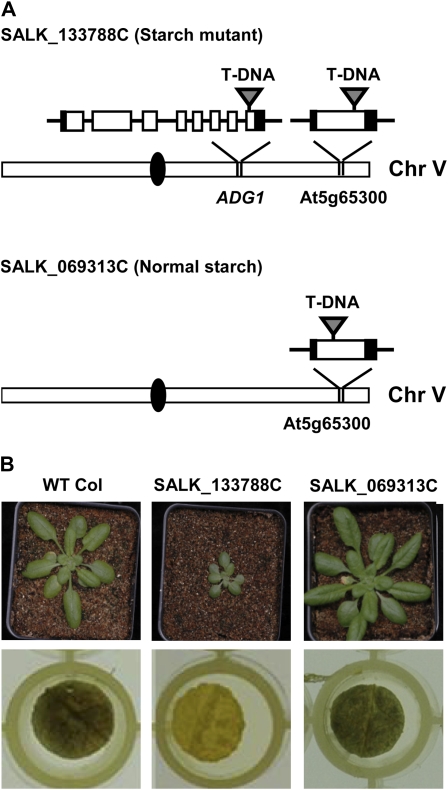
Results with SALK_133788C reinforce the importance of confirming the association of mutant genotype and phenotype with a second line of evidence. This SALK line contains an insertion in the single exon of At5g65300, a gene of unknown function. Seedlings for SALK_133788C are small and slow growing, with reduced accumulation of leaf starch in the afternoon under the standard 12-h photoperiod used in the pipeline (Fig. 7B). However, SALK_069313C, a mutant also carrying a homozygous insertion in the exonic region of At5g65300, had normal morphology and starch accumulation (Fig. 7B). This led to the hypothesis that the starch deficiency and slow growth phenotype of the first mutant line was caused by a secondary mutation in a gene involved in starch biosynthesis. This was confirmed by demonstration of a T-DNA insertion in the last exon of the small subunit of ADP-Glc pyrophosphorylase (ADG1; At5g48300; Fig. 7A), a key enzyme of starch biosynthesis (Wang et al., 1998). Taken together, these data argue that the low afternoon starch and slow growth under the 12-h photoperiod observed for SALK_133788C are due to a lesion at ADG1 and not to the locus of unknown function, At5g65300. The ADG1 mutation has been named adg1-2.
Figure 7.
Molecular basis of the reduced starch phenotype of mutant SALK_133788C. A, Two T-DNA insertions were detected in the starch-deficient mutant SALK_133788C: one insertion lies in the single exon of At5g65300 and the other in the last exon of ADG1. Starch-proficient SALK_069313C carries an insertion in At5g65300, with no T-DNA in ADG1. Schematic representation of chromosomes (Chr), gene models, and mutations is as described in the Figure 1 and 6 legends. B, The top row shows whole plant images for the wild type (WT Col), SALK_133788C, and SALK_069313C. The bottom row shows iodine staining of leaf discs (from the same plants as in the top row) harvested 8 h into the 12-h light period.
SALK_134724C: a Mutant with High Seed Thr and Cys
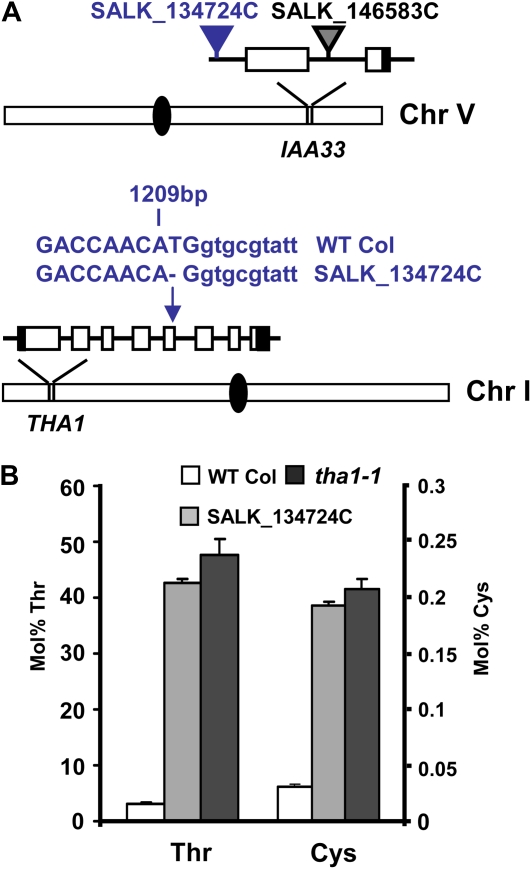
SALK_134724C is annotated as mutant at locus At5g57420 because it contains a T-DNA insertion within 1,000 bp upstream from the inferred start of translation. This line was found to have seeds containing abnormally high levels of the amino acids Thr and Cys (Fig. 8B). At5g57420 encodes IAA33, annotated as a member of the auxin/indole-3-acetic acid (AUX/IAA) gene family. Characterized members of this gene family are nucleus-targeted transcriptional repressors that play a role in regulating auxin-mediated gene expression (Overvoorde et al., 2005). We were unaware of examples where mutants of the AUX/IAA gene family cause changes in seed metabolism; therefore, this was an intriguing phenotype for IAA33. However SALK_146583C, a second insertion mutant of IAA33, did not accumulate high Thr and Cys, suggesting that a secondary mutation caused this phenotype.
