Abstract
Mitogen-activated protein kinase (MAPK) cascades have been identified in various signaling pathways involved in plant development and stress responses. We identified a drought-hypersensitive mutant (drought-hypersensitive mutant1 [dsm1]) of a putative MAPK kinase kinase (MAPKKK) gene in rice (Oryza sativa). Two allelic dsm1 mutants were more sensitive than wild-type plants to drought stress at both seedling and panicle development stages. The dsm1 mutants lost water more rapidly than wild-type plants under drought stress, which was in agreement with the increased drought-sensitivity phenotype of the mutant plants. DSM1-RNA interference lines were also hypersensitive to drought stress. The predicted DSM1 protein belongs to a B3 subgroup of plant Raf-like MAPKKKs and was localized in the nucleus. By real-time PCR analysis, the DSM1 gene was induced by salt, drought, and abscisic acid, but not by cold. Microarray analysis revealed that two peroxidase (POX) genes, POX22.3 and POX8.1, were sharply down-regulated compared to wild type, suggesting that DSM1 may be involved in reactive oxygen species (ROS) signaling. Peroxidase activity, electrolyte leakage, chlorophyll content, and 3,3′-diaminobenzidine staining revealed that the dsm1 mutant was more sensitive to oxidative stress due to an increase in ROS damage caused by the reduced POX activity. Overexpression of DSM1 in rice increased the tolerance to dehydration stress at the seedling stage. Together, these results suggest that DSM1 might be a novel MAPKKK functioning as an early signaling component in regulating responses to drought stress by regulating scavenging of ROS in rice.
Plants are exposed to diverse abiotic stresses including drought, salt, and adverse temperatures throughout their life cycles. For stress response and tolerance, plants have evolved a variety of biochemical and physiological mechanisms. Among the stresses, drought is a major environmental factor limiting productivity and distribution of plants (Boyer, 1982). Many stress-responsive genes have been identified and altered gene expression plays an important role in plant drought resistance (Zhu, 2002; Li et al., 2008; Lee et al., 2009); however, the biological functions of most of the stress-responsive genes are largely unknown.
Mitogen-activated protein kinase (MAPK) cascades are conserved signal transduction modules in all eukaryotes. These modules typically consist of three protein kinases: MAPK kinase kinase (MAPKKK), MAPK kinase (MAPKK), and MAPK. Upstream signals activate MAPKKKs, which then phosphorylate MAPKKs; MAPKKs in turn activate a specific MAPK. The downstream targets of MAPKs can be transcription factors, phospholipases, or cytoskeletal proteins (Sturgill and Ray, 1986; Lin et al., 1993; Jonak et al., 2002; Cheong et al., 2003). In plants, MAPK cascades have been identified in signaling pathways including cell division, hormone responses, development, disease resistance, and abiotic stress (Tena et al., 2001; Nakagami et al., 2005). The first complete MAPK cascade, MEKK1-MKK4/MKK5-MPK3/MPK6-WRKY22/WRKY29, was proposed as being downstream of the flagellin receptor FLS2 (Asai et al., 2002). Another MAPK cascade, YDA-MKK4/MKK5-MPK3/MPK6, plays an important role in regulating stomatal development (Bergmann et al., 2004; Wang et al., 2007).
In Arabidopsis (Arabidopsis thaliana), there are 20 MAPKs, 10 MAPKKs, and 80 MAPKKKs (Colcombet and Hirt, 2008). Compared with MAPKs and MAPKKs, MAPKKKs have more complex and variable primary structures and domain compositions. MAPKKKs from higher plants can be divided into two distinct families, the MEKK family and the Raf family. All Raf family proteins have a C-terminal kinase domain (KD) and a long N-terminal regulatory domain (RD). In contrast, MEKK family members have less conserved protein structure (Jouannic et al., 1999). In recent years, the roles of individual plant MAPKKK proteins in diverse plant signal transduction pathways have been explored. The NPK1 gene from tobacco (Nicotiana tabacum) has previously been shown to play an important role in regulating cell division (Banno et al., 1993; Nishihama et al., 2001, 2002; Soyano et al., 2003). More recently, Shou et al. (2004) reported that overexpression of the KD of NPK1 enhances abiotic stress tolerance in transgenic maize (Zea mays). The Arabidopsis ANP1 gene is homologous to the tobacco NPK1 gene. ANP1 is responsive to oxidative stress and is involved in negative regulation of the auxin signal transduction pathway (Kovtun et al., 1998, 2000). OMTK1, a MAPKKK activated by hydrogen peroxide (H2O2) in alfalfa (Medicago sativa), can specifically activate the MAPK MMK3 in the protoplast, resulting in cell death (Nakagami et al., 2004). To date, only two Raf family MAPKKKs, namely CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) and ENHANCED DISEASE RESISTANCE1 (EDR1), have been published. The CTR1 gene negatively regulates ethylene responses. The Arabidopsis mutant ctr1 exhibits characteristic constitutive ethylene responses in the absence of ethylene (Kieber et al., 1993). Furthermore, CTR1 has been found to interact with the His-KD of ETR1 and the ethylene response sensor (ERS1) in vitro (Clark et al., 1998). The EDR1 was identified and the edr1 mutant was shown to be resistant to powdery mildew (Frye and Innes, 1998). In addition, EDR1 negatively regulated salicylic acid-inducible defense responses in Arabidopsis (Frye et al., 2001), but its substrates are still unknown.
Rice (Oryza sativa) is one of the most important crops in the world, especially in Asia. One of the main environmental factors that affects rice yield is water deficit or drought stress. Drought resistance is a complex quantitative trait. Although genetic mapping of drought resistance-related traits of rice has been reported (Yue et al., 2005, 2006) and many drought stress-related genes in rice have been identified, the molecular basis of drought response and adaptation in rice is largely unclear. Recent studies have suggested that many protein kinases are involved in drought resistance in rice. Among them, members of the calcium-dependent protein kinase (CDPK), calcineurin B-like protein-interacting protein kinase (CIPK), and MAPK families have been identified as conferring stress resistance in rice. For example, overexpression of OsCDPK7 in rice results in enhanced stress tolerance at the seedling stage (Saijo et al., 2000); overexpression of three CIPK genes (OsCIPK03, OsCIPK12, and OsCIPK15) enhances tolerance to cold, drought, and salt stress, respectively, in transgenic rice (Xiang et al., 2007); overexpression of a MAPK family gene OsMAPK5a in rice leads to increased OsMAPK5a kinase activity and enhanced tolerance to drought, salt, and cold stresses (Xiong and Yang, 2003). OsMAPK44, another rice MAPK gene, is highly inducible by salt and drought treatment and OsMAPK44 overexpression in rice plants generates increased tolerance to salt stress (Jeong et al., 2006b). However, evidence for MAPKKK genes being involved in stress resistance of rice has not been reported.
