Abstract
To maintain homeostasis in an ever-changing environment organisms have evolved mechanisms to reprogram gene expression. One central mechanism regulating gene expression is messenger RNA (mRNA) degradation, which is initiated by poly(A) tail shortening (deadenylation). The carbon catabolite repressor 4-CCR4 associated factor1 (CCR4-CAF1) complex is the major enzyme complex that catalyzes mRNA deadenylation and is conserved among eukaryotes. However, the components and functions of this global regulatory complex have not been well characterized in plants. Here we investigate the CAF1 family in Arabidopsis (Arabidopsis thaliana). We identify 11 AtCAF1 homologs and show that a subset of these genes are responsive to mechanical wounding, among them are AtCAF1a and AtCAF1b whose expression levels are rapidly and transiently induced by wounding. The differential expression profiles of the various AtCAF1s suggest that not all AtCAF1 genes are involved in stress-responsive regulation of transcript levels. Comparison of misexpressed genes identified via transcript profiling of Atcaf1a and Atcaf1b mutants at different time points before and after wounding suggests that AtCAF1a and AtCAF1b target shared and unique transcripts for deadenylation with temporal specificity. Consistent with the AtPI4Kγ3 transcript exhibiting the largest increase in abundance in Atcaf1b, AtCAF1b targets AtPI4Kγ3 mRNA for deadenylation. Stress-tolerance assays demonstrate that AtCAF1a and AtCAF1b are involved in mediating abiotic stress responses. However, AtCAF1a and AtCAF1b are not functionally redundant in all cases, nor are they essential for all environmental stresses. These findings demonstrate that these closely related proteins exhibit overlapping and distinct roles with respect to mRNA deadenylation and mediation of stress responses.
Communication of developmental and environmental information is mediated through alteration of gene expression. The steady-state level of messenger RNA (mRNA) within a cell is determined by the combination of the rate of transcription and the rate of posttranscriptional processes regulating mRNA decay. Genome-wide approaches are revealing that in response to environmental stimuli there is a large reprogramming of gene expression (Eulgem, 2005; Swindell, 2006; Kilian et al., 2007; Ma and Bohnert, 2007; Walley et al., 2007; Walther et al., 2007; Shalem et al., 2008). While changes in the rate of mRNA degradation provide a rapid mechanism to alter mRNA abundance, research has largely focused on changes in transcriptional regulation as a means to control mRNA levels (Gutierrez et al., 2002; Yamashita et al., 2005; Narsai et al., 2007; Belostotsky and Sieburth, 2009; Chiba and Green, 2009; Lee and Glaunsinger, 2009).
In eukaryotes, degradation of mRNA begins with shortening of the poly(A) tail, referred to as deadenylation, which is the rate-limiting step (Meyer et al., 2004; Belostotsky and Sieburth, 2009; Chiba and Green, 2009). The process of deadenylation has been best studied in yeast (Saccharomyces cerevisiae) where it has been shown that the carbon catabolite repressor 4-CCR4 associated factor1 (CCR4-CAF1; also called Pop2p) complex serves as the major deadenylase complex (Tucker et al., 2001). In addition to the CCR4-CAF1 complex, both the poly(A) ribonuclease (PARN), for which mutants in Arabidopsis (Arabidopsis thaliana) are embryo lethal, and poly(A) nuclease are also active deadenylases (Chiba et al., 2004; Meyer et al., 2004; Reverdatto et al., 2004; Belostotsky and Sieburth, 2009). Following deadenylation, mRNA decay proceeds via two distinct pathways. In one pathway mRNA is degraded in a 3′-to-5′ manner via the exosome (Chekanova et al., 2007; Belostotsky and Sieburth, 2009). Alternatively, deadenylated mRNA can also undergo 5′-to-3′ decay, which first requires that the 5′ cap is removed from the deadenylated mRNA (Goeres et al., 2007; Belostotsky and Sieburth, 2009).
The biological role of CAF1 has been examined in a range of eukaryotic species. In yeast, caf1 deletion strains are sensitive to high temperatures and caffeine (Hata et al., 1998). Analysis of CAF1 in Caenorhabditis elegans using RNAi and deletion alleles demonstrates that CAF1 is essential for embryonic and larval development (Molin and Puisieux, 2005). Additionally, CAF1 is required for normal growth of trypanosomes (Schwede et al., 2008). Finally, mice lacking CAF1 are sterile (Nakamura et al., 2004).
In plants, limited experiments have investigated components of the CCR4-CAF1 complex. However, the studies that have been conducted focus on CAF1 and demonstrate a role for CAF1 in both development as well as response to biotic stresses. In Arabidopsis, AtCAF1 homologs have been shown to be induced by hormone treatment as well as abiotic and biotic stresses (Lee et al., 2005; Zheng et al., 2006; Ferrari et al., 2007; Walley et al., 2007; Liang et al., 2009). Additionally, one study found that overexpression of a yeast CAF1 homolog from pepper (Capsicum annuum; CaCAF1) in tomato (Solanum lycopersicum) resulted in growth enhancement and resistance to the oomycete pathogen Phytophthora infestans. Conversely, when CaCAF1 is silenced in pepper there is significant growth retardation and plants are susceptible to the pathogen Xanthomonas axonopodis pv vesicatoria (Sujon et al., 2007). Furthermore, in Arabidopsis it has been demonstrated that two yeast CAF1 homologs, AtCAF1a (At3g44260) and AtCAF1b (At5g22250), are active deadenylases in vitro and partially complement growth defects of a yeast caf1 deletion strain. Finally, Arabidopsis mutant lines Atcaf1a-1 and Atcaf1b-1 are susceptible to Pseudomonas syringae pv tomato DC3000 (Liang et al., 2009).