Figure 8.
Molecular basis of the high free Thr and Cys in mutant SALK_134724C. A, Mutations identified in SALK_134724C include a T-DNA insertion in the promoter region of IAA33 and deletion (illustrated with a dash) of a T residue 1,209 nucleotides downstream from the translational start site of THA1 (based on gene model At1g08630.1). Exonic nucleotides are in uppercase, while intronic nucleotides are in lowercase. Schematic representation of chromosomes (Chr), gene models, and mutations is as for Figure 6. B, Liquid chromatography-mass spectrometry analysis of Thr and Cys levels (mol %) in wild-type (WT Col), SALK_134724C, and tha1-1 seeds. Error bars represent se values of four replicates. Based upon these results, the At1g08630 allele found in SALK_134724C is now designated tha1-3 and the insertional mutation in IAA33 is referred to as iaa33-2.
High seed Thr phenotype (Jander et al., 2004; Joshi et al., 2006) and high Thr plus Cys (Lu et al. 2008) were observed for the loss-of-function tha1-1 Thr aldolase mutant (Fig. 8B). THA1 enzyme (At1g08630) degrades Thr to Gly and is highly expressed in seeds and seedlings. Therefore, accumulation of Thr, the THA1 substrate, is expected for a tha1 mutant that is blocked in Thr degradation. The THA1 gene was sequenced from SALK_134724C and found to contain a single nucleotide deletion (T), predicted to result in translational frameshift, loss of the last 83 amino acids of the normal protein, and incorporation of 20 spurious amino acids (Fig. 8A). We conclude that this mutation likely causes the altered seed amino acid profile and designate this allele tha1-3.
Chlorophyll Fluorescence as a Case Study
The examples described above illustrate how this large-scale reverse genetic screen yields mutants defective in the annotated gene or unlinked mutations, both of which can contribute to our understanding of plant biology. This principle is observed beyond the metabolite phenotypes described above (fatty acids, starch, and amino acids). Query of the database for mutant lines with consistently decreased Fv/Fm before a 3-h high-light treatment, associated with altered PSII function (Maxwell and Johnson, 2000), revealed examples of the various scenarios encountered in the project (Table I). SALK_067017C has a T-DNA insertion upstream from the predicted mRNA-coding sequence of At5g52440, the homolog of the maize High Chlorophyll Fluorescence106 (HCF106) gene (Voelker and Barkan, 1995; Mori et al., 1999). The reduction in Fv/Fm in SALK_067017C is the result of an increase in minimal chlorophyll fluorescence. We are unaware of published phenotypic characterization of this or other Arabidopsis hcf106 mutants, and this mutant supports the hypothesis that reduced HCF106 function has a similar physiological consequence as in maize. SALK_011143C, annotated as containing an insertion upstream of the mRNA-coding sequence of At5g13770, which encodes a pentatricopeptide repeat-containing protein predicted to be chloroplast localized, reinforces the fact that multiple alleles are necessary to equate genotype with phenotype. This line consistently has a lower Fv/Fm before high light, but a second insertion allele that maps very close to the low Fv/Fm mutant insertion (SALK_051012C) did not show a strong phenotype. Further experiments are required to distinguish between the hypothesis that the inconsistency in chlorophyll fluorescence phenotypes is due to differences in severity of the alleles or that a secondary mutation is responsible for the phenotype of SALK_011143C. Lines SALK_043116C, SALK_098173C, SALK_151984C, and SALK_096388C all have lower Fv/Fm before high light (Table I), but lack of available second alleles precluded confirmation. These mutants are candidates for validation by alternative approaches such as confirmation with other alleles, segregation analysis, or analysis of candidate genes.