In this study, we identified a rice drought-sensitive mutant (drought-hypersensitive mutant1 [dsm1]). The dsm1 mutant showed enhanced drought sensitivity in both the seedling stage and the panicle development stage. DSM1 encodes a putative MAPKKK belonging to the Raf family of protein kinases. Our results demonstrate that DSM1 may act as an early signal regulating the responses to drought stress in rice. Our data also indicate that drought hypersensitivity of dsm1 is partially due to a decrease in reactive oxygen species (ROS) scavenging.
RESULTS
Identification of a Drought-Sensitive Mutant in Rice
To reveal the potential functions of MAPK cascades in drought-stress signaling in rice, we collected a total of 15 T-DNA insertion mutants (japonica rice Zhonghua11 [ZH11] background) corresponding to six putative MAPKKK genes (Supplemental Table S1) available in the T-DNA mutant library in the Rice Mutant Database (Wu et al., 2003). These mutants were prescreened for drought resistance under field conditions. Under normal growth conditions, most of the mutant lines showed no obvious phenotype change (or segregation) compared to the wild-type ZH11. Under drought stress (water withheld from 4-week-old plants for 10 d), one of the mutant lines for a predicted MAPKKK gene (LOC_Os02g50970) showed increased sensitivity to drought stress, and this mutant, dsm1-1 (dsm1, allele 1), was selected for further analysis. We collected a second T-DNA mutant allele of this gene (dsm1-2; Hwayoung [HY] background) from the Pohang University of Science and Technology Rice T-DNA Insertion Sequence Database (POSTECH RISD; An et al., 2003; Jeong et al., 2006a). Sequence analysis indicated that the insertion site of the dsm1-1 mutant is located in exon 12, 7,174 bp downstream of ATG, and the T-DNA of the dsm1-2 mutant is inserted in intron 10, 6,734 bp downstream of ATG (Fig. 1A). Transcription of DSM1 in dsm1 mutants was checked by semiquantitative reverse transcription (RT)-PCR using RNA templates isolated from homozygous mutants. The mRNA was not detectable in either mutant, suggesting that the expression of DSM1 was abolished in the mutants (Fig. 1B).
Figure 1.
Identification of dsm1 T-DNA insertion mutants. A, Schematic diagram of DSM1 gene and two alleles of T-DNA insertion mutants, dsm1-1 and dsm1-2. LB, Left border; RB, right border. Exons and introns are indicated in blue and white, respectively. B. RT-PCR analysis of DSM1 in ZH11, HY, dsm1-1, and dsm1-2. Actin1 was used as internal control. [See online article for color version of this figure.]
When grown under normal growth conditions, neither of the dsm1 mutants displayed any obvious phenotype change (Fig. 2A, top). To verify the drought-sensitivity phenotype, homozygous mutant (dsm1/dsm1) and wild-type (DSM1/DSM1) lines derived from DSM1/dsm1 genotype plants for both mutant alleles were grown in sandy-mixed soil along with wild-type rice ZH11 or HY. The 4-week-old plants were subjected to drought treatment by stopping irrigation for 10 to 15 d. Under the moderate stress condition, the dsm1 mutant lines showed more severe wilting and a lesser degree of recovery after rewatering than the wild-type genotypes (Fig. 2A, bottom), and the phenotype cosegregated with the genotype (data not shown). After severe drought-stress treatment, we found that only about 17% of the mutant plants survived, while wild-type plants had a significantly higher survival rate (approximately 63%; Fig. 2B). Subsequently, we measured water loss rates of detached leaves from the wild type and the dsm1 mutant lines. Results showed that the detached leaves of the dsm1 mutants lost water slightly faster than wild type (Fig. 2C). Drought sensitivity of dsm1 mutants was also tested at the panicle development stage. The dsm1-1 and wild type were planted in a paddy field with a removable rain-off shelter and in PVC tubes filled with sandy soil as described previously (Hu et al., 2006). During drought stress, dsm1-1 plants showed earlier leaf rolling and wilting than wild-type plants (Fig. 2D). Statistical analysis of the yield and spikelet fertility also suggested that dsm1-1 exhibited hypersensitivity to drought stress at the panicle development stage (Fig. 2, E and F). In the field, the total grain yield of dsm1-1 was reduced by about 22% when compared to the wild type; the spikelet fertility of dsm1-1 (45%) was also significantly (P < 0.01) lower than that of wild-type plants (60%; Fig. 2E). In the PVC tubes (more severe drought stress applied), the mutant showed 60% less yield than the wild type (DSM1/DSM1), and the spikelet fertility of the mutant was only about half that in the wild type (Fig. 2F). Nevertheless, no significant difference in yield and fertility was observed between the mutant and wild-type plants under normal growth conditions. We also checked the sensitivity of the dsm1-1 mutant to salt stress, and results showed that the mutant was more sensitive to salt stress than the wild type (Supplemental Fig. S1), but no significant difference was observed for cold stress (data not shown).
Figure 2.
