Abstract
The endogenous trans-acting small interfering RNA (ta-siRNA) pathway plays a conserved role in adaxial-abaxial patterning of lateral organs in simple-leafed plant species. However, its function in compound-leafed species is largely unknown. Using the compound-leafed species Lotus japonicus, we identified and characterized two independent mutants, reduced leaflet1 (rel1) and rel3, whose most conspicuous defects in compound leaves are abaxialized leaflets and reduction in leaflet number. Concurrent mutations in REL genes also compromise flower development and result in radial symmetric floral organs. Positional cloning revealed that REL1 and REL3 encode the homologs of Arabidopsis (Arabidopsis thaliana) SUPPRESSOR OF GENE SILENCING3 and ARGONAUTE7/ZIPPY, respectively, which are key components of the ta-siRNA pathway. These observations, together with the expression and functional data, demonstrated that the ta-siRNA pathway plays conserved yet distinct roles in the control of compound leaf and flower development in L. japonicus. Moreover, the phenotypic alterations of lateral organs in ta-siRNA-deficient mutants and the regulation of downstream targets by the ta-siRNA pathway in L. japonicus were similar to those in the monocots but different from Arabidopsis, indicating many parallels between L. japonicus and the monocots in the control of lateral organ development by the ta-siRNA pathway.
Plant endogenous small RNAs can be categorized into microRNAs (miRNAs) and small interfering RNAs (siRNAs) according to their mechanism of biogenesis (Vaucheret, 2006). trans-Acting siRNAs (ta-siRNAs) are one type of siRNA, and their biogenesis requires several key components, such as SUPPRESSOR OF GENE SILENCING3 (SGS3), RNA-DEPENDENT RNA POLYMERASE6 (RDR6), DICER-LIKE4 (DCL4), ARGONAUTE7 (AGO7)/ZIPPY (ZIP), and dsRNA-BINDING4 (Peragine et al., 2004; Vazquez et al., 2004; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Adenot et al., 2006; Nakazawa et al., 2007). Recent studies revealed that the ta-siRNA pathway is integrated into different processes of plant development, such as vegetative phase transition in Arabidopsis (Arabidopsis thaliana; Hunter et al., 2003; Peragine et al., 2004; Xie et al., 2005; Nakazawa et al., 2007) and shoot apical meristem (SAM) initiation in rice (Oryza sativa; Satoh et al., 1999; Itoh et al., 2000; Nagasaki et al., 2007). Parallel studies of this pathway in simple-leafed species also showed that the ta-siRNA pathway plays critical roles in patterning of leaves and floral organs.
In flowering plants, leaves and flowers are produced on the periphery of the apical meristem. These lateral organs are structurally asymmetric with regard to the apical meristem. The adaxial side is adjacent to the meristem, while the abaxial side is away from the meristem. The ta-siRNA pathway was found to play a conserved role in specifying the adaxial identity of lateral organs in both monocots and dicots, but defects in the ta-siRNA pathway caused more severe phenotypes in monocots than in dicot Arabidopsis. In Arabidopsis, no clear leaf polarity defects were detected in the ta-siRNA-defective mutants. However, blocking the ta-siRNA pathway in asymmetric1 (as1) or as2 background, which are regulators of leaf adaxial identity (Lin et al., 2003; Xu et al., 2003), results in enhanced adaxial-abaxial leaf defects (Li et al., 2005; Xu et al., 2006; Garcia et al., 2006). In addition, the as2rdr6 double mutants also display aberrant flowers with sepals failing to enwrap the inner whorl organs and some sepals and petals becoming needle-like structures (Li et al., 2005). In maize (Zea mays), mutations in LEAFBLADELESS1 (LBL1), which encodes the Arabidopsis SGS3 ortholog, give rise to abnormal leaves with partial or complete loss of adaxial cell identity (Timmermans et al., 1998; Nogueira et al., 2007). In severe lbl1 mutants, leaf-like lateral organs of inflorescences and flowers develop as symmetric, thread-like organs, and the immature ear is exposed and arrested in development (Timmermans et al., 1998). In rice, the osdcl4-1 mutants display an abaxialized epidermis in coleoptiles and in the first leaf, and knockdown of OsDCL4 can lead to the awn-like lemma with a radial abaxialized identity and the stamens and carpel not enwrapped by the lemma and pelea (Liu et al., 2007). Transgenic rice plants with ectopic expression of SHOOTLESS4 (SHL4), the homolog of Arabidopsis AGO7, exhibit partially adaxialized leaves (Nagasaki et al., 2007; Shi et al., 2007).
In addition to the ta-siRNA pathway, other components have also been shown to be involved in the adaxial-abaxial patterning of lateral organs. The Antirrhinum majus PHANTASTICA (PHAN) gene (Waites et al., 1998; Byrne et al., 2000; Xu et al., 2003; Qi et al., 2004), which is the ortholog of Arabidopsis AS1, and CLASS III HOMEODOMAIN-LEUCINE ZIPPER (HD-ZIP III) gene family members (McConnell et al., 2001; Emery et al., 2003) contribute to adaxial pattern formation of lateral organs, whereas members of YABBY (YAB; Sawa et al., 1999; Siegfried et al., 1999) and KANADI (Eshed et al., 2001; Kerstetter et al., 2001) gene families, AUXIN RESPONSE FACTOR3 (ARF3) and ARF4 (Pekker et al., 2005), and the miRNAs miR165/166 (Emery et al., 2003; Eshed et al., 2004; Mallory et al., 2004) are required for specifying abaxial identity. How the activities of these adaxial and abaxial determinants are coordinated has been extensively studied. It was found that ARF3 and ARF4 are regulated by the TAS3 ta-siRNA, and this regulation is conserved in both monocots and dicots (Allen et al., 2005; Williams et al., 2005). Recent studies in Arabidopsis suggest that ta-siRNAs act in a non-cell-autonomous manner to spatially restrict ARF activity (Chitwood et al., 2009; Schwab et al., 2009).