Table I.
SALK lines with lower Fv/Fm before high light
| Locus | Top Line Annotation | Allele | Insertiona | Genotype |
|---|---|---|---|---|
| At5g52440 | HCF106 (high chlorophyll fluorescence 106) | SALK_067017C | 5′-300 | Homozygous |
| At5g13770 | Pentatricopeptide repeat-containing protein | SALK_011143C | 5′-300 | Homozygous |
| At1g54580 | ACP2 (acyl carrier protein 2) | SALK_043116C | 5′-1,000 | Homozygous |
| At1g71500 | Rieske (2Fe-2S) domain-containing protein | SALK_098173C | 5′-300 | Homozygous |
| At5g57210 | Microtubule-associated protein-related | SALK_151984C | Exon | Homozygous |
| At2g42620 | MAX2 (more axillary branches 2) | SALK_096388C | Exon | Wild typeb |
5′-300 and 5′-1,000 indicate that the insertion is located within 300 and 1,000 nucleotides from the start of translation, respectively.
SALK_096388C did not show a mutant allele and thus is not found in the Chloroplast 2010 Project database.
DISCUSSION
Understanding a role for every gene in the genome of one or more reference plant species is a major challenge for the international community of plant scientists. There are increasing numbers of resources for experimental (quantitative trait locus analysis, artificial microRNAs, whole genome association genetic analysis, large-scale gene expression analysis resources, and large-scale proteomics, among others) and Web-based gene function analysis (Lu and Last, 2008) available for analysis of Arabidopsis gene function. The tens of thousands of sequence-indexed homozygous Arabidopsis T-DNA insertion mutants in the “unimutant” collection (Alonso et al., 2003) is among the most useful sets of tools for plant functional genomics, because it enables rapid and inexpensive access to mutants altered in expression of the majority of genes in the Columbia ecotype.
The Chloroplast 2010 Project (www.plastid.msu.edu; Lu et al., 2008) makes extensive use of this Arabidopsis mutant resource. Several thousand nuclear genes are being targeted for broad phenotypic analysis of loss-of-function mutants, and most of these genes are predicted to encode plastid-targeted proteins. Due to the large size of the project, a “one size fits all” pipeline approach is used to interrogate gene function. Because having two lines of evidence is essential for high confidence that a mutant phenotype is associated with alteration of a specific gene, our approach is to strive to assay two or more independent homozygous mutants for each target gene. Available mutants on the target list of more than 5,300 genes are ordered from the stock center, and two plants are tested for the genotype. Out of 3,673 lines tested at the time of this writing, 2,733 mutants were found to be homozygous for the insertion and 70% of these tested as having a single homozygous mutant line (Fig. 5B; Supplemental Table S1). In contrast, 26% of genes have two independent homozygous mutants and 4% of genes have more than two homozygous mutants.
Limitations of High-Throughput Genotyping
It is important to note the limitations of the genotyping data in Supplemental Table S1. First, only two plants typically are assayed for each line; thus, if the population of seeds obtained is segregating for the allele or contaminated with a line wild type for the target locus, we may not find a homozygous line in the tested individuals. Second, our high-throughput genotyping approach tests for the presence of the wild-type allele; while a control sample is run with wild-type tissue to confirm the ability of the assay to detect a wild-type allele, lack of an amplification product in the mutant line could be due to a problem with the PCR rather than to homozygosity of the mutant. This could cause us to misscore some heterozygous or homozygous wild-type plants. Finally, while we employ a sophisticated laboratory information management system to minimize tracking errors, no process is error free. For these reasons, it is suggested that the data in Supplemental Table S1 be used as a starting place for investigations of specific lines. Experiments should be conducted on lines obtained from the Arabidopsis Biological Resource Center or the Nottingham Arabidopsis Stock Centre/European Arabidopsis Stock Centre to test the genotype of the each stock.