Increased drought sensitivity of dsm1 at both seedling and heading stages. A, Five-leaf stage wild-type (ZH11 or HY) and dsm1 mutant plants were growing in barrels filled with a mixture of soil and sand (1:1). The left and right half of each barrel was planted with wild-type and mutant plants, respectively. Plants were not watered for 12 d, followed by rewatering for 7 d. The photograph was taken before drought stress and after recovery. B, Comparison of survival rate of wild type (ZH11 or HY) and dsm1 mutants (dsm1-1 [ZH11 background] or dsm1-2 [HY background]) after drought stress. Values are means ± se (n = 3). **P < 0.01 (t test). C, Water loss from detached leaves of wild type (ZH11 or HY) and dsm1 mutants (dsm1-1 [ZH11 background] or dsm1-2 [HY background]) at indicated time points. D, ZH11 and dsm1 plants were subjected to drought stress at heading stage. The photograph was taken on day 20 after rewatering. E, Spikelet fertility and yield of ZH11 and dsm1-1 under drought stress in paddy field with a removable rain-off shelter. Values are means ± sd (n = 32). **P < 0.01 (t test). F, Spikelet fertility and yield of wild type (WT; DSM1/DSM1 [ZH11 background]), heterozygous, and homozygous dsm1-1 under drought stress in PVC tubes filled with sandy soil. Values are means ± sd (n = 12). **P < 0.01 (t test).
Suppression of DSM1 Resulted in Enhanced Sensitivity to Drought Stress
To further examine the functions of DSM1 in rice, DSM1-RNA interference (RNAi) rice plants were produced with a 400 bp open reading frame region of DSM1 as the targeted interference region. Among 24 T0 RNAi plants checked, five showed significantly decreased expression of DSM1 (Fig. 3A). Three independent RNAi lines (RNAi-10, RNAi-11, and RNAi-20) were selected for drought testing at the seedling stage. Results showed that the DSM1-RNAi lines were also hypersensitive to drought stress (Fig. 3B), which was in agreement with the phenotype of dsm1 mutant plants. The survival rate of DSM1-RNAi lines was only 10% to 15%, whereas 60% to 70% of wild-type plants recovered (Fig. 3C). Consistent with this result, detached leaves of DSM1-RNAi lines lost water more quickly than the wild-type leaves (Fig. 3D). Taken together, these results suggest that DSM1 plays essential roles in drought resistance in rice.
Figure 3.
Suppression of DSM1 showed increased sensitivity to drought stress. A, Transcript level of DSM1 was measured by RT-PCR in DSM1-RNAi plants (ZH11 background). Numbers 8, 10, 11, 17, and 20 indicated independent RNAi transgenic plants. Actin1 was used as internal control. B, Five-leaf stage ZH11 and DSM1-RNAi plants were subjected to no watering for 12 d, followed by rewatering for 7 d. Surviving seedlings were photographed. C, Comparison of survival rate of ZH11 and DSM1-RNAi plants after drought stress. Values are means ± se (n = 3). **P < 0.01 (t test). D, Water loss from detached leaves of ZH11 and DSM1-RNAi plants at indicated time points.
DSM1 Expression Profile and Subcellular Localization of DSM1
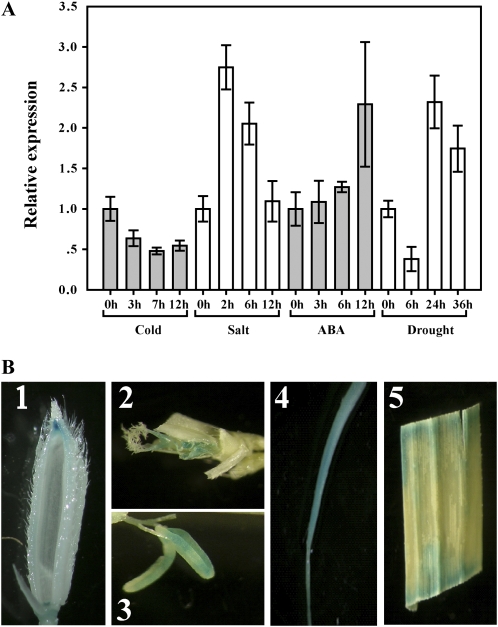
As dsm1 showed hypersensitivity to drought stress, we examined the expression level of DSM1 under drought as well as other treatments including cold, salt, and abscisic acid (ABA) at the seedling stage. As shown in Figure 4A, the DSM1 transcript level increased after drought and salt treatment, albeit not very strongly (about 2- to 3-fold). DSM1 was only slightly induced by ABA. Contrary to the induction of DSM1 expression by salt and drought stresses, DSM1 was not induced by cold stress but rather slightly suppressed. These results seem to be consistent with the dsm1 phenotypes in stress responses and indicate that DSM1 may function mainly in drought- and salt-stress responses.
Figure 4.
Expression pattern of DSM1. A, Expression of DSM1 under abiotic stress. Two-week-old seedlings were subjected to cold (4°C), salt (200 mm NaCl in the hydroponic culture medium), ABA (100 μm), and drought stress (removing the water supply from the plants). Relative expression levels of DSM1 were examined by quantitative real-time PCR. B, DSM1 promoter∷GUS expression patterns in transgenic rice plants (ZH11 background). GUS staining was shown in hull (1), pistil (2), stamen (3), root (4), and mature leaves (5).
To investigate the tissue-specific expression pattern of DSM1 in rice, we generated transgenic rice plants with a construct (PDSM1∷GUS) in which the GUS reporter gene was under the control of a DSM1 promoter. Histochemical staining for GUS activity was performed in five independent transgenic plants. Results showed that strong GUS expression was observed in the stamen, pistil, mature leaves, and roots (Fig. 4, C–F), and weak GUS expression was detected in the hull (Fig. 4B), suggesting a constitutive express pattern of DSM1 in rice. This result was supported by microarray data of the gene in the genomic expression profiles surveyed for an entire life cycle of rice (L. Wang and Q. Zhang, unpublished data).
To determine the subcellular localization of DSM1, the DSM1 cDNA was fused in frame to the GFP marker gene under the control of cauliflower mosaic virus 35S promoter. In the transgenic rice expressing DSM1-EGFP, the fluorescence was observed only in the nuclei (Fig. 5A). To confirm this result, rice protoplasts prepared from an etiolated shoot were cotransformed with 35S:sGFP-DSM1 and 35:CFP-GHD7 by polyethylene glycol treatment. GHD7 was used as a marker since it has been reported as a transcription factor localized in the nucleolus (Xue et al., 2008). As shown in Figure 5B, green fluorescence produced by sGFP-DSM1 overlapped with blue fluorescence produced by CFP-GHD7, which further confirmed that DSM1 is a nuclear protein.