In this study we examine the role of the Arabidopsis CAF1 family of deadenylases in abiotic stress responses. We identify a total of 11 AtCAF1 homologs that exhibit differential patterns of expression in response to wounding. Transcriptional profiling of Atcafa-1 and Atcaf1b-2 suggests that AtCAF1a and AtCAF1b target unique transcripts for deadenylation with temporal specificity. Additionally, we show that AtCAF1b targets a putative phosphatidylinositol 4-kinase mRNA for deadenylation. Finally, stress-tolerance assays suggest that while AtCAF1a and AtCAF1b are involved in mediating responses to a number of environmental stresses, they do not act redundantly in all cases nor are they required for all stresses.
RESULTS
Identification of the CAF1 Family in Arabidopsis
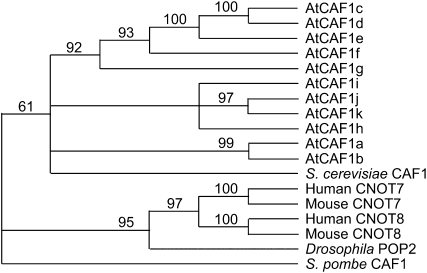
We previously identified two Arabidopsis homologs of the yeast CAF1 protein, AtCAF1a and AtCAF1b, via transcript profiling, as rapid wound response (RWR) genes (Walley et al., 2007). To determine the full complement of AtCAF1 homologs present in the Arabidopsis genome, we used the AtCAF1a sequence to identify all proteins with sequence similarity using a basic local alignment search tool (BLAST; Altschul et al., 1990) at Phytozome (www.phytozome.net). This search yielded nine additional sequences, which we named AtCAF1c to AtCAF1k (Figs. 1 and 2), resulting in a small family of 11 members. Alignment the AtCAF1 proteins with CAF1 sequence from other eukaryotes demonstrates that the RNaseD domain (Daugeron et al., 2001; Sujon et al., 2007) is well conserved in the AtCAF1 family (Fig. 1). We also found two closely related sequences in moss (Physcomitrella patens), eight sequences in rice (Oryza sativa), and 10 sequences in poplar (Populus trichocarpa; data not shown), demonstrating that there has been a significant expansion of the CAF1 gene family in angiosperms, possibly due to gene duplication events. In other eukaryotes such as yeast, fly, mouse, and human there are only one or two CAF1 genes (Fig. 2; Albert et al., 2000; Daugeron et al., 2001; Nakamura et al., 2004; Temme et al., 2004; Yamashita et al., 2005).
Figure 1.
ClustalW generated alignment of CAF1 protein sequences from Arabidopsis, pepper (CaCAF1), yeast (SpCAF1), human (HsCNOT7/8), and Drosophilia (DmPOP2). Conserved RNase D-domain residues are shaded in color. [See online article for color version of this figure.]
Figure 2.
Phylogenetic tree of the CAF1 family from Arabidopsis. Related sequences from yeast (S. cerevisiae and S. pombe), human, mouse, and fly are also included. While most eukaryotes contain only one or two CAF1 proteins, Arabidopsis has 11, which group into three well-supported clades. Clade I includes AtCAF1a and AtCAF1b; clade II includes AtCAF1c to AtCAF1g; and clade III includes AtCAF1j and AtCAF1k. Bootstrap values on nodes were generated in PAUP*.
To determine which of the AtCAF1 sequences are most closely related to the yeast CAF1 protein we first performed an alignment of all 11 AtCAF1 protein sequences with the yeast, fly, mouse, and human CAF1 sequences using Se-Al and CLUSTALW. The alignment was trimmed to include only conserved regions (data not shown). Using this alignment we ran a parsimony analysis in PAUP* (Harrison and Langdale, 2006). We obtained 10 trees from this analysis, which were then computed into a consensus tree using the strict option in PAUP* (Fig. 2). The tree was then rooted with Saccharomyces pombe CAF1 and bootstrap values were calculated in PAUP*. The obtained consensus tree places all of the AtCAF1s as sister to yeast CAF1, showing that while AtCAF1a to AtCAF1k are related to the yeast protein orthology is equivocal (Fig. 2). The Arabidopsis homologs group into three well-supported clades: AtCAF1c to AtCAF1g; AtCAF1h to AtCAF1k; and AtCAF1a and AtCAF1b (Fig. 2). These phylogenetic relationships suggest that there could be functional overlap between clade members. Given the large number of AtCAF1 homologs it is also possible that either sub- or neofunctionalization has occurred among some of the family members.