Lessons from the Phenotypic Pipeline Studies
A forward genetic ethyl methanesulfonate mutant leaf FAMEs screen was performed exhaustively by Browse et al. (1985), and discovering mutants in other genes that cause changes in this phenotype reveals the promise of the high-throughput reverse genetics approach. One likely reason why this approach identified genes not discovered in the forward genetic screen is that the pipeline process involves running FAMEs on duplicate samples from each plant (process replicates) from two or more plants grown in separate flats (biological replicates) of each mutant line. This allows detection of more subtle changes in fatty acids. At1g10310, annotated as a pterin aldehyde reductase member of the large SDR family of proteins (Fig. 1), and At4g25050, encoding ACP4 (Fig. 3), are two such examples. The discovery that reduction (Fig. 1C) and overexpression (Fig. 2) of the SDR family protein mRNA causes changes in desaturation patterns of 18-carbon fatty acids extends our understanding of potential roles for this gene product. Support for the hypothesis that this enzyme functions in fatty acid or lipid metabolism was obtained by discovery that bacterial homologs of At1g10310 genetically cluster with known genes of fatty acid biosynthesis, including acyl carrier proteins and fatty acid synthase subunits (http://www.functionalnet.org/aranet/). Our results are not sufficient to differentiate between a direct or indirect role for this enzyme in determining 18:1 and 18:3 fatty acid levels. An intriguing (but unlikely) possibility is that the protein directly participates in reduction of polyunsaturated fatty acid double bonds. Alternatively, the protein could have enzymatic activity against medium- to long-chain fatty aldehydes, thereby indirectly affecting 18-carbon substrate pools. Our results with acp4 mutants validate published phenotypes of antisense ACP4 transgenic lines and show that loss of ACP4 function alone is sufficient for small light green plants with altered fatty acid metabolism and photosynthetic electron transport. The acp4 mutant results also underline the value of measuring multiple types of phenotypes for each mutant: the changes in whole plant morphology and chlorophyll fluorescence call attention to these mutants and make the relatively subtle differences in FAME profiles more noticeable during analysis of the high-throughput data.
Second lines of evidence supporting the results of phenotypic analysis of a single mutant allele can come from a variety of sources other than independent mutations in Arabidopsis. The T-DNA insertion allele SALK_067017C is an example where a second line of evidence comes from functional analysis of another organism. While we are not aware of phenotypic mutant results from any other Arabidopsis At5g52440 alleles, reduction in expression of the homologous maize gene HCF106 results in a similar phenotype (Voelker and Barkan, 1995). This strongly suggests that the T-DNA insertion upstream from the At5g52440 coding region is responsible for the defect in PSII function of the Arabidopsis mutant.
Not surprisingly, we discovered several examples of phenotypes caused by mutations outside of the annotated gene. Because the average number of insertions per T-DNA line was estimated to be more than one (Alonso et al., 2003), we anticipated finding examples of second site T-DNA insertions that cause the observed phenotypes. Indeed, the reduced afternoon starch mutant SALK_133788C contains a second insertion in the starch biosynthetic enzyme gene ADG1 (At5g48300). Two other mutants whose phenotypes were not confirmed by independent T-DNA mutations were found to have small deletions in previously described genes (fad5-2 [Fig. 6A] and tha1-3 [Fig. 7A]). Other mutants whose phenotypes did not confirm in second insertion alleles presumably have secondary mutations in other genes (e.g. the light green and altered 16-carbon FAME pattern of SALK_027707C [Fig. 4] and the altered Fv/Fm trait found in SALK_011143C [Table I]). While the existence of tagged and untagged secondary mutations complicates the process of identifying genes responsible for specific phenotypes, these mutants also create opportunities for discovering novel genes affecting the phenotypes that are being assayed in the Chloroplast 2010 Project or new alleles of previously characterized genes. This would require the identification of secondary insertion mutations, map-based cloning, or whole genome sequence analysis to discover the genetic changes responsible for the phenotype.