Figure 5.
Subcellular localization of DSM1. A, Subcellular localization of DSM1 in transgenic rice (ZH11 background). DSM1 cDNA was fused to GFP and the construct was expressed in transgenic rice under the control of the cauliflower mosaic virus 35S promoter. Confocal image of the root is shown. B, Subcellular localization of DSM1 in rice protoplast (ZH11 background). 35S:sGFP-DSM1 and 35:sCFP-GHD7 were cotransformed into rice etiolated shoot protoplasts. 35S:sGFP was transformed as control.
DSM1 Features a Raf-Like MAPKKK
DSM1 encodes a putative MAPKKK of 1,111 amino acids with a molecular mass of 123 kD. Based on domain scanning against the Pfam database (http://pfam.sanger.ac.uk/) and comparison with other plant MAPKKKs, DSM1 consists of a C-terminal KD (amino acids 838–1,090) and an N-terminal putative RD (Fig. 6A). DSM1 was compared with reported MAPKKKs from other plants (Supplemental Fig. S2). The results showed that DSM1 contains all 11 subdomains common to all known protein kinases (Hanks and Quinn, 1991). Alignment of the full-length protein sequence of DSM1 with the well-characterized plant MAPKKKs suggests that DSM1 has high sequence similarity in both the KD and the RD to EDR1 (73% identity in KD and 36% identity in RD) and CTR1 (65% identity in KD and 42% identity in RD). Both EDR1 and CTR1 belong to the B3 subgroup of plant Raf-like MAPKKKs (Ichimura, 2002). In contrast, DSM1 showed relatively low sequence identity with other plant MAPKKK proteins. For example, DSM1 shared only 31% and 30% identity, respectively, with AtMEKK1 and NPK1 even in the conserved KD. A phylogenetic tree based on the conserved KD and the position of the KD also placed DSM1 into the Raf-like MAPKKK family (Fig. 6B).
Figure 6.
Protein sequence analysis of DSM1. A, Schematic diagram of functional domains. B, Phylogenetic tree of MAPKKK members in plants. The phylogenetic tree was created in MEGA4 software with the neighbor-joining method. Numbers indicate percentage values after 1,000 replications. Scale indicates amino acid substitutions per position. Prefixes on protein names indicate species of origin: At, Arabidopsis; Bn, Brassica napus; Nt, N. tabacum; Os, O. sativa; Le, S. lycopersicum var. lycopersicum; Cm, Cucumis melo; Fs, Fagus sylvatica; Ah, Arachis hypogaea.
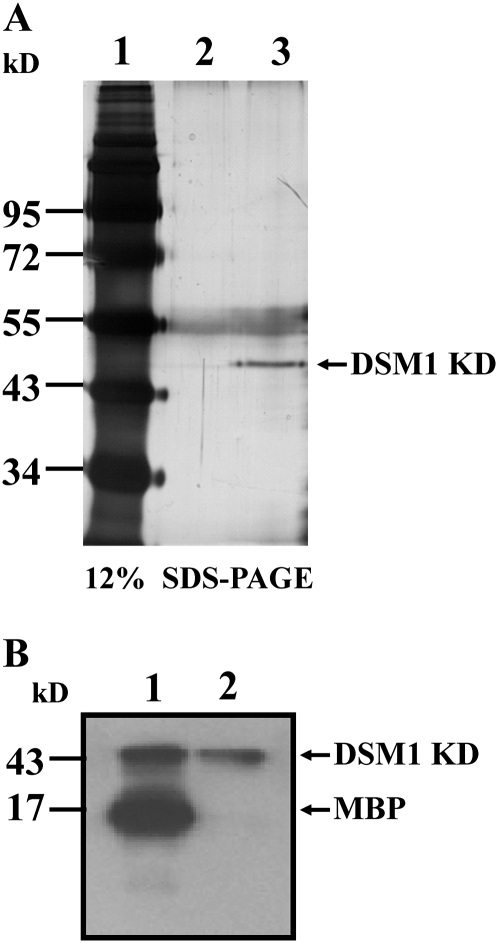
Raf-like MAPKKKs in plants have been reported to possess kinase activities. CTR1 and its KD display autophosphorylation activity, as well as the ability to phosphorylate myelin basic protein (MBP) in vitro. The EDR1 KD also can autophosphorylate (Covic et al., 1999; Tang and Innes, 2002; Huang et al., 2003). To determine if DSM1 has kinase activity, the KD was expressed in Escherichia coli strain BL21, but the expressed protein was highly insoluble (data not shown). To overcome this problem, transgenic rice were generated expressing the DSM1 KD with a FLAG tag (provided by Sun and Zhou, 2008) fused at the carboxyl terminus. The KD-FLAG protein was immunoprecipitated from a total protein extract of the KD-FLAG transgenic plants with anti-FLAG M2 affinity gel. Immunoprecipitated proteins were separated by SDS-PAGE with immunoprecipitated protein extract from wild type as a negative control. Silver staining of the SDS-PAGE gel showed a clearly visible band (slightly larger than the predicted 43-kD fusion protein) only in the transgenic plants (Fig. 7A). The band was further confirmed to be KD of DSM1 by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) following tryptic digestion (Supplemental Fig. S3). The purified protein was subjected to the kinase assay as described previously (Huang et al., 2003). MBP is a universal substrate that has been widely used to examine kinase activity (Eichberg and Iyer, 1996). Our results demonstrated that MBP was strongly phosphorylated by the DSM1 KD and that the DSM1 KD had autophosphorylation activity regardless of the presence or absence of MBP (Fig. 7B). These results together suggest that DSM1 functions as a typical Raf-like MAPKKK in rice.
Figure 7.
DSM1 KD displayed kinase activity. A, Silver-stained SDS-PAGE analysis of immunopurified DSM1 KD protein (with FLAG) from ZH11 (lane 2) and transgenic rice expressing DSM1 KD (lane 3). Molecular weight marker (lane 1) is indicated on the left. B, Immunopurified DSM1 KD protein was mixed with MBP (lane 1) or buffer alone (lane 2) in kinase reaction. Phosphorylated protein was analyzed by SDS-PAGE, then followed by autoradiography. The arrows indicate the position of the proteins.