In contrast to simple leaves with their single lamina, compound leaves are composed of one petiole and several leaflets. It is found that genes required for the adaxial-abaxial patterning of lateral organs in simple-leafed species also play critical roles in compound-leafed species, but these genes play multiple roles in compound leaf development. In tomato (Solanum lycopersicum), down-regulation of PHAN ortholog disturbs the leaf polarity as well as leaflet formation (Kim et al., 2003). Extensive studies of the PHAN expression in diverse compound-leafed species suggest that the function of PHAN in maintaining leaf adaxial identity is associated with leaflet formation in compound leaves and reduced adaxial identity of leaf primordia by down-regulation of PHAN could change pinnate compound leaves into palmate leaves (Kim et al., 2003). In pea (Pisum sativum), the role of PHAN in compound leaf development has also been elucidated by characterization of the phan mutant crispa (cri; Tattersall et al., 2005). However, unlike antisense PHAN transgenic tomato leaves, the cri mutant has the individual leaflet abaxialized, rather than the whole leaf. The number of lateral organs on the cri mutant compound leaves, including leaflets, is not altered, and the leaves remain pinnate. Apart from leaf development, the cri mutation also affects flower development. Although the floral organ identity and organ number are not altered, the laminar floral organ display abaxialized identity (Tattersall et al., 2005).
The ta-siRNA pathway plays a critical role in simple-leafed species, but its role in compound-leafed species is not understood. Here, we address this question by analyzing loss-of-function reduced leaflet (rel1) and rel3 mutants in the compound-leafed species Lotus japonicus. Phenotypic characterization shows compound leaves of rel mutants exhibit a conspicuous disturbance in leaflet polarity as well as reduction in leaflet number. Besides the abnormal compound leaves, flower development is also severely affected in rel mutants, showing radial symmetric petals. REL1 and REL3 were identified by map-based cloning and were shown to be homologs of Arabidopsis SGS3 and AGO7, respectively. REL1 and REL3 act in the same genetic pathway and are both required for the biogenesis of TAS3 ta-siRNA. Further investigation reveals that the homolog of the Arabidopsis ARF3 is duplicated in the L. japonicus genome and that the duplicate ARF3 homologs and the ARF4 homolog are all negatively regulated by the ta-siRNA pathway. Furthermore, we found that the expression of LjYAB1, a homolog of Arabidopsis YAB1, was decreased in rel mutants, which may be associated with the reduced lamina.
Taken together, our data reveal that the ta-siRNA pathway is integrated into the regulatory networks in the control of lateral organ development in L. japonicus and further emphasize the importance of the ta-siRNA pathway in compound leaf development. Moreover, our results also indicate many parallels between L. japonicus and monocots for the ta-siRNA pathway in the regulation of lateral organs.
RESULTS
Phenotypic Characterization of Compound Leaves and Flowers in rel1 and rel3
From large scale mutagenesis screens in L. japonicus (Feng et al., 2006), we isolated a series of mutants with disrupted leaf polarity and reduction in leaflet number in compound leaves, designated as rel. Apart from the conspicuous leaf abnormality, the mature rel plants also display aberrant flowers.
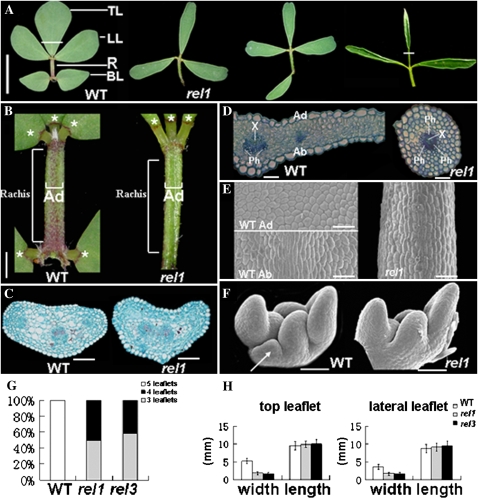
Wild-type L. japonicus plants display classical compound leaves, with one rachis and five leaflets: one top and two lateral leaflets are clustered at the tip of petiole, and two basal ones are arranged at the base of rachis (Fig. 1A). In rel1 mutants, loss of one or two basal leaflets occurs in nearly all mature compound leaves (Fig. 1, A and G), and the leaflets are narrower than those of the wild type (Fig. 1, A and H). In addition, needle-like leaflets are found occasionally (Fig. 1A). However, the length of the rachis is not altered, and the distinct adaxial domain is still present throughout the rachis region (Fig. 1, B and C). Anatomical analysis of the needle-like leaflet showed that the cell type around the central vasculature was homogeneous and that phloem tissue surrounded the xylem tissue as a ring (Fig. 1D). This is in contrast to the wild-type leaflet, where palisade mesophyll cells and xylem are on the adaxial side, and spongy mesophyll cells and phloem are on the abaxial side (Fig. 1D). Scanning electron microscopy (SEM) analysis showed that the needle-like leaflets were covered with long and narrow epidermal cells, which are similar to the epidermal cells observed on the abaxial side of the wild-type leaflet (Fig. 1E). These data clearly indicate that in rel1 mutants, specification of the leaflet adaxial-abaxial polarity is disturbed and leaflets are abaxialized. SEM was also used to examine vegetative shoots and revealed the absence of the basal leaflets commenced at the early stage when leaflet primordia were initiated, and the basal leaflet primordia were normally not found in rel1 mutants (Fig. 1F).