Relevance of the Results to the Chloroplast 2010 Project
The results in this study can be used to guide investigators using the phenotypic data in the Chloroplast 2010 Project database (http://bioinfo.bch.msu.edu/2010_LIMS). Genes with two mutants of similar phenotypes offer the most attractive candidates for analysis. The next best case scenario is one in which only a single mutant is available but other types of evidence can be used to support the hypothesis that mutation of the annotated gene causes the observed phenotype. Examples include mutations in other organisms that give rise to a similar phenotype (e.g. HCF106 and At5g52440), coregulation with other genes of known function (Gao et al., 2009), physical linkage with genes of known function in prokaryotes (At1g10310), and biochemical activity of a homologous enzyme from Arabidopsis or another organism. Examples where mutants with independent alleles yield conflicting phenotypes can have a variety of causes, including the existence of secondary mutations or incorrect genotyping, phenotyping, or tracking by our high-throughput pipeline. In any case, where one or more mutants have a phenotype of interest, it is prudent to order the lines from the relevant stock center (Arabidopsis Biological Resource Center or Nottingham Arabidopsis Stock Centre/European Arabidopsis Stock Centre) and test the genotype and phenotype. If a reproducible phenotype is detected in one of the lines, identification of the secondary mutation can be pursued by appropriate methods.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Phenotyping
SALK lines and fad5-1 of Arabidopsis (Arabidopsis thaliana) were obtained from the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm). Seed for tha1-1 was provided by Monsanto, and SAIL line SAIL_868_G02 was obtained from John Browse and Hua Weng. The transposon-tagged RIKEN line 15-1699-1 and a wild-type segregant control were kindly provided by Alexandre Noiriel and Andrew Hanson. Growth conditions and phenotypic assays for plants grown in the Chloroplast 2010 pipeline were described in detail by Lu et al. (2008), unless otherwise indicated. Detailed protocols used in the Chloroplast 2010 Project are available at http://plastid.msu.edu/about/protocols.html. Seed FAMES were extracted as described by Li et al. (2006). Seed triacylglycerols were extracted and separated by thin-layer chromatography as described by Focks and Benning (1998). Bands corresponding to triacylglycerols were scraped from thin-layer chromatography plates, and FAMES were analyzed by GC-FID. Seeds for lines described in this paper have been deposited to the Arabidopsis Biological Resource Center (Supplemental Table S2).
Genotyping SALK Lines
DNA samples were archived onto Whatman FTA Plantsaver cards and prepared following the manufacturer's recommendations. SALK lines were genotyped using two different PCR-based strategies. The first approach followed the guidelines recommended by the SALK Institute (http://signal.SALK.edu/tdnaprimers.2.html). Primers were designed utilizing the SALK T-DNA primer design tool with the Ext5 value changed to 200 and using LBa1 as the T-DNA-specific primer. Briefly, each archived sample was subjected to two PCR procedures: an amplification using two specific genomic primers, and an amplification using a specific genomic primer and a T-DNA insert-specific primer (LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′). The specific genomic primers were also used to amplify a wild-type plant sample in parallel to assess primer quality. PCR products were then visualized using agarose gels with ethidium bromide.
To process the large number of archived samples, more than half of those shown in Supplemental Table S1 were genotyped using a high-throughput “first-pass” SYBR Green method, with only those samples not appearing to be homozygous subjected to further analysis to distinguish between heterozygous and wild-type genotypes using the two-reaction method and agarose gel visualization. In this method, two sibling samples were placed in the same reaction well to genotype with two gene-specific genomic primers (LP and RP), designed to amplify the wild-type allele. One wild-type sample was also genotyped in the adjacent well with the same two specific genomic primers as a positive control. Samples were amplified using SYBR Green PCR Master Mix following the manufacturer's recommended cycling conditions (Applied Biosystems) except that reactions were supplemented with one additional unit of Ampli-Taq Gold (Applied Biosystems) and primers were added to a final concentration of 0.1 μm. End-point values were read at 72°C by a 7900 HT Sequence Detection System (Applied Biosystems). End-point fluorescence values of wild-type and mutant samples were compared, and mutant samples that did not appear to contain PCR product were considered to be homozygous. If the pooled mutant samples appeared to contain amplified product, each sibling was genotyped again separately on agarose gels using the two-reaction approach described above. Regenotyping eliminated false positives caused by primer-dimer detection and allowed us to infer whether a line was heterozygous for the insertion mutation or homozygous for the wild-type allele. However, it is likely that occasionally low-yield PCRs were incorrectly interpreted as being homozygous.