Microarray Analysis of dsm1 Plants
It is known that MAPK cascade-mediated signaling often leads to regulation of gene expression through phosphorylation of transcriptional regulators. To identify genes with altered expression levels in the dsm1 mutant, microarray (Affymetrix Rice GeneChip) analysis was performed to compare the expression profiles between the dsm1 mutant and wild type (the DSM1/DSM1 genotype in the progenies of DSM1/dsm1 plants) under both normal and drought-stress conditions. Results suggest that only a small portion of genes in the rice genome show significant expression level changes in the dsm1 mutant compared to the wild type under normal conditions. Only 12 genes (0.02% of all rice genes) were down-regulated by more than 2.5-fold in the mutant (Supplemental Table S2). However, the transcription levels of many well-studied abiotic stress-related marker genes (such as RD22, RAB21, and RAB16c) were not changed in the dsm1 mutant compared to the wild type. Among the down-regulated genes in the dsm1 mutant, two peroxidase (POX) genes POX22.3 and POX8.1, which showed predominant expression during resistant interactions (Chittoor et al., 1997), were dramatically down-regulated (15- and 5.3-fold, respectively). Several genes annotated as encoding expression proteins and retrotransposon proteins were also down-regulated in the dsm1 plants, but their relevance to stress resistance is not clear. Even though we noticed that 47 genes (0.08% of all rice genes) were up-regulated (more than 2-fold) in the mutant under normal conditions (Supplemental Table S3), no obvious link or relevance to abiotic stress was found for these genes based on their predicted function and expression features. By contrast, the dsm1 mutant showed significantly altered gene expression compared with wild-type plants under drought stress. A total of 678 genes (1.18% of all rice genes) and 126 genes (0.22% of all rice genes) showed up- and down-regulated expression, respectively, in dsm1 mutants compared to wild-type plants under stress conditions (Supplemental Tables S4 and S5). Among these, a number of stress-responsive genes were altered in the dsm1 mutants, and these genes encode proteins with diverse functions such as transcription factors, protein kinases, calcium-binding proteins, stress-inducible proteins, antioxidant enzymes, and cell wall biosynthetic proteins. To confirm the microarray results and to have a closer view of the two POX genes, the expression of POX22.3 and POX8.1 were assessed by real-time PCR in the dsm1 and wild-type seedling plants treated with or without drought stress for 72 h. Consistent with the microarray results, the transcript levels of POX22.3 and POX8.1 were significantly lower in the mutants than in wild type under both normal and drought-stress conditions (Fig. 8). A number of other genes, including two genes encoding protein Brittle-1 (LOC_Os09g32200) and ATP-binding protein (LOC_Os01g54420), were also confirmed by real-time PCR for their expression level changes in the mutant (Fig. 8).
Figure 8.
Real-time PCR analysis of four genes in the seedlings of wild type (WT) and dsm1-1 under normal and drought stress (with water depleted for 72 h). The rice Actin1 gene was amplified as the internal control. Primers used in PCR are listed in Supplemental Table S4.
The dsm1 Mutant Is Sensitive to Oxidative Stress
Peroxidases are involved in defense against abiotic stresses through scavenging of ROS (Hiraga et al., 2001). In the dsm1 mutants, the expression levels of POX22.3 and POX8.1 and a few other genes encoding putative oxidoreductases were significantly reduced. This suggests that the hypersensitivity of the dsm1 mutant to drought stress may result from reduced capacity for ROS scavenging. To test this, POX activities in the soluble protein extracts from the drought-stressed and nonstressed leaves of dsm1 and wild-type plants were measured. Peroxidase activity was increased after drought stress in both dsm1 and wild-type plants; however, significantly lower POX activity was detected in the dsm1 plants than in the wild-type plants (Fig. 9A).
Figure 9.
The dsm1 mutants showed hypersensitivity to oxidative stress. A, The POX activity in ZH11 and dsm1-1 during drought stress. Values are means ± se (n = 3). **P < 0.01 (t test). FW, Fresh weight. B, Analysis of MV stress-related electrolyte leakage in ZH11 and dsm1-1 at indicated time. Values are means ± se (n = 4). **P < 0.01 and *P < 0.05 (t test). C, Leaf phenotype of ZH11 and dsm1-1 plants (three-leaf stage) after MV stress (10 μm MV for 4 d). D, Phenotype of ZH11 and dsm1-1 (three-leaf stage) plants after MV stress (30 μm MV for 4 d and recovery for 2 d). E, Total chlorophyll contents at 96 h after 30 μm MV stress. Values are means ± se (n = 3). **P < 0.01 (t test). WT, Wild type. F, DAB staining for H2O2 in unstressed and MV-stressed leaves of ZH11 and dsm1-1 plants.
It has been determined previously that the POX activity in plants is correlated with increased tolerance to oxidative stresses (Kim et al., 2008). Therefore, the response of dsm1 plants to oxidative stress was analyzed. Methyl viologen (MV) is a well-known oxidative stress inducer that inhibits electron transport during photosynthesis and generates H2O2 in chloroplasts under light (Chen et al., 2006). The leaves from plants grown in soil were floated on 4 μm MV and electrolyte leakage, an indicator of membrane damage, was measured every 4 h. The results showed that the dsm1-1 leaves exhibited significantly higher electrolyte leakage than the wild-type leaves (Fig. 9B). After 24 h of MV treatment, more than 49% of ions leaked from cells in the dsm1-1 mutant, whereas ion leakage of the wild type was less than 36%. The sensitivity to MV was also checked in intact plants. Two-week-old seedlings of dsm1-1 and wild type were exposed to 10 μm MV. At 1 d after MV treatment, necrosis was observed on the leaves of dsm1-1, while all wild-type leaves were still green (Fig. 9C). At 3 d, the dsm1-1 mutants showed much more severe chlorosis and damage than wild-type plants (Fig. 9C). After severe oxidative stress (2-week-old seedlings treated with 30 μm MV for 3 d and followed by recovery for 2 d), most of the mutant plants died while the wild-type plants recovered very quickly (Fig. 9D). A severe decrease of chlorophyll content (35% chlorophyll retained compared to untreated plants) was observed in the dsm1-1 plants exposed to 30 μm MV for 3 d, whereas no obvious decrease of chlorophyll content was noted in wild-type plants after the same treatment (Fig. 9E).