The AtCAF1 Gene Family Exhibits Differential Expression Patterns in Response to Stress
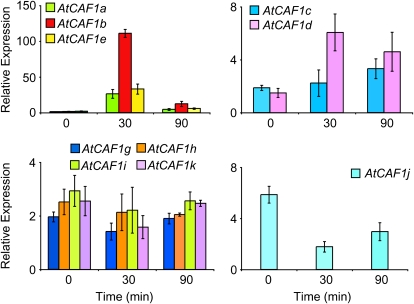
It has previously been shown that AtCAF1a and AtCAF1b respond rapidly and transiently to a range of stimuli including mechanical wounding, methyl jasmonate, abscisic acid, 1-aminocyclopropane-1-carboxylic acid, salicylic acid, and Pseudomonas syringae pv tomato DC3000 (Walley et al., 2007; Liang et al., 2009). To determine if stress-induced transcriptional activation of AtCAF1a and AtCAF1b was unique or a general property of all 11 family members, we examined the expression of AtCAF1a to AtCAF1k in response to mechanical wounding by quantitative reverse transcription (qRT)-PCR. These data show that five of the 11 AtCAF1 genes are up-regulated in response to mechanical stress, while four of the 11 AtCAF1 genes do not respond to mechanical wounding and one is down-regulated (Fig. 3). Finally, we could not detect expression of AtCAF1f under the conditions we examined. The differential expression patterns exhibited by the AtCAF1 family members suggests that a subset of the AtCAF1 genes may be involved in rapid stress-responsive regulation of transcript levels while others may regulate deadenylation of mRNAs responsive to various developmental and environmental cues.
Figure 3.
Expression of the AtCAF1 gene family in response to wounding. Total RNA was extracted from 3-week-old rosette leaves before and at intervals after mechanical wounding and subjected to qRT-PCR analysis. Transcript levels were normalized to internal control genes (At4g34270 and At4g26410) measured in the same samples. Data are means of three independent biological replicates ±sem.
Transcript Profiling of AtCAF1a and AtCAF1b Mutants
For further investigation into the role of the Arabidopsis CAF1 homologs in regulating gene expression and stress tolerance we focused on AtCAF1a and AtCAF1b. These two AtCAF1 family members were selected because (1) they were identified from previous studies as RWR genes (Walley et al., 2007) and (2) they form a monophyletic clade, suggesting that they may exhibit functional overlap. Many of the RWR genes are also categorized as abiotic and biotic stress-responsive genes, suggesting that they likely comprise a set of core components in the general stress-response network (Walley et al., 2007). We were therefore interested in elucidating what role(s) AtCAF1a and AtCAF1b may play during signaling upon environmental stress. Towards this aim we obtained Salk T-DNA insertion lines for AtCAF1a and AtCAF1b and confirmed that they do not produce full-length transcripts and are likely functional nulls (Supplemental Fig. S1). These mutant lines are visually indistinguishable form the wild-type plants.
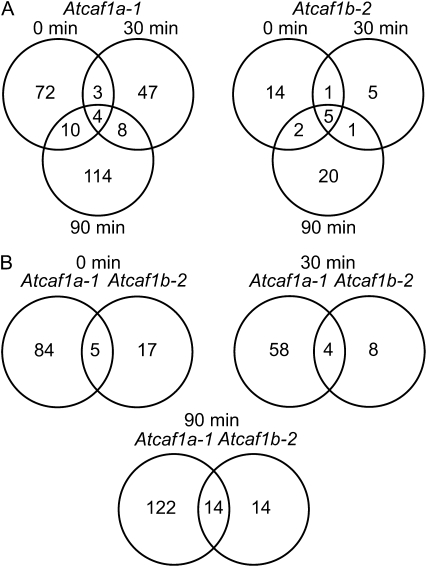
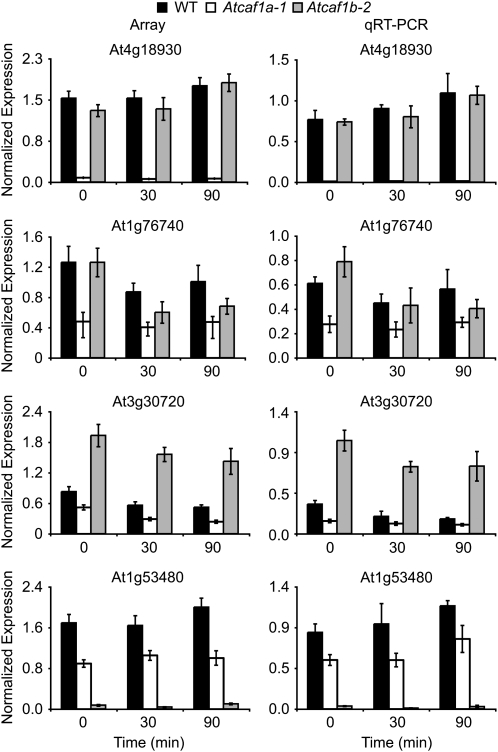
We hypothesized that AtCAF1a and AtCAF1b could interact with target mRNAs to result in altered gene expression profiles. To test this hypothesis we performed transcript profiling using the Agilent V3 oligo microarray, which contains probes for 28,500 genes from The Institute for Genomic Research's ATH1 v.5 annotation as well 10,000 probes based on Massively Parallel Signature Sequencing data (Meyers et al., 2004) and identified genes that are misexpressed in the absence of AtCAF1a and AtCAF1b. For this experiment we examined 3-week-old rosette leaves from wild-type, Atcaf1a-1, and Atcaf1b-2 plants, both before and intervals after wounding, using three independent biological replicates for each genotype and time point. The full set of the raw intensity microarray data are deposited at http://www.ncbi.nlm.nih.gov/geo/, under Gene Expression Omnibus (GEO) accession GSE17022. Misexpressed genes were classified as probes that exhibited an increase or decrease in signal of at least 2-fold and had a P value ≤ 0.05. Using this approach, we found that between 62 and 136 genes were misexpressed in Atcaf1a-1 compared to wild type, depending on the time point (Fig. 4; Supplemental Table S1). While a smaller number of genes (12–28) were misexpressed in Atcaf1b-2 compared to wild type (Fig. 4; Supplemental Table S1). The number of up- and down-regulated genes in each of the caf mutant lines at various intervals post wounding are shown (Supplemental Table S2). Atcaf1a-1 and Atcaf1b-2 misexpressed genes were classified according to gene ontology (GO) terms to provide insight into their biological function (Berardini et al., 2004). The two largest defined classes of GO terms involve response to stress or abiotic/biotic stimuli (Supplemental Fig. S2), suggesting that these two AtCAF1 genes are involved in stress responses. Comparison of the misexpressed genes revealed a limited temporal overlap for both Atcaf1a-1 as well as Atcaf1b-2 (Fig. 4A). Additionally, there is limited overlap in transcripts that are misexpressed in the absence of either AtCAF1a or AtCAF1b at 0, 30, or 90 min post wounding (Fig. 4B). While there is minimal overlap in the transcript profiles, the overlap that is observed is greater than would be expected by random chance (Fig. 4). The microarray results were next validated by qRT-PCR for five selected misexpressed genes (Figs. 5 and 6; Supplemental Fig. S3). The expression levels determined by qRT-PCR data are in good agreement with those observed by transcript profiling with a spearman rank order correlation coefficient of 0.938 (P value = 0.000; Sokal and Rohlf, 1995). These results collectively indicate that AtCAF1a and AtCAF1b target shared and unique transcripts for deadenylation with temporal specificity.