Identification of Secondary Mutations
Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen) following the manufacturer's protocol. The following primers were used to amplify and sequence the FAD5 genomic locus in SALK_006881C: 5′-ATAAGTTAAGGGTTTAAGCC-3′ and 5′-AAAGCAGAACAAAATCCTTT-3′. The following primers were used to further sequence FAD5: 5′-TTGATACCAATACAATCACCC-3′, 5′-GACGAAAATCAAATCAGAAACC-3′, and 5′-GAACCAAAACCCATCAAGTG-3′. The THA1 genomic locus was amplified from SALK_134724C and sequenced using the following primers: 5′-GGTGTTGGTACTCAAAAGTGTTCC-3′ and 5′-TTAATTAAAATGTACTTTAGACAAC-3′. The following primers were used to further sequence THA1: 5′-ATTAGAGATCCTAAAGGAAGCACG-3′, 5′-GATAAGAGTCATTCACACACACTC-3′, and 5′-GTGACTCAGTGATGCATCAATCAC-3′. Presence of an insertion in the At5g48300 locus was identified in SALK_133788C using a gene-specific primer (5′-GGACTCCGTTCCTGCATATC-3′) and a T-DNA right border primer (5′-GGTTCTGTCAGTTCCAAACG-3′) for PCR confirmation and sequence verification.
Semiquantitative Reverse Transcription-PCR Analysis
Total RNA was extracted from rosette leaves using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of RNA with SuperScript III (Invitrogen). ReadyMix redtaq (Sigma-Aldrich) was used for the PCR using the following thermal cycling conditions: 95°C for 2 min; 36 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 45 s; and 72°C for 10 min. Primers specific to the At1g10310 SDR were 5′-ATGACGATGGCGACGCCTTT-3′ and 5′-CAGTAAGGTGCAACCAGGGC-3′. Primers for the control gene elongation factor1-α (At5g60390; Ajjawi et al., 2007) were 5′-CATGGGTGTTGGACAAACTT-3′ and 5′-CTCCTTGATGATTTCATCGT-3′.
Generation of the 35S:At1g10310 Construct and Plant Transformation
Total RNA was isolated and cDNA was synthesized as described above. The SDR open reading frame (gene model At1g10310.1) was amplified using primers 5′-ATGACGATGGCGACGCCTTT-3′ and 5′-TTAGACTGTGAGAGATCCGC-3′. The resulting PCR fragment was cloned into pCR8/GW/TOPO (Invitrogen) according to the manufacturer's recommendations. The SDR open reading frame was inserted into the Gateway-compatible vector pMDC32 (Curtis and Grossniklaus, 2003) using LR Clonase (Invitrogen) as recommended by the manufacturer. The binary plant transformation vector carrying the SDR coding sequence designated 35S:At1g10310 was introduced into Agrobacterium tumefaciens strain C5851 and transformed into wild-type Columbia plants by floral dip (Clough and Bent, 1998). The T1 seeds were screened for hygromycin B (25 μg mL−1) resistance, and the resulting T2 seeds were again screened for hygromycin resistance to identify lines with antibiotic segregation ratios consistent with the presence of a single transgene locus.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. FAMEs of seed triacylglycerols for a wild-type segregant (WT segregant) and transposon-tagged line 15-1699-1 (Ds).
Supplemental Figure S2. Evidence for cytosolic localization of At1g10310-GFP protein.
Supplemental Table S1. Chloroplast 2010 Project genotyping results.
Supplemental Table S2. Mutant lines submitted to the Arabidopsis Biological Resource Center (ABRC).
Supplementary Material
Acknowledgments
This project is the result of the efforts of a very large number of people. We thank the co-Principal Investigators of the Chloroplast 2010 Project at Michigan State University for guiding various aspects of the project: Christoph Benning, Dean DellaPenna, John Ohlrogge, Katherine Osteryoung, Yair Shachar-Hill, Andreas Weber, Bill Wedemeyer, and Curtis Wilkerson. Numerous undergraduate assistants were instrumental in growing plants, harvesting tissue, capturing genotypic and phenotypic data, and performing data analysis. Special thanks are due to longtime undergraduate contributors Kayla Kerr, David Hall, and Claire Moore. Kathleen Imre has been central to assay development and data acquisition and analysis. Matt Larson has played a crucial role in development of the database, without which this project would not be possible. Technical support was provided by Amanda Charbonneau, Amanda Ellsworth, Eric Jones, Jessica Reif, and Val Reisen. We thank the Arabidopsis Biological Resource Center for thousands of seed stocks as well as John Browse, Andrew Hanson, Alexandre Noiriel, Hua Weng, and Monsanto Co. for seeds and DNA clones. Thanks to Ivan Baxter for helpful discussions on the project.