ROS are known to cause damage to cellular membranes. It is possible that the membrane damage in the dsm1-1 plants caused by oxidative stress may be associated with accumulation of ROS. Therefore, we determined the accumulation of H2O2 by 3,3′-diaminobenzidine (DAB) staining and superoxide by nitroblue tetrazolium staining. Under normal growth conditions, no obvious H2O2 was detected in either wild-type or dsm1-1 plants. After MV stress (30 μm MV treatment for 72 h), H2O2 was detected in both the wild type and the mutant, but accumulation of brown precipitate (DAB staining) in the mutant was much larger than in the wild type (Fig. 9F). However, no significant difference in nitroblue tetrazolium staining was observed between the mutant and wild type (data not shown). Together, these observations demonstrate the enhanced sensitivity of the dsm1 mutant to oxidative stress resulted from reduced ability of ROS (at least H2O2) scavenging.
Overexpression of DSM1 Improves Tolerance to Dehydration Stress
Genetic and physiological analysis of the dsm1 mutant implies that DSM1 may play a positive role in the regulation of stress resistance in rice. We predicted that overexpression of DSM1 might result in an increased stress tolerance. To test this hypothesis, transgenic plants were generated by overexpressing the full-length DSM1 fused with the FLAG tag under the control of the maize ubiquitin promoter. A total of 43 independent transgenic plants were obtained and the expression level of DSM1 was verified by western blot with antibody against the FLAG tag. Three independent transgenic lines (denoted with numerals 9, 38, and 42) with higher expression (Fig. 10A) were selected for further analyses.
Figure 10.
Overexpression of DSM1 showed tolerance to mannitol stress. A, Western blot showed expression of DSM1-FLAG protein in the overexpression plants. Numbers 2, 9, 38, and 42 are independent overexpression transgenic plants (ZH11 background). The Coomassie Brilliant Blue-stained blot is shown as the loading control. B, Performance of ZH11 and DSM1 overexpression plants subjected to mannitol stress in MS medium with 125 mm mannitol. C, Relative shoot length of ZH11 and DSM1 overexpression plants under mannitol stress. Values are means ± se (n = 3). *P < 0.05 (t test; unstressed controls = 100).
We first tested the tolerance of DSM1-overexpression plants under mannitol (120 mm) treatment, which mimics drought stress in terms of dehydration and oxidative outcomes. We observed that the shoots of the overexpression plants were slightly shorter than those of wild-type plants in normal Murashige and Skoog (MS) medium, but the wild-type plants exhibited more serious growth inhibition than the overexpression plants under the stress conditions (Fig. 10B). Statistical analysis suggested that the overexpression lines had significantly less suppression of shoot growth (58% to 65% of normal growth) than wild type (only about 45% of the normal growth) under the mannitol treatment (Fig. 10C). These results suggest that overexpression of DSM1 has a positive effect on stress tolerance.
DISCUSSION
Plants have developed sophisticated signaling pathways to deal with environmental stress. Protein kinases such as CDPKs, CIPKs, and MAPK cascades have been shown to have important roles in abiotic stress signaling as well as in many other signaling processes. MAPK cascades are evolutionarily conserved and involved in signaling pathways related to biotic and abiotic stress responses. Compared with those in yeast (Saccharomyces cerevisiae) and mammalian, however, only a limited number of genes in MAPK cascades have been identified in plants (Ichimura, 2002).
Here, we report the identification and characterization of a drought-sensitive mutant (dsm1) in rice. A database search and sequence analysis suggests that DSM1 contains a conserved protein KD and features a B3 subgroup of plant Raf-like MAPKKK. In Arabidopsis, six genes encoding MAPKKK proteins have been identified (Ichimura, 2002). But there are only two members of the B3 subgroup of plant Raf-like MAPKKKs reported with known function: (1) CTR1 and its tomato (Solanum lycopersicum var. lycopersicum) ortholog negatively regulate ethylene responses and play a role in defense response (Kieber et al., 1993; Leclercq et al., 2002; Lin et al., 2008), and (2) EDR1, which is involved in salicylic acid-inducible defense response. The edr1 mutant showed resistance to powdery mildew and senesced early in response to exogenous ethylene (Frye and Innes, 1998; Frye et al., 2001). Protein kinase activity has been found for many reported MAPKKKs (Covic et al., 1999; Tang and Innes, 2002; Huang et al., 2003). Here we also demonstrated that the DSM1 KD can phosphorylate itself and MBP. To the best of our knowledge, DSM1 is the first member of the MAPKKK family to be identified as having drought-stress signaling function in plants.
To study the function of DSM1, we identified two T-DNA insertion mutant alleles for the DSM1 gene. Meanwhile, DSM1-RNAi plants were also generated to genetically confirm the mutant phenotype. The results revealed that both of the dsm1 mutants and the DSM1-RNAi plants displayed hypersensitivity to drought stress in seedling and panicle developmental stages. Even though the mutants showed slightly increased sensitivity to salt stress at the seedling stage, no significant difference in seed germination or seedling growth was observed between wild-type and mutant plants when they were treated with ABA, salt, cold, and heat stress (data not shown). These findings suggest that DSM1 may mainly function in drought-stress signaling through an ABA-independent pathway. In many previous studies, knock out of MAPKKK genes often resulted in aberrant morphologies when the mutants were grown under normal growth conditions. For example, a mutant of the Arabidopsis CTR1 gene exhibited constitutive triple responses in the absence of ethylene (Kieber et al., 1993). Double mutant (anp2 anp3) for two MAPKKK genes displayed an overall reduction in total plant size and disrupted cell division, but the mutant of a single gene was not significantly different from the wild type (Krysan et al., 2002). Most recently, a mekk1 mutant was reported with abnormal phenotypes including extremely dwarfed, curled leaves and early senescence (Nakagami et al., 2006). We also examined the phenotypes of the two dsm1 mutants under normal growth conditions. However, neither of the dsm1 mutants displayed any obvious phenotypic change throughout the life cycle of rice. Although it is possible that functional redundancy may exist for DSM1 and other rice MAPKKK genes with additional function, our results suggested that DSM1 plays essential roles in drought-stress signaling in rice since a single mutation of this gene caused hypersensitivity to drought stress.