Figure 1.
Different effects of rel mutations on compound leaf. A, In the wild type (WT), the mature compound leaf consists of one top (TL), two lateral (LL), and two basal leaflets (BL). In rel mutants, the leaflets are narrow, and one or two BL leaflets are absent. Needle-shaped leaflets can be found. B, Adaxial domain on the rachis of wild-type and rel1 compound leaves. The adaxial domain could be observed throughout the rachis of both wild-type and rel1 compound leaves. Asterisks, leaflets; Ad, adaxial domain. C, Transverse sections of rachis in the wild type and rel1. D, Transverse sections of TL leaflets in the wild type and rel1 (the sectioning regions in the TL leaflets are indicated with white lines in A). E, Wild-type adaxial and abaxial epidermal patterns of leaves and epidermal patterns of needle-shaped leaflets in rel1 mutants. Ad, Adaxial; Ab, abaxial. F, SEM analysis of initiation of leaflet primordia. The initiation of BL primordia commences in the wild type but is blocked in rel1 (arrows). Asterisks, meristem; TL, top leaflet primordium; LL, lateral leaflet primordium; BL, basal leaflet primordium. Bars = 1 cm in A, 1 mm in B, and 100 μm in C to F. G, Quantitative analysis of leaflet numbers on wild type (n = 12) and rel mutants (n = 12) between nodes 6 and 10. Bars show sd. H, Quantitative analysis of the top and lateral leaflet width and length on the wild type (n = 12) and rel mutants (n = 12) between nodes 6 and 10. Bars show sd.
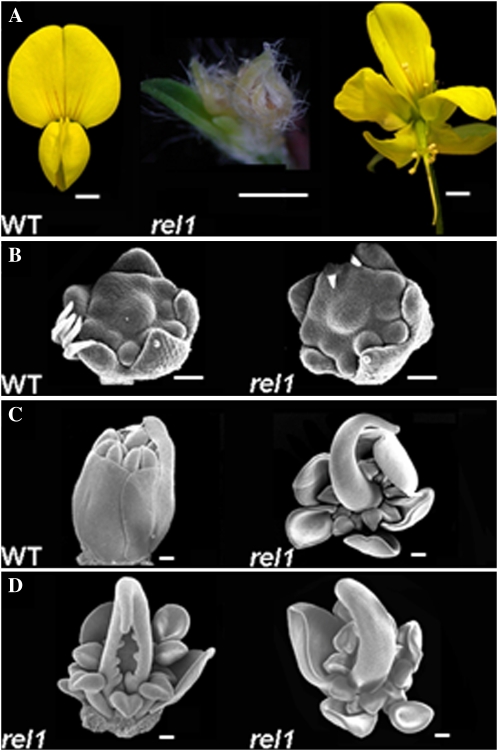
Another prominent phenotype of rel1 mutants was their infertile flowers (Fig. 2A). In comparison with floral organs in wild-type L. japonicus plants, most rel1 flowers were dramatically reduced in size. We examined more than 300 rel1 flowers, and only two re11 flowers were as large as wild-type flowers, but these two flowers opened prematurely (Fig. 2A). The phenotypic abnormalities of rel1 flowers were further analyzed by SEM. During early stages of flower development, the initiation of floral organ primordia in the floral meristem was normal and no defects in floral organ number and organ identity were detected (Fig. 2B). In the later stage when floral organs were further differentiated, the carpel and stamens were enclosed by the petals in the wild type (Fig. 2C). However, all petals in rel1 mutants were open, and the inner two reproductive organs were exposed (Fig. 2C). In addition, the carpel were frequently split (Fig. 2D), and some petal primordia formed a nearly symmetric and trumpet-shaped structure containing small undifferentiated epidermal cells (Fig. 2E), indicating that the adaxial-abaxial patterning of floral organ was also disturbed in rel1 mutant. The phenotypic analysis of rel3 mutant leaves and flowers showed that the developmental defects in rel3 were very similar to those in rel1 (data not shown).
Figure 2.
Flowers of the wild type (WT) and the rel1 mutant. A, In the rel1 mutant, only a few infertile flowers reached the full size as does the wild type, and most are degenerate. B, No detectable difference between the wild type and the rel1 mutant is found during the primordium initiation of floral organs. C to E, SEM analysis of floral organ differentiation in the wild type and the rel1 mutant. At the stage when the petals enclose stamens and carpel in the wild type (C), the inner two whorls of reproductive organs were exposed (C–E). In the rel1 mutant, the carpel is frequently split (D), and symmetric petals are occasionally observed (E). Bars = 1 mm in A and 100 μm in C to E.