This work was supported by the U.S. National Science Foundation 2010 Project (grant no. MCB–0519740).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert L. Last (lastr@msu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams DJ, van der Weyden L (2008) Contemporary approaches for modifying the mouse genome. Physiol Genomics 34 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J 41 859–874 [DOI] [PubMed] [Google Scholar]
- Ajjawi I, Rodriguez Milla MA, Cushman J, Shintani DK (2007) Thiamin pyrophosphokinase is required for thiamin cofactor activation in Arabidopsis. Plant Mol Biol 65 151–162 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7 524–536 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al (2003) Arabidopsis genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a Web-based database. Plant Physiol 132 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Ohlrogge JB (2002) Differential regulation of mRNA levels of acyl carrier protein isoforms in Arabidopsis. Plant Physiol 128 223–235 [PMC free article] [PubMed] [Google Scholar]
- Branen JK, Shintani DK, Engeseth NJ (2003) Expression of antisense acyl carrier protein-4 reduces lipid content in Arabidopsis leaf tissue. Plant Physiol 132 748–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, McCourt P, Somerville CR (1985) A mutant of Arabidopsis lacking a chloroplast-specific lipid. Science 227 763–765 [DOI] [PubMed] [Google Scholar]
- Cho Y, Walbot V (2001) Computational methods for gene annotation: the Arabidopsis genome. Curr Opin Biotechnol 12 126–130 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA (1991) T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J 1 71–82 [Google Scholar]
- Focks N, Benning C (1998) Wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Ajjawi I, Manoli A, Sawin A, Xu C, Froehlich JE, Last RL, Benning C (2009) FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J 60 832–839 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391 [DOI] [PubMed] [Google Scholar]
- Heilmann I, Mekhedov S, King B, Browse J, Shanklin J (2004. a) Identification of the Arabidopsis palmitoyl-monogalactosyldiacylglycerol delta7-desaturase gene FAD5, and effects of plastidial retargeting of Arabidopsis desaturases on the fad5 mutant phenotype. Plant Physiol 136 4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I, Pidkowich MS, Girke T, Shanklin J (2004. b) Switching desaturase enzyme specificity by alternate subcellular targeting. Proc Natl Acad Sci USA 101 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlousek-Radojcic A, Post-Beittenmiller D, Ohlrogge JB (1992) Expression of constitutive and tissue-specific acyl carrier protein isoforms in Arabidopsis. Plant Physiol 98 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JL, Porter J, Gray CA, Andersen RJ, et al (2009) A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol 27 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K (2002) A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol 129 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu S, Li L, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening: evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39 465–475 [DOI] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G (2006) Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, et al (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J 47 640–651 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67 904–915 [DOI] [PubMed] [Google Scholar]
- Lu Y, Last RL (2008) Web-based Arabidopsis functional and structural genomics resources. In The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, http://www.aspb.org/publications/arabidopsis/, doi/10.1199/tab.0118 [DOI] [PMC free article] [PubMed]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, Dellapenna D, Ohlrogge JB, Osteryoung KW, Weber AP, et al (2008) New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol 146 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51 659–668 [DOI] [PubMed] [Google Scholar]
- Mori H, Summer EJ, Ma X, Cline K (1999) Component specificity for the thylakoidal Sec and Delta pH-dependent protein transport pathways. J Cell Biol 146 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiriel A, Naponelli V, Bozzo GG, Gregory JF III, Hanson AD (2007) Folate salvage in plants: pterin aldehyde reduction is mediated by multiple non-specific aldehyde reductases. Plant J 51 378–389 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U (2002) The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet 3 124–136 [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ (2000) RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol 10 1619–1622 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Wood V, Dolinski K, Draghici S (2008) Use and misuse of the Gene Ontology annotations. Nat Rev Genet 9 509–515 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schnurr JA, Shockey JM, de Boer GJ, Browse JA (2002) Fatty acid export from the chloroplast: molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol 129 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Barkan A (1995) Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J 14 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SM, Lue WL, Yu TS, Long JH, Wang CN, Eimert K, Chen J (1998) Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J 13 63–70 [DOI] [PubMed] [Google Scholar]
- Wendland J (2003) PCR-based methods facilitate targeted gene manipulations and cloning procedures. Curr Genet 44 115–123 [DOI] [PubMed] [Google Scholar]
- Wu S, Ying G, Wu Q, Capecchi MR (2007) Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet 39 922–930 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.