Figure 4.
Comparison of Atcaf1a and Atcaf1b dependent transcript profiles. A, Overlap between genes misexpressed at 0, 30, and 90 min post wounding in either Atcaf1a-1 or Atcaf1b-2. B, Comparison of Atcaf1a-1 versus Atcaf1b-2 misregulated genes at each time point. The minimal overlap observed for all comparisons is statistically significant at a P value of at least 3.04 × 10−8. Misexpressed genes in each mutant were determined based on an increase or decrease in expression level of at least 2-fold (P ≤ 0.05) relative to wild type at each respective time point.
Figure 5.
Validation of Atcaf1a and Atcaf1b misexpressed genes. Normalized expression values from microarray (left) and qRT-PCR (right) for selected genes. Data are means of three independent biological replicates ±sem. For qRT-PCR, transcript levels were normalized to internal control genes (At4g34270 and At4g26410) measured in the same samples. WT, Wild type.
Figure 6.
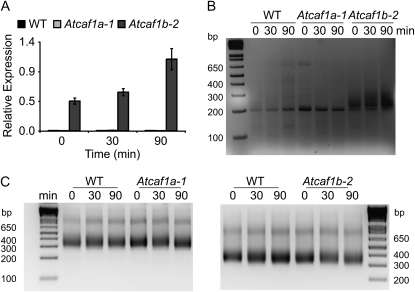
AtPI4Kγ3 PAT is stabilized in Atcaf1b. A, qRT-PCR of AtPI4Kγ3 in response to wounding, normalized to internal control genes (At4g34270 and At4g26410) measured in the same samples (n = 3 ± sem). WT, Wild type. B, PAT assay measuring the PAT of AtPI4Kγ3 in wild type, Atcaf1a-1, and Atcaf1b-2 before and in response to mechanical wounding. C, PAT assay measuring the PAT of UBQ10 in wild type, Atcaf1a-1, and Atcaf1b-2 before and in response to mechanical wounding.
AtCAF1b Is Required for Deadenylation of AtPI4Kγ3 mRNA
We hypothesized that direct mRNA targets of AtCAF1s would not be deadenylated in caf1 mutants and would therefore show an increase in abundance in the absence of AtCAF1a or AtCAF1b. The gene that exhibited the greatest increase in abundance observed for either of the Atcaf1 mutants encodes a putative phosphatidylinositol 4-kinase (AtPI4Kγ3; Mueller-Roeber and Pical, 2002). In Atcaf1b-2 transcript levels of AtPI4Kγ3 were increased 42- to 63-fold in our microarray, suggesting that it may be a direct target of AtCAF1b (Supplemental Fig. S3). We confirmed by qRT-PCR that AtPI4Kγ3 levels were indeed increased exclusively in Atcaf1b-2 (Fig. 6A). To test if AtCAF1b is acting directly on AtPI4Kγ3 mRNA we performed poly(A) tail length (PAT) assays (Salles and Strickland, 1999). As shown in Figure 6B the length of AtPI4Kγ3 mRNA increased specifically in Atcaf1b-2. Additionally, the increase in length of AtPI4Kγ3 mRNA in Atcaf1b-2 is not a result of a general increase in PAT, as other transcripts tested, including UBQ10, did not increase in size (Fig. 6C; data not shown). Taken together these results strongly suggest that AtCAF1b targets AtPI4Kγ3 mRNA for deadenylation. We also tested, via the PAT assay, a number of transcripts that were increased in abundance in Atcaf1-a, albeit to a much lower level than AtPI4Kγ3 was in Atcaf1b-2, and were unable to uncover any potential targets of CAF1a (data not shown), likely due to the low sensitivity of the PAT assay.