Cloning of REL1 and REL3
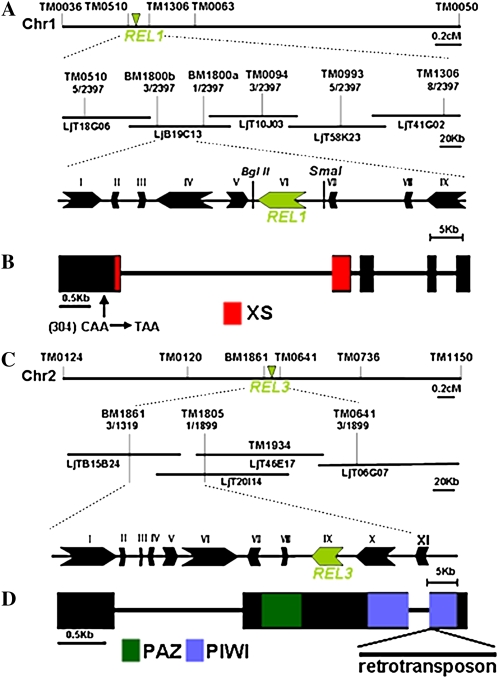
A map-based cloning strategy was utilized to clone the REL1 and REL3 loci. The REL1 locus was positioned on chromosome 1 between markers BM1800a and BM1800b, which contains nine putative genes by annotation analysis (Fig. 3A). Sequence comparison of the wild type and rel1 mutants in this region revealed a nucleotide substitution in rel1, leading to a premature stop codon in the coding region of the homolog of Arabidopsis SGS3 (Fig. 3B). The REL3 locus was mapped to chromosome 2 and then was narrowed down to a region between markers BM1861 and TM1805, where eleven 11 putative genes were identified (Fig. 3C). Further analysis revealed that rel3 carried a retrotransposon insertion in the coding region of the gene encoding an ARGONAUTE protein with PAZ and PIWI domains (Fig. 3D). The coding sequence of REL1 and REL3 was obtained by reverse transcription (RT)-PCR. Phylogenetic analysis further indicated that REL1 is a likely homolog of Arabidopsis SGS3 and maize LBL1, while REL3 was closely related with Arabidopsis ZIP/AGO7 and rice SHL4 (Supplemental Fig. S1, A and B).
Figure 3.
Cloning of REL1 and REL3 genes. A, Fine structure mapping to localize REL1 on chromosome 1 between markers BM1800a and BM1800b; the BglII-SmaI genomic fragment containing the REL1 gene was used in the complementation experiment. B, The REL1 gene structure and position of nucleotide changes in rel1 mutants. The red box represents a region encoding the XS domain. C, Fine structure mapping to localize REL3 on chromosome 2 between markers BM1861 and TM1805. D, The REL3 gene structure and position of insertion in rel3 mutants. The green and blue boxes indicate the region encoding the PAZ domain and PIWI domain, respectively.
A 10-kb genomic fragment containing the L. japonicus SGS3 homolog in the REL1 region was transformed into the rel1 mutant by stable Agrobacterium tumefaciens-mediated gene transfer. In resulting transformants, the mutant compound leaf and flower phenotypes were fully recovered (Supplemental Fig. S2). To test the possible genetic interaction between rel1 and rel3, genetic analysis was conducted, and the rel1rel3 double mutants were obtained. No detectable difference was found between the double mutants and the single mutant (data not shown), indicating that REL1 and REL3 act in the same genetic pathway. Accordingly, we conclude that REL genes should encode different components in the same ta-siRNA pathway, and their mutations are responsible for the rel phenotypes.
Molecular Characterization of REL1 and REL3
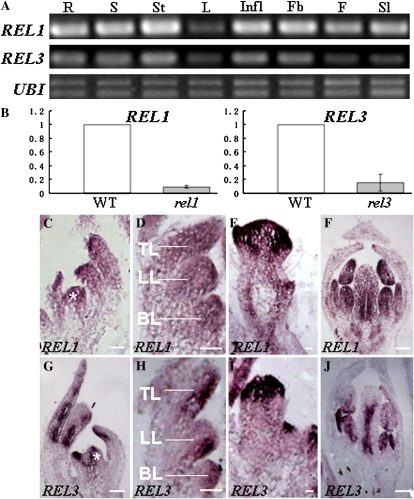
We analyzed the expression of REL1 and REL3 by RT-PCR, and both transcripts were detected in all tissues tested, including aerial organs and roots (Fig. 4A). The broad mRNA expression profiles of REL genes were consistent with their roles in regulating multiple developmental processes. Transcript abundance of REL1 and REL3 was dramatically reduced in rel1 and rel3 mutants, respectively (Fig. 4B), indicating that the mutations in rel1 and rel3 plants may affect both protein function and mRNA stability.
Figure 4.
Expression analysis of REL1 and REL3. A, RT-PCR analysis of REL1 and REL3 expression in all plant tissues examined. R, Root; S, seedling; St, stem; L, leaf; Infl, inflorescence; Fb, floral bud; F, flower; Sl, silique. B, Real-time PCR analysis of the transcript levels of REL1 and REL3 in the wild type (WT) and mutants. Quantification was normalized to that of UBI and then to the value of wild-type Gifu plants, whose value was arbitrarily fixed at 1. Bars show se. C to F, An antisense probe of REL1 is used in the RNA in situ hybridization on longitudinal sections of vegetative and reproductive shoot apices of the wild type. G to J, An antisense probe of REL3 is used. Asterisks, meristem; TL, top leaflet primordium; LL, lateral leaflet primordium; BL, basal leaflet primordium. Bars = 50 μm.
RNA in situ hybridization was conducted to further examine spatial expression patterns of REL1 and REL3 in the wild type. At the vegetative stage, transcripts of REL1 were detected throughout the SAM, young leaf primordial, and developing leaflets (Fig. 4, C and D). However, the domain of REL3 expression was largely limited to central region of the SAM (Fig. 4G). In the newly emerging leaf primordium, REL3 accumulated preferentially on the adaxial side, and the expression was retained on the adaxial side of emerging leaflet primordium (Fig. 4, G and H). At the reproductive stage, higher levels of expression of both REL1 and REL3 were found in the floral meristem and developing floral organs (Fig. 4, E, F, I, and J), but in the carpel, transcripts of REL1 and REL3 localized mainly on the inner side (Fig. 4, F and J).