Phylogenetic analysis suggests that DSM1 belongs to a subfamily of MAPKKK in plants in which only two proteins, CTR1 and EDR1, have been functionally characterized. Tang and Innes failed to obtain overexpression plants on transformation of the full-length cDNA of EDR1 in either the wild type or an edr1 mutant of Arabidopsis (Tang and Innes, 2002). It was proposed that overexpression of EDR1 was either lethal to Arabidopsis or blocked some step in the transformation process. A similar lethal phenotype was observed in the overexpression of MEKK1 in Arabidopsis under the 35S Cauliflower mosaic virus or a dexamethasone-inducible promoter (Nakagami et al., 2006). Unlike the overexpression of EDR1 and MEKK1, overexpression of DSM1 in rice yielded no obvious phenotypic changes under normal growth conditions, suggesting that DSM1 has evolved with different function in rice compared to its closely related homologs in Arabidopsis. Indeed, overexpression of DSM1 enhanced tolerance to dehydration stress caused by mannitol. Western-blot analysis with anti-FLAG antibody showed that the increased DSM1 protein level was linked to increased mannitol tolerance. Together with the mutant phenotypes, these results suggest that DSM1 participates in positive control of drought resistance in rice. Even though the survival rates of the DSM1 overexpression lines were similar to those of the wild type after drought stress at the seedling stage under soil conditions (data not shown), the effect of DSM1 overexpression on drought stress under different conditions remains to be determined since drought resistance is a complex trait that is affected by developmental stages. In addition, constitutively increasing transcript or protein level of a protein kinase may not necessarily lead to significant phenotypic gains since most protein kinases need to be activated to exert functions.
It was previously shown that overexpression of NPK1, a MAPKKK family gene, increased tolerance to freezing, heat shock, and salt stress (Kovtun et al., 2000). DSM1 is significantly different from NPK1, not only in functional domain composition but also in the spectrum of functions in stress resistance. Our results suggest that DSM1 functions specifically in drought and oxidative stresses. To elucidate the mechanisms of dsm1 in drought resistance, we measured Pro content, stomata number, and stomatal conductance of dsm1 mutants. However, no significant difference was found for these measurements between the mutants and wild-type plants in response to drought stress (data not shown). Therefore, we further checked the global expression profiles of dsm1 and wild-type plants. Even though the expression of a number of genes was significantly increased or decreased in the mutant compared to wild type, most of these genes appear to be irrelevant to stress resistance based on annotations or expression patterns of these genes. However, we noticed that the expression of two POX-encoding genes, POX22.3 and POX8.1, were markedly reduced in the dsm1 mutant. This clue led us to test oxidative stress response and ROS scavenging of the mutant.
ROS homeostasis is well known to be associated with oxidative stresses. When ROS is low in cells, they act as signal molecules. Under serious biotic and abiotic stress, ROS can be overproduced and cause damage of cell growth by oxidizing proteins, lipids, and DNA (Polle, 2001; Mittler et al., 2004). Therefore, plants have developed sophisticated systems to protect themselves from oxidative stress by adjusting ROS homeostasis. These systems include numerous scavenging enzymes such as superoxide dismutase, POX, ascorbate peroxidase, glutathione peroxidase, catalase, and low Mr antioxidants (Noctor and Foyer, 1998; Apel and Hirt, 2004; Mittler et al., 2004). Peroxidases scavenge ROS by catalyzing oxidoreduction between H2O2 and reductants. Plant POXs exist as a large family of isozymes and are involved in a broad range of physiological processes (Lagrimini et al., 1990; Young et al., 1995; Quiroga et al., 2000; Passardi et al., 2005). Peroxidases have been implicated in defense against abiotic stresses through increased scavenging of ROS (Hiraga et al., 2001). For example, overexpression of sweet potato (Ipomoea batatas) POX gene swpa4 led to 50-fold higher POX activity compared with the wild-type plant. The transgenic plants showed a significantly enhanced tolerance to a variety of abiotic and biotic stresses (Kim et al., 2008). Most recently, a metallothionein protein in rice was reported to confer enhanced tolerance to drought and oxidative stress by increasing activities of antioxidants catalase, POX, and ascorbate peroxidase (Yang et al., 2009). POX22.3 and POX8.1 are POX genes that are predominantly expressed during resistant interactions (Chittoor et al., 1997). Here, we found that the expression levels of these two genes were dramatically down-regulated in the dsm1 mutant compared to the wild type, and the POX activity in dsm1 was also significantly decreased. In accordance with the reduced POX activity, dsm1 showed markedly increased sensitivity to oxidative stress as indicated by the significant increase of chlorophyll degradation, chlorosis, and ion leakage. Overall, our results indicated that dsm1 mutant plants are more sensitive to drought and oxidative stresses due to an increase in ROS damage caused by the reduced POX activity. Several studies have demonstrated that MAPK cascades are involved in ROS signaling or homeostasis regulation. OMTK1, a MAPKKK activated by H2O2 in alfalfa, channels oxidative stress signaling by direct MAPK interaction (Nakagami et al., 2004). Studies of MEKK1 revealed that MEKK1 functions in integrating ROS homeostasis with plant development and hormone signaling (Ichimura et al., 2006; Nakagami et al., 2006). A recent study suggested that overexpression of a maize MAPKK gene, ZmMPK7, in transgenic tobacco can enhance protection provided by the POX defense systems against ROS-mediated injury during osmotic stress (Zong et al., 2009). Here we show that DSM1 plays a critical role in mediating oxidative stress signaling and drought resistance in rice. We have attempted to identify DSM1-interacting proteins by yeast two-hybrid screening and pair-wise interaction testing with rice MAPKKs, but neither experiment resulted in positive DSM1-interaction proteins. Of special note, DSM1 is localized in the nucleus, which contrasts to previously reported MAPKKKs thought to be localized in cytoplasm. Although the direct phosphorylation targets or interacting proteins of DSM1 remain to be revealed, our results suggest that DSM1 may be a novel nuclear protein kinase mediating oxidative stress signaling by a different pathway from traditional MAPK cascades.