AtCAF1a and AtCAF1b Regulate Abiotic Stress Responses
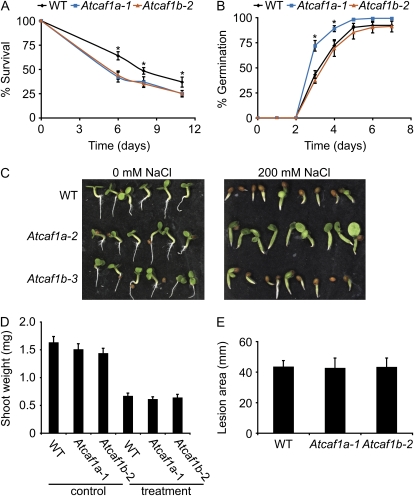
AtCAF1a and AtCAF1b were originally identified as RWR genes, which are enriched for abiotic and biotic stress-responsive genes, suggesting that these CAF1s may be involved in mediating multistress resistance. Additionally, it was recently shown that both AtCAF1a and AtCAF1b are required for resistance to the pathogen Pseudomonas syringae (Liang et al., 2009). We therefore chose to test both Atcaf1a and Atcaf1b mutants for tolerance to various environmental stresses. We first assayed survival rate of seedlings germinated on the reactive oxygen species inducer methyl viologen (MV; paraquat). This experiment demonstrated that both Atcaf1a and Atcaf1b are more sensitive to MV than wild type (Fig. 7A; Supplemental Fig. S4). We also tested their ability to cope with salt stress by germinating seeds on salt plates. Atcaf1a plants show an increased germination rate on 200 mm NaCl compared to wild type and Atcaf1b (Fig. 7, B and C). No differences in survival or germination were observed for plants grown on control plates (Fig. 7C; data not shown). However, Atcaf1a and Atcaf1b mutants did not differ in tolerance to waterlogging or the pathogen Botrytis cinerea when compared to wild type (Fig. 7, D and E). These data suggest that while AtCAF1a and AtCAF1b are involved in mediating response to both abiotic and biotic stresses they do not act redundantly in all cases nor are they required for all stresses.
Figure 7.
AtCAF1a and AtCAF1b mediate response to abiotic stresses. A, Survival of wild type (WT), Atcaf1a-1, and Atcaf1b-2 on 5 μm MV (paraquat). Data are means of nine independent biological replicates ±sem. Asterisks denote a significant difference of both Atcaf1a and Atcaf1b from wild type (P < 0.05) as determined by t tests. B, Germination of wild type, Atcaf1a-1, and Atcaf1b-2 on 200 mm NaCl. Data are means of four independent biological replicates ±sem. Asterisks denote a significant difference of Atcaf1a from wild type and Atcaf1b (P < 0.05) as determined by t tests. C, Wild type, Atcaf1a-2, and Atcaf1b-3 grown on either 0 mm NaCl for 2 d or 200 mm NaCl for 12 d. D, Waterlogging tolerance of wild type, Atcaf1a-1, and Atcaf1b-2. Three-week-old plants were submerged for 7 d and then allowed to recover for 5 d prior to measurement of fresh weight. Data are means of 32 independent biological replicates ±sem. E, Lesion area 4 d after spot inoculation with spores of B. cinerea isolate B05.10. Similar results were obtained using the B. cinerea isolate DN (data not shown). Data are means of 15 independent biological replicates ±sem. All abiotic and biotic stress-tolerance assays were independently repeated one to four times with similar results.
DISCUSSION
In eukaryotes control of mRNA degradation is an essential component in the regulation of transcript abundance (Meyer et al., 2004; Belostotsky and Sieburth, 2009; Chiba and Green, 2009). As deadenylation is the rate-limiting step in mRNA decay it is critical for maintaining appropriate levels of mRNA. The CCR4-CAF1 complex forms the major deadenylation complex in yeast, where it has been extensively studied (Tucker et al., 2001; Chiba and Green, 2009). In eukaryotes, such as yeast, fly, mouse, and human, CAF1 homologs have been identified and shown to be important for deadenylation. Furthermore, in these species there are only one or two CAF1 genes (Albert et al., 2000; Daugeron et al., 2001; Nakamura et al., 2004; Temme et al., 2004; Yamashita et al., 2005). In contrast, our results demonstrate that there has been an expansion of the CAF1 gene family in angiosperms (Fig. 2; data not shown). Given the large number of AtCAF1 homologs in Arabidopsis it is possible that sub- or neofunctionalization has occurred among some of the family members, resulting in distinct CAF1 proteins functioning under specific conditions and/or targeting unique transcripts. In support of this idea qRT-PCR analysis of the AtCAF1 family revealed that AtCAF1 genes exhibit differential expression patterns in response to mechanical wounding (Fig. 3). This differential expression behavior exhibited by AtCAF1 family members is consistent with the processes of sub- and/or neofunctionalization.
In a recent review Belostotsky and Sieburth (2009) hypothesized that deadenylation complexes in plants may act nonredundantly and target specific subsets of transcripts. This hypothesis is based on the finding that Arabidopsis parn (a second type of deadenylase) mutants are embryo lethal and that only a subset of embryo-expressed genes have increased PAT in parn mutants (Reverdatto et al., 2004). In support of this hypothesis our results suggest that AtCAF1a and AtCAF1b are also able to target specific transcripts. First, the transcript-profiling experiment reveals that loss of AtCAF1a or AtCAF1b leads to largely distinct sets of altered transcripts. Additionally, there is limited overlap between the three time points assayed for both Atcaf1a as well as Atcaf1b (Fig. 4), suggesting that these deadenylases act with temporal specificity. Second, the expression of AtPI4Kγ3 is increased specifically in Atcaf1b. The increased expression of AtPI4Kγ3 is consistent with the observed increase in PAT specifically in the absence of AtCAF1b, further demonstrating that AtCAF1a and AtCAF1b are capable of targeting specific transcripts (Fig. 6). Lastly, while Atcaf1a and Atcaf1b exhibit similar responses to most environmental stresses, Atcaf1a is more tolerant to salt treatment. These results demonstrate a functional specificity between these two paralogs, which would be consistent with a sub (or neo) functionalization event (Fig. 7). Taken together these expression and stress analyses show that not only do the different types of deadenylation (PARN versus CCR4-CAF1) complexes target unique transcripts but also that closely related deadenylases are able to target unique mRNAs, resulting in unique biological responses.