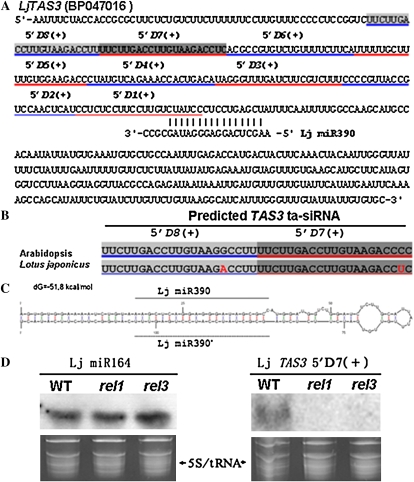
Previous studies showed that TAS3 ta-siRNA and miR390, which direct the cleavage of TAS3 ta-siRNA precursor, are conserved between monocots and dicots (Allen et al., 2005; Liu et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007). Using the Arabidopsis TAS3 ta-siRNA sequence as a query, we BLAST-searched the current available L. japonicus genome database (www.kazusa.or.jp/lotus/) and identified sequence BP047016, which matched the criteria to be a precursor of TAS3 ta-siRNA and should be a putative TAS3 gene in L. japonicus (Fig. 5A). The predicted L. japonicus TAS3 ta-siRNA carried only one nucleotide mismatch with the Arabidopsis TAS3 ta-siRNA (Fig. 5B). The precursor of miR390 with the ability to form stem-loop structures was also found (Fig. 5C). The accumulation of TAS3 ta-siRNA in the wild type and rel mutants was compared using small RNA blots. The accumulation of TAS3 ta-siRNA was evident in the wild type but was abolished in rel mutants (Fig. 5D). By comparison, the expression level of a miRNA, namely miR164, did not show any difference between the wild type and rel mutants (Fig. 5D). These results confirmed that REL1 and REL3 were the key components required for the biogenesis of L. japonicus TAS3 ta-siRNA.
Figure 5.
Characterization of the TAS3 ta-siRNA precursor and TAS3 ta-siRNA in L. japonicus (Lj). A, Predicted ta-siRNAs in LjTAS3 (BP047016; underlined in red or blue). B, Alignment of the sequences of AtTAS3 ta-siRNA and LjTAS3 ta-siRNA (one base mismatch is indicated in red). C, Predicted stem-loop structure of LjmiR390. The positions of mature LjmiR390 and complementary LjmiR390* are shown. D, Northern-blot analysis of the accumulation of LjTAS3 ta-siRNA in the wild type (WT), rel1, and rel3. L. japonicus miR164 is used as a control.
Isolation and Expression Analysis of Direct and Indirect Targets Regulated by REL1 and REL3
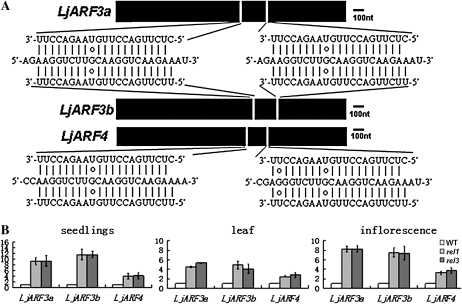
ARF3 and ARF4 are direct targets of TAS3 ta-siRNA, and the mechanism of this regulation is well conserved in both monocots and dicots (Allen et al., 2005; Williams et al., 2005). In L. japonicus, several partial sequences homologous to ARF3 were identified from the current L. japonicus genome database. Based on this sequence information, we cloned two copies of ARF3 homolog using RACE PCR. Further identification and analysis of the corresponding TAC clones revealed that the ARF3 homologs in L. japonicus genome were duplicated, and the two ARF3 homologs shared 69.1% similarity and 64.0% identity, respectively. One putative ARF4 homolog in L. japonicus was also identified. Phylogenetic analysis confirmed their close relationship with ARF3 and ARF4 in Arabidopsis, respectively (Supplemental Fig. S1C). Therefore, these ARF genes were named LjARF3a, LjARF3b, and LjARF4, respectively. Both LjARF3a and LjARF3b contain two adjacent sequences in the coding region that are complementary to TAS3 ta-siRNA, whereas LjARF4 carries only one complementary sequence (Fig. 6A). We then compared the expression level of these three ARF genes in the wild type and rel mutants. We found that the transcript levels of these genes were all up-regulated in seedlings, leaves, and inflorescence of rel mutants (Fig. 6B), though the up-regulation of LjARF3a and LjARF3b was more dramatic than that seen for LjARF4 (Fig. 6B).
Figure 6.
Isolation and expression analysis of LjARF3a, LjARF3b, and LjARF4. A, Diagrammatic representation of coding sequence of LjARF3a, LjARF3b, and LjARF4 with TAS3 ta-siRNA complementary sites. B, Real-time PCR analysis of transcript levels for LjARF3a, LjARF3b, and LjARF4 in the wild type (WT) and rel mutants. Quantification was normalized to that of UBI and then to the value of wild-type Gifu plants, whose value was arbitrarily fixed at 1. Bars indicate se.