The ability of AtCAF1a and AtCAF1b to target specific mRNAs raises the question of how this specificity is achieved. In Drosophila, the interaction of CAF1 with the RNA-binding protein Smaug allows for the specific targeting of the CCR4-CAF1 complex to Hsp83 and nanos mRNAs (Semotok et al., 2005; Zaessinger et al., 2006). Additionally, in yeast the RNA-binding protein Mpt5p directly interacts with CAF1 and recruits the CCR4-CAF1 complex specifically to HO mRNA (Goldstrohm et al., 2006). Given the high degree of conservation among eukaryotic CAF1 homologs we hypothesize that AtCAF1 mRNA targets may be identified in a similar fashion via association with specific RNA-binding proteins. These characteristic CAF1-RNA-binding protein complexes would then enable deadenylation of distinct mRNA substrates.
The limited studies investigating CAF1 function in plants have focused on the role of these genes in mediating biotic stress responses. Initially, it was reported that a pepper CAF1 homolog, CaCAF1, is required for resistance to the pathogen Xanthomonas axonopodis pv vesicatoria (Sujon et al., 2007). Additionally, Liang et al. (2009) reported recently that AtCAF1a and AtCAF1b act redundantly in defense against the pathogen P. syringae pv tomato DC3000, which correlates with reduced levels on PR1 expression in nontreated Atcaf1 mutant plants. Consistently, we also assayed the steady-state levels of PR1 via qRT-PCR and found reduced levels in both Atcaf1a-1 and Atcaf1b-2 plants (data not shown). The results of our stress assays add a number of important observations toward the understanding of the biological role CAF1 proteins play in plants. First, we found that AtCAF1a and AtCAF1b do not act redundantly, in all cases, in response to stress (Fig. 7, B and C; Supplemental Fig. S4). In addition, AtCAF1s are not simply required for stress tolerance but in certain cases they can actually act as a negative regulator of stress tolerance (Fig. 7, B and C). We also show that AtCAF1a and AtCAF1b play a role in abiotic stress responses (Fig. 7, A–C; Supplemental Fig. S4). Finally, while AtCAF1a and AtCAF1b mediate resistance toward both abiotic and biotic stresses, they are not involved in mediating the response toward all environmental stresses (Fig. 7, D and E).
The observed increase in salt tolerance exhibited specifically by Atcaf1a is supported at the molecular level by our microarray profiling experiments. We observed that 29% of transcripts that increase in abundance in nonwounded Atcaf1a-1 plants are genes previously shown to be induced by salt treatment (Ma et al., 2006). This enrichment in salt-induced genes is highly significant at a P value of 5.5 × 10−11, as calculated by the hypergeometric test (Sokal and Rohlf, 1995; Covington and Harmer, 2007).
Included in this common gene set is ALDH7B4, which has been shown to confer salt tolerance when overexpressed (Kotchoni et al., 2006). Consistent with Atcaf1b mutants behaving like wild-type plants in response to salt treatment, there was no overlap between transcripts increased in Atcaf1b-2 and salt-induced transcripts. The transcript profiling experiment also provides insight into the increased sensitivity of both Atcaf1a and Atcaf1b to MV. One gene, galactinol synthase2 (GolS2), which is decreased in abundance in nonwounded Atcaf1 as well as Atcaf1b, confers tolerance to MV when overexpressed (Nishizawa et al., 2008).
In conclusion, we show that the CAF1 family has expanded in angiosperms and that a subset of the AtCAF1 family responds to mechanical stress. Significantly, we also demonstrate that both AtCAF1a and AtCAF1b directly or indirectly mediate abiotic stress tolerance. However, they are not functionally redundant in all cases, at either the molecular or biological level. We therefore hypothesize that the AtCAF1 family members may have undergone sub- or neofunctionalization subsequent to gene duplication. In the future, employment of more sensitive techniques such as RNA immunoprecipitation followed by sequencing will be useful in identification of the complete suite of AtCAF1a and AtCAF1b direct mRNA targets. Additionally, biochemical characterization of AtCAF1a and AtCAF1b associated mRNA-binding proteins will provide further insight into how these deadenylases act with such a high degree of specificity.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 plants were grown in a 16-h light/8-h dark photoperiod at 22°C. All experiments were conducted on 3-week-old soil-grown plants unless otherwise noted. Atcaf1a-1 (Salk_070336; Liang et al., 2009), Atcaf1a-2 (Salk_050279), Atcaf1b-2 (Salk_121555), and Atcaf1b-3 (Salk_044043) T-DNA insertional lines (Alonso et al., 2003) were obtained from the Arabidopsis Biological Resource Center. For wounding treatments all rosette leaves were mechanically wounded one to two times with a hemostat, resulting in approximately 20% of the leaf being damaged, as previously described (Walley et al., 2007). Mechanical wounding was performed 4 to 6 h after dawn.