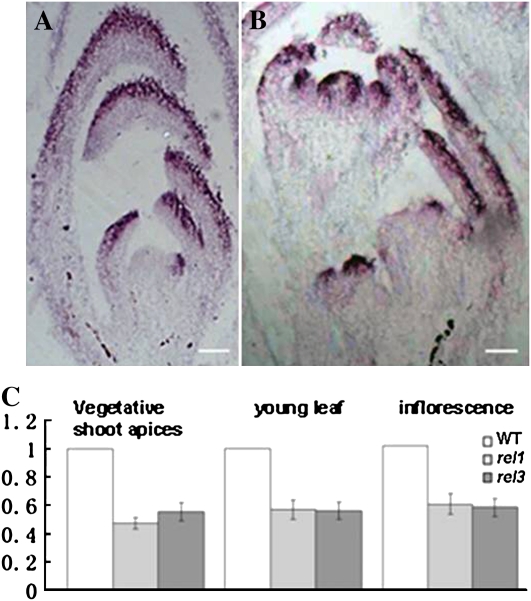
Among the genes involved in the adaxial-abaxial patterning of lateral organs in simple-leafed species, YAB genes are the common downstream targets of other polarity genes (Lin et al., 2003; Eshed et al., 2004; Li et al., 2005; Garcia et al., 2006; Xu et al., 2006) and had been reported to be regulated indirectly by ta-siRNA pathway in both Arabidopsis and maize (Juarez et al., 2004; Li et al., 2005; Garcia et al., 2006). The expression of homologs of YAB genes was also analyzed in wild-type and rel mutant plants in L. japonicus. Based on the sequence information from the current L. japonicus genome database, a full-length cDNA of LjYAB1, which shares the highest similarity with Arabidopsis YAB1/FILAMENTOUS (Supplemental Fig. S1D), was first isolated to allow expression analysis. In the vegetative stage, LjYAB1 accumulated predominantly in the abaxial region of leaf primordia and leaflet primordia (Fig. 7A). In reproductive stage, transcripts of LjYAB1 were detected in floral organ primordia, but in the sepal and petal primordia, they were mainly confined to the abaxial region (Fig. 7B). In rel mutants, the LjYAB1 expression pattern was not altered in the primordia of lateral organs (Supplemental Fig. S3); however, its expression level was dramatically reduced (Fig. 7C).
Figure 7.
Expression analysis of LjYAB1. A and B, RNA in situ hybridization on longitudinal sections of vegetative and reproductive shoot apices in the wild type (WT) using an antisense LjYAB1 probe. Bars = 50 μm. C, Real-time PCR analysis of the transcript levels of LjYAB1 in the wild type and rel mutants. Quantification was normalized to that of UBI and then to the value of wild-type Gifu plants, whose value was arbitrarily fixed at 1. Bars indicate se.
DISCUSSION
Many Parallels between the Dicot L. japonicus and the Monocots in the Regulation of Lateral Organs by the ta-siRNA Pathway
In this study, we cloned two REL genes in L. japonicus (Fig. 3, A–D) and showed that in these mutants, the production of TAS3 ta-siRNAs was affected (Fig. 5D). We conclude that REL1 and REL3 are the functional orthologs of SGS3 and ZIP in Arabidopsis, respectively. Both REL1 and REL3 are key components of the ta-siRNA pathway in L. japonicus. It has been shown that the ta-siRNA pathway is involved in different developmental processes in both monocots and dicots. In Arabidopsis, it is found that SGS3 and ZIP play an important role in the control of vegetative phase change but have subtle effect on lateral organ morphology (Hunter et al., 2003; Peragine et al., 2004). However, when the ta-siRNA pathway is blocked in monocots, dramatic and pleiotropic effects on plant development, including disruption of SAM formation and patterning of lateral organs, were observed (Timmermans et al., 1998; Satoh et al., 1999; Itoh et al., 2000; Liu et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007). In L. japonicus, the rel mutants also displayed severe developmental defects in lateral organs, suggesting similarity between L. japonicus and the monocots for the ta-siRNA pathway in the control of lateral organ development.
ARF3 and ARF4 are the key effectors in the ta-siRNA pathway. In this study, two copies of ARF3 homolog were identified in the genome of L. japonicus, and both were regulated by the ta-siRNA pathway (Fig. 6, A and B). In rice and maize, ARF3 homologs are also duplicated (Liu et al., 2007; Nogueira et al., 2007), while the Arabidopsis genome contains only one copy of ARF3. It is possible that the severe developmental defects in the ta-siRNA-deficient mutants are partially due to the increased dosage effects of the duplicated ARF3 homologs in maize, rice, and L. japonicus. In Arabidopsis ta-siRNA-deficient mutants, the up-regulation of endogenous single-copy ARF3 accounts for the altered vegetative phase transition but causes only subtle aberrant phenotype in lateral organs (Fahlgren et al., 2006; Hunter et al., 2006). However, severe developmental defects in leaf and floral organs, which are similar to those observed in maize, rice, and L. japonicus ta-siRNA-deficient mutants, could be found when extra copies of ARF3 were introduced into the rdr6 mutant (Fahlgren et al., 2006). Therefore, duplication of ARF3 homologs during evolution may be one of the molecular bases underlying the functional diversity of the ta-siRNA pathway in different species.
In addition to directly regulating the ARF genes, the ta-siRNA pathway also regulates members of the YAB gene family indirectly, but the regulatory mode appears to be different in different species. Arabidopsis YAB genes are expressed on the abaxial side of the developing leaf and contribute to the specification of abaxial identity and laminar expansion (Siegfried et al., 1999; Eshed et al., 2004). Although YAB expression is not altered in single ta-siRNA-defective mutants, nor in the as1 or as2 single mutants, the expression of YAB gene is up-regulated in mutants in which AS1 or AS2 and key components of the ta-siRNA pathway are simultaneously inactivated, suggesting that the ta-siRNA pathway negatively regulates YAB genes (Li et al., 2005; Garcia et al., 2006). In maize, the function of YAB-like genes may be associated with lateral blade outgrowth rather than specifying adaxial cell fate. Expression of YAB-like genes is limited to the adaxial side of leaf primordia and is repressed in lbl1 mutants, indicating that YAB genes are positively regulated by the ta-siRNA pathway (Juarez et al., 2004). In L. japonicus, we found that LjYAB1 is predominantly expressed in the abaxial side of both vegetative and reproductive organ primordia (Fig. 7, A and B). In rel mutants, the expression level of the LjYAB1 gene is dramatically reduced in both narrow leaves and tiny flowers (Fig. 7C). The reduction in LjYAB1 expression in rel mutants may be associated with the reduced lamina.
Multiple Roles for the ta-siRNA Pathway in Controlling Compound Leaf Development in L. japonicus
The ta-siRNA pathway plays a conserved role in specifying leaf adaxial identity in Arabidopsis, maize, and rice (Garcia et al., 2006; Xu et al., 2006; Liu et al., 2007; Nagasaki et al., 2007; Nogueira et al., 2007). However, all these species possess simple leaves, and the contribution of the ta-siRNA pathway in controlling compound leaf development is largely unknown. L. japonicus, like most legume species, has compound leaves consisting of one petiole and several leaflets (Fig. 1A). In rel mutants, narrow and needle-like leaflets with loss of adaxial identity are observed (Fig. 1, A, D, and E). Analysis of expression patterns of REL genes demonstrates that REL1 is expressed more broadly than REL3, but both genes accumulate in the adaxial domain of leaf primordia, where the transcripts of REL3 are predominantly accumulated (Fig. 4, C, D, G, and H). These data indicate that REL genes are required for specifying leaf adaxial identity; thus, the ta-siRNA pathway has a conserved function in the establishment of leaf adaxial-abaxial polarity in both simple and compound leaf development.
The conspicuous loss of basal leaflets in rel mutants suggests that the ta-siRNA pathway is also involved in the determination of leaflet formation during compound leaf development in L. japonicus and the basal leaflet formation seems to be more sensitive to the ta-siRNA pathway than the top leaflet and lateral leaflet formation. Unlike most compound-leafed species, in which Class 1 KNOTTED1-like (KNOX1) genes play a critical role in the determination of leaflet formation (Bharathan et al., 2002), legume species such as pea and alfalfa (Medicago sativa) utilize the FLORICAULA (FLO)/LEAFY (LFY) ortholog in place of the KNOX1 genes to control leaflet formation (Champagne et al., 2007). Loss-of-function mutations in the FLO/LFY ortholog, such as UNI of pea and SGL1 of Medicago truncatula, cause dramatic reduction in leaflet number (Hofer et al., 1997; Wang et al., 2008). Slightly reduced leaflet number is also observed in soybean (Glycine max) RNA interference transgenic lines with decreased expression of FLO/LFY ortholog (Champagne et al., 2007). Consistently, the proliferating floral meristem (pfm) mutant of FLO/LFY ortholog in L. japonicus gives rise to aberrant compound leaves lacking one or two basal leaflets (Dong et al., 2005). Thus, it raises a question whether the reduced leaflet formation in rel mutants is associated with altered action of the PFM. Our preliminary data indicated that no detectable alteration of both expression level and expression pattern of PFM was found in rel mutants (data not shown). Furthermore, subtle differences in the petiole can be found between rel mutants and the pfm mutant. The petiole length of rel mutants is not altered, and this is in contrast to the shortened petiole phenotype observed in pfm (Dong et al., 2005). This indicates that regulation of compound leaf development by the ta-siRNA pathway may be independent of the FLO/LFY pathway in L. japonicus.
Recent work revealed that apart from the typical KNOXI and FLO/LFY pathways, other factors were also found to be required for the leaflet formation in compound leaf development. In tomato, TCP transcription factors play a central role in the modulation of compound leaves and prevent leaflet formation via limiting growth (Ori et al., 2007). In Cardamine hirsuta, the promotion of leaflet formation was shown to be correlated with auxin activity (Barkoulas et al., 2008). It has been shown that the NO APICAL MERISTEM/CUP-SHAPED COTYLEDON boundary genes could have a non-cell-autonomous effect on leaflet formation within different eudicot compound-leafed species, including legume (Blein et al., 2008). Thus, future work to discover possible connections of these factors with the ta-siRNA pathway will provide new insight into the function of the ta-siRNA pathway in the control of compound leaf development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Lotus japonicus Gifu B-129 was used as the wild-type control and the parental line in the mutagenesis experiments. L. japonicus Miyakojima MG-20 was used as another parental line in the crossing to construct mapping populations. All plants were grown at 20°C to 22°C with a 16-h-light/8-h-dark photoperiod at 150 mE m−2 s−1. The rel1 and rel3 mutants were isolated from an ethyl methanesulfonate-mutagenized M2 population. Offspring segregating in the M4 generations with 3:1 ratios of the wild type and mutants were backcrossed thrice to Gifu B-129.
Map-Based Cloning
Mapping of the REL1 and REL3 loci was performed by analysis of F2 populations generated by crossing the Rel1rel1 and Rel3rel3 heterozygotes with MG-20. F2 homozygous mutant plants were selected, and genotyping was conducted using simple sequence repeats markers from the L. japonicus genome Web site.
Transgenic and Complementation Experiments
For genetic complementation, a SmaI-BglII fragment (10.0 kb) containing the entire REL1 gene was excised from BAC clone LjB19C13 (BM1800) and ligated into the SmaI-SpeI site of binary vector pCAMBIA1301. A derived cleaved amplified polymorphic sequence marker was designed to isolate rel1 homozygotes using primers 5′-CTTCGGGAAATCCCTGGCATGCA-3′ and 5′-CATCCATGCTCCAACGGAGGTCGA-3′ and the enzyme EcoT22I. The resultant construct was introduced into rel1 plants. Transformation was carried out as described (Feng et al., 2006). Twelve independent transformants were obtained, of which eight displayed the complemented phenotype.
Microscopy
SEM was conducted according to the methods described previously (Chen et al., 2000). The tissues for histology analysis were fixed and sectioned as described previously (Luo et al., 2005).