BLAST Analyses to Identify CAF1 Homologs in Plants
Unless otherwise stated, all computer analyses were performed on a single Macintosh iMac running Mac OS 9 or OS X. Using the Arabidopsis CAF1a (AtCAF1a) sequence as a query we performed a BLAST search to the Viridiplantae node at Phytozome (www.phytozome.net). After filtering out any repetitive sequences we downloaded all of the retrieved sequences. Yeast (Saccharomyces cerevisiae), fly, mouse, and human CAF1 sequences were identified by performing a BLAST search at the National Center for Biotechnology Information. Locally installed versions of Se-Al and CLUSTALW were used to generate alignments using all the obtained amino acid sequences, which were subsequently edited to include only conserved regions of the proteins.
Phylogenetic Analysis
All steps for phylogeny reconstruction were performed according to Harrison and Langdale (2006). Using the amino acid sequence alignment generated above we performed a parsimony and heuristic search to generate a phylogenetic tree in PAUP* (Swofford, 1999). The 10 trees generated by this analysis were subsequently computed into a strict consensus tree and bootstrap values were calculated in PAUP*.
RT-PCR
Total RNA from rosette leaves was isolated by TRIzol extraction (Life Technologies) and further purified using Qiagen RNeasy kit with on-column DNase treatment (Qiagen Inc.). RNA was reverse transcribed using Superscript III (Invitrogen). PCR for RT-PCR were conducted in 25 μL reactions containing 20 ng cDNA, 1.5 mm MgCl2, 0.2 mm each dNTP, 0.05 μm each primer, and 1 unit Choice-Taq blue (Denville Scientific) and amplified for 29 cycles. Quantitative RT-PCR was conducted in 50-μL reactions containing 10 ng cDNA, 1× iQ SYBR Green supermix (Bio-Rad Laboratories), and 200 or 250 nm each primer. Amplification and data analysis were carried out as previously described (Walley et al., 2008). The internal controls At4g34270 and At4g26410 previously described were used for transcript normalization (Czechowski et al., 2005). Primers are listed in Supplemental Table S3.
Microarray Analyses
The Agilent Arabidopsis V3 oliogoarray chip containing 60-mer oligos probes for 28,500 genes from The Institute for Genomic Research's ATH1 v.5 annotation as well as 10,000 probes based on Massively Parallel Signature Sequencing data (Meyers et al., 2004) were utilized for transcript profiling (Agilent Technologies). Total RNA was prepared from three independent biological replicates for each genotype × treatment combination as described for RT-PCR. Prior to hybridizations, the quality and quantity of the total RNA sample was confirmed by running on an Agilent 2100 bioanalyzer, and by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc.). Labeled cRNA was produced using the Agilent low RNA input linear amp kit according to the manufacturer's protocol. Briefly, total RNA was used as a template for cDNA synthesis primed with oligo(dT) followed by production of cRNA labeled with cyanine dyes. Each experimental sample was labeled with cy3. Additionally, aliquots from each experimental sample were pooled and labeled with cy5, serving as the reference sample for normalization. Labeled cy3 and cy5 cRNA was then mixed and hybridized according to Agilent protocol. Arrays were washed and dried, then scanned on an Agilent DNA microarray scanner. The raw TIFF images were analyzed and normalized using the Agilent Feature Extraction software v. 8.1 using the recommended default settings. The normalized data was then imported into GeneSpring GX7.3 (Agilent Technologies) to analyze for misexpressed genes according to the manufacturer's protocol. Genes with a fold-change ≥ 2.0 and a P value ≤ 0.5 were selected as misexpressed.
Comparison of Transcript Profiles
The statistical significance of the observed overlap in transcript profiles, as well as enrichment of GO terms, was analyzed using hypergeometric tests (Sokal and Rohlf, 1995; Covington and Harmer, 2007; Rivals et al., 2007).
PAT Assay
PAT assays were performed as previously described (Salles and Strickland, 1999) on RNA isolated for qRT-PCR. PCR was conducted in 25 μL reactions containing 1 μL of PAT cDNA as template, 1.5 mm MgCl2, 0.2 mm each dNTP, 0.5 μm each primer, and 1 unit Choice-Taq blue (Denville Scientific) and amplified for 35 cycles. PCR products were run on a 2% agarose gel and stained with ethidium bromide for visualization.
Stress-Tolerance Assays
For salt (NaCl) and reactive oxygen species (MV) stress assays seeds were planted on plates containing 1× Murashige and Skoog basal media with Gamborg's vitamins (Sigma Aldrich). Additionally, 200 mm NaCl or 5 μm MV (Sigma Aldrich) was added to the media as indicated. For salt assays 25 seeds were planted per plate and each plate was scored for percent germination (n = 4). For MV assays 15 seeds per genotype were plated on each plate (all three genotypes on the same plate) and each plate was scored for percent survival (n = 9). Waterlogging experiments were based upon a previous study (Huynh et al., 2005). Plants for waterlogging were grown in individual pots. Three-week-old plants were then treated by submerging each pot individually in water 1 cm above the soil surface. After 7 d the pots were removed from submersion and plants were given 5 d to recover prior to weighing. Botrytis cinerea detached leaf assays were performed using the B. cinerea isolates B05.10 and DN (Rowe and Kliebenstein, 2007). Arabidopsis leaves were inoculated with 5 μL of spores at a concentration of 50,000 spores/mL (Denby et al., 2004; Rowe and Kliebenstein, 2007; Walley et al., 2008). Statistical differences between wild type and mutants were detected using t tests.