Gene Cloning and RT-PCR
RACE PCR, semiquantitative RT-PCR, and real-time PCR were performed as described (Feng et al., 2006). Sequences of all primers are given in Supplemental Table S1.
In Situ Hybridization
Tissues for in situ hybridization was fixed, sectioned, and hybridized to digoxigenin-labeled RNA probes as described previously (Coen et al., 1990). The probes were generated from the relevant cDNA fragment with gene-specific primers (sequences of all primers are given in Supplemental Table S1).
Small RNA Filter Hybridization
Vegetative shoot apices were used to extract RNA. RNA extraction and filter hybridization were conducted as described previously (Liu et al., 2007). 32P-end labeled LNA probe complementary to TAS3 ta-siRNA (5′-TTCTTGACCTTGTAAGACCTC-3′) and probe to miR164 (5′-TGGAGAAGCAGGGCACGTCA-3′) were used to detect ta-siRNA and miRNA, respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ617269 to FJ617274.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1 Phylogenetic tree of protein sequences.
Supplemental Figure S2. Complementation of rel1 mutants.
Supplemental Figure S3. Expression analysis of LjYAB1 in wild-type and rel mutants leaves.
Supplemental Table S1. Oligonucleotide primers used in our experiments.
Supplementary Material
Acknowledgments
We acknowledge H. Huang, X. Cao, and D. Jackson for helpful discussion of this work. We acknowledge Z. Zhao, Z. Dong, Z. Wang, L. Wang, H. Yin, and current members of our laboratory for critical reading of the manuscript and support for this experiment. We also thank X. Song, B. Luo, Y. Wang, Z. Liu, M. Xing, B. Xu, L. Pi, Q. Liu, W Liu, X. Li, and C. Xu from other laboratories for encouragement and support for this experiment.
This work was supported by the National Natural Science Foundation of China (grant no. 30528016) and the National Basic Research Priorities (973) Programs of China (2010CB126501). P.M.G. thanks the Australian Research Council for a Centre of Excellence grant and Strategic Funds from the University of Queensland.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Da Luo (dluo@sibs.ac.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221 [DOI] [PubMed] [Google Scholar]
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M (2008) A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet 40 1136–1141 [DOI] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR (2002) Homologies in leaf form inferred from KNOXI gene expression during development. Science 296 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P (2008) A conserved molecular framework for compound leaf development. Science 322 1835–1839 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 [DOI] [PubMed] [Google Scholar]
- Champagne CE, Goliber TE, Wojciechowski MF, Mei RW, Townsley BT, Wang K, Paz MM, Geeta R, Sinha NR (2007) Compound leaf development and evolution in the legumes. Plant Cell 19 3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang S, Huang H (2000) LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26 42–54 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC (2009) Pattern formation via small RNA mobility. Genes Dev 23 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R (1990) floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63 1311–1322 [DOI] [PubMed] [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, Luo JH, Yang J, Huang WH, Hu XH, Wang TL, Luo D (2005) Floral patterning in Lotus japonicus. Plant Physiol 137 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16 939–944 [DOI] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA (2006) Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16 933–938 [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15 1494–1500 [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N (1997) UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7 581–587 [DOI] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13 1734–1739 [DOI] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh JI, Kitano H, Matsuoka M, Nagato Y (2000) Shoot organization genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12 2161–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Twigg RW, Timmermans MC (2004) Specification of adaxial cell fate during maize leaf development. Development 131 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709 [DOI] [PubMed] [Google Scholar]
- Kim M, McCormick S, Timmermans M, Sinha N (2003) The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424 438–443 [DOI] [PubMed] [Google Scholar]
- Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H (2005) The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen Z, Song X, Liu C, Cui X, Zhao X, Fang J, Xu W, Zhang H, Wang X, et al (2007) Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JH, Yan J, Weng L, Yang J, Zhao Z, Chen JH, Hu XH, Luo D (2005) Different expression patterns of duplicated PHANTASTICA-like genes in Lotus japonicus suggest their divergent functions during compound leaf development. Cell Res 15 665–677 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T (2007) The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol Biol 63 777–785 [DOI] [PubMed] [Google Scholar]
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC (2007) Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev 21 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39 787–791 [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Sun Y, Xu L, Xu Y, Huang H (2004) ERECTA is required for protection against heat-stress in the AS1/AS2 pathway to regulate adaxial-abaxial leaf polarity in Arabidopsis. Planta 219 270–276 [DOI] [PubMed] [Google Scholar]
- Satoh N, Hong SK, Nishimura A, Matsuoka M, Kitano H, Nagato Y (1999) Initiation of shoot apical meristem in rice: characterization of four SHOOTLESS genes. Development 126 3629–3636 [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Maizel A, Ruiz-Ferrer V, Garcia D, Bayer M, Crespi M, Voinnet O, Martienssen RA, Bendahmane M (2009) Endogenous tasiRNAs mediate non-cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS One 4 e5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Wang J, Wan X, Shen G, Wang X, Zhang J (2007) Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226 99–108 [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128 [DOI] [PubMed] [Google Scholar]
- Tattersall AD, Turner L, Knox MR, Ambrose MJ, Ellis TH, Hofer JM (2005) The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell 17 1046–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Schultes NP, Jankovsky JP, Nelson T (1998) Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125 2813–2823 [DOI] [PubMed] [Google Scholar]
- Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20 759–771 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69–79 [DOI] [PubMed] [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779–789 [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Wen J, Tadege M, Li G, Liu Y, Mysore KS, Ratet P, Chen R (2008) Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol 146 1759–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA 102 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Huang H (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107 [DOI] [PubMed] [Google Scholar]
- Xu L, Yang L, Pi L, Liu Q, Ling Q, Wang H, Poethig RS, Huang H (2006) Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol 47 853–863 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.