The microarray data discussed in this publication have been deposited in the National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession GSE17022. AtCAF1a: At3g44260, AtCAF1b: At5g22250, AtCAF1c: At1g27820, AtCAF1d: At1g27890, AtCAF1e: At1g61470, AtCAF1f: At3g44240, AtCAF1g: At1g06450, AtCAF1h: At1g15920, AtCAF1i: At5g10960, AtCAF1j: At1g80780, AtCAF1k: At2g32070, AtPI4Kγ3: At5g24240, GolS2: At1g56600, Atcaf1a-1: Salk_070336, Atcaf1a-2: Salk_050279, Atcaf1b-2: Salk_121555, and Atcaf1b-3: Salk_044043.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of Atcaf1a and Atcaf1b Salk T-DNA alleles.
Supplemental Figure S2. GO annotation of Atcaf1a and Atcaf1b misexpressed genes.
Supplemental Figure S3. Normalized microarray expression values of AtPI4Kγ3.
Supplemental Figure S4. Survival of wild type, Atcaf1a-2, and Atcaf1b-3 on 5 μm MV (paraquat).
Supplemental Table S1. List of misexpressed genes in Atcaf1a-1 and Atcaf1b-2 compared to wild-type plant, at different time points post wounding.
Supplemental Table S2. Numbers of misexpressed genes in each mutant relative to wild type at each time point post wounding.
Supplemental Table S3. List of primers used in RT-PCR analyses.
Supplementary Material
Acknowledgments
We thank Michael Covington for assistance with the hypergeometric tests. We would also like to thank Mandy Lee and Jessica Weant for assistance with the waterlogging experiments.
This work was supported by the National Science Foundation (grant nos. 0543904 and 0606838 to K.D.) and a National Institutes of Health training grant in cellular and molecular biology (grant no. T32 GM070377 to J.W.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Katayoon Dehesh (kdehesh@ucdavis.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HTM (2000) Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res 28 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Belostotsky DA, Sieburth LE (2009) Kill the messenger: mRNA decay and plant development. Curr Opin Plant Biol 12 96–102 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131 1340–1353 [DOI] [PubMed] [Google Scholar]
- Chiba Y, Green PJ (2009) mRNA degradation machinery in plants. J Plant Biol 52 114–124 [Google Scholar]
- Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ (2004) AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 328 95–102 [DOI] [PubMed] [Google Scholar]
- Covington M, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5 e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Mauxion F, Seraphin B (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby KJ, Kumar P, Kliebenstein DJ (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38 473–486 [DOI] [PubMed] [Google Scholar]
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10 71–78 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE (2007) Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19 1549–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M (2006) PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13 533–539 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Langdale JA (2006) A step by step guide to phylogeny reconstruction. Plant J 45 561–572 [DOI] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh LN, VanToai T, Streeter J, Banowetz G (2005) Regulation of flooding tolerance of SAG12:ipt Arabidopsis plants by cytokinin. J Exp Bot 56 1397–1407 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Kotchoni S, Kuhns C, Ditzer A, Kirch H, Bartels D (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29 1033–1048 [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Glaunsinger BA (2009) Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7 e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Li C, Liu F, Jiang H, Li S, Sun J, Wu X, Li C (2009) The Arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res 19 307–316 [DOI] [PubMed] [Google Scholar]
- Ma S, Bohnert H (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8 R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57 1097–1107 [DOI] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 39 197–216 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS: an online resource for quantitative expression analysis. Plant Physiol 135 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin L, Puisieux A (2005) C. elegans homologue of the Caf1 gene, which encodes a subunit of the CCR4-NOT complex, is essential for embryonic and larval development and for meiotic progression. Gene 358 73–81 [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yao R, Ogawa T, Suzuki T, Ito C, Tsunekawa N, Inoue K, Ajima R, Miyasaka T, Yoshida Y, et al (2004) Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat Genet 36 528–533 [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J (2007) Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19 3418–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA (2004) mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 10 1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals I, Personnaz L, Taing L, Potier MC (2007) Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23 401–407 [DOI] [PubMed] [Google Scholar]
- Rowe HC, Kliebenstein DJ (2007) Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol Plant Microbe Interact 20 1126–1137 [DOI] [PubMed] [Google Scholar]
- Salles FJ, Strickland S (1999) Analysis of poly(A) tail lengths by PCR: the PAT assay. In S Haynes, ed, Methods in Molecular Biology: RNA-Protein Interaction Protocols, Vol 118. Humana Press, Totowa, NJ, pp 441–448 [DOI] [PubMed]
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C (2008) A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res 36 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA (2005) Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 15 284–294 [DOI] [PubMed] [Google Scholar]
- Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y (2008) Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol 4 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R, Rohlf F (1995) Biometry: The Principles and Practice of Statistics in Biological Research, Ed 3. W.H. Freeman and Co, New York
- Sujon S, Hyun Woo O, Hye Sun C, Kwang-Hyun B, Eun Soo S, Young Hee J, Gyung Ja C, Sanghyeob L, Doil C (2007) Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J 51 792–802 [DOI] [PubMed] [Google Scholar]
- Swindell WR (2006) The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics 174 1811–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D (1999) PAUP*: Phylogenetic Analysis Using Parsimony (And Other Methods), Version 4.0. Sinauer Associates, Sunderland, MA
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E (2004) A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104 377–386 [DOI] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3 e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog 4 e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D, Brunnemann R, Selbig J (2007) The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet 3 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CYA, Shyu AB (2005) Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12 1054–1063 [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M (2006) Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133 4573–4583 [DOI] [PubMed] [Google Scholar]
- Zheng W, Zhai Q, Sun J, Li CB, Zhang L, Li H, Zhang X, Li S, Xu Y, Jiang H, et al (2006) Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol 141 1400–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.