In conclusion, this study establishes that DSM1 is a novel nuclear protein kinase with sequence similarity to Raf-like MAPKKKs that plays critical roles in drought and oxidative stress resistance in rice by directly or indirectly regulating POX22.3 and POX8.1 expression and ROS scavenging. Efforts are ongoing to identify the target proteins of DSM1 to reveal the complete pathway of DSM1 in drought and oxidative stress signaling.
MATERIALS AND METHODS
Plant Materials
The japonica rice (Oryza sativa) cultivars HY and ZH11 were used in this study. The dsm1-1 (ZH11 background) and dsm1-2 (HY background) mutant seeds were obtained from the Rice Mutant Database (http://rmd.ncpgr.cn) and POSTECH RISD (http://www.postech.ac.kr/life/pfg/risd/), respectively. Homozygous mutants were identified and used in further analyses.
Stress Treatments
To measure the transcript level of the DSM1 under various stresses, ZH11 plants were grown in the greenhouse with a 14-h-light/10-h-dark cycle. Plants at the four-leaf stage were treated with drought (removal of the water supply from the plants), salt (addition of 200 mm NaCl in the hydroponic culture medium), cold (exposure of the plants to 4°C), and 100 μm ABA followed by sampling at the designated time.
DSM1 overexpression and RNAi transgenic plants were selected by germinating seeds on MS medium containing 50 mg L−1 hygromycin. Wild-type and homozygous mutants were grown on MS medium or grown in soil.
For drought stress at the seedling stage, mutant/transgenic plants and wild-type plants were growing in the same barrels filled with a mixture of soil and sand (1:1). At the five-leaf stage, the water supply was depleted to allow drought stress to develop. After recovery, surviving seedlings were photographed and analyzed. Water-loss rates of detached leaves from the wild type and the mutant were measured by monitoring the fresh weight loss at indicated time points. The drought stress at the panicle development stage was conducted by growing plants in PVC tubes (1 m in height and 20 cm in diameter) and drought-stress method was the same as described previously (Hu et al., 2006; Yue et al., 2006).
To evaluate mannitol stress tolerance, geminated seeds were transplanted in MS medium supplemented with 125 mm mannitol. After 10 d of growth, shoot length was measured.
For the oxidative treatment, rice seedlings at the two-leaf stage were submerged into a 10 or 30 μm MV solution. After 3 d, the seedlings were transferred to distilled water for recovery and photographed.
RNA Isolation and RT-PCR
Total RNA was isolated from rice leaves using TRIzol reagent (Invitrogen). cDNA templates were synthesized using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time quantitative RT-PCR was performed on a 7500 real-time PCR system (Applied Biosystems) using SYBR Premix Ex Taq (TaKaRa) according to the manufacturer's protocol. Rice Actin1 gene was used as the endogenous control. The relative quantization method (ΔΔCT) was used to evaluate quantitative variation of replicates examined. For semiquantitative RT-PCR, PCR amplifications were performed in 50 μL volumes with the following protocol: one cycle of 94°C for 4 min, 25 to 30 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min. The gene-specific primers are listed in Supplemental Table S6.
Plasmid Construction and Rice Transformation
A full-length cDNA of DSM1 was obtained from the Knowledge-based Oryza Molecular biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/). To generate the DSM1 full-length and KD overexpression constructs, a DNA fragment containing the entire coding region or only the KD was amplified from the full-length cDNA clone by PCR. The fragments were inserted into the p1301U-FLAG vector as described previously (Sun and Zhou, 2008). To investigate DSM1 gene expression, a genomic DNA sequence corresponding to 2,100 bp upstream of the predicted ATG codon of the open reading frame was cloned into pCAMBIA-1391Z binary vector. For subcellular localization, the coding region of DSM1 cDNA was prepared by PCR, and cloned into the GATEWAY destination vector pH7WGF2,0 binary vector by recombination reaction (Invitrogen; Karimi et al., 2005). DSM1-RNAi construct was made by introducing a fragment targeting 400 bp open reading frame region of DSM1 into the pDS1301 vector (Chu et al., 2006). The constructs were introduced into japonica rice ZH11 by Agrobacterium-mediated transformation (Hiei et al., 1994). The GUS assay was performed using the histochemical staining method as described previously (Wu et al., 2003). Rice organ, tissue, or young shoots were incubated in staining buffer (50 mm sodium phosphate at pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 1 mg mL−1 of X-Gluc, 100 μg mL−1 of chloramphenicol, 1 mm potassium ferricyanide, 1 mm potassium ferrocyanide, and 20% methanol) at 37°C overnight and then washed with 70% ethanol to remove chlorophyll. GUS images were taken with a fluorescence stereomicroscope (Leica MZ FLIII). To determine the subcellular localization of DSM1, more than 20 DSM1-GFP transgenic plants were obtained. Young roots of 2-week-old transgenic plants were examined for GFP fluorescence under a Leica TCS SP2 florescence. Rice protoplast transform was performed according to the method of Zhou et al. (2009) with minor modifications (Zhou et al., 2009).
Immunoprecipitation and Kinase Assay
Rice leaves (2 g) were ground into powder in liquid nitrogen and mixed with 4 mL of protein extraction buffer (100 mm HEPES, 5 mm EDTA, 5 mm EGTA, 10 mm dithiothreitol [DTT], 10 mm Na3VO4, 10 mm NaF, 50 mm P-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, 5 pg mL−1 aprotinin, 5 pg mL−1 leupeptin, 10% [v/v] glycerol). The extract was centrifuged at 12,000 rpm for 20 min at 4°C, and the supernatant was incubated with 40 μL anti-FLAG M2 affinity gel for overnight at 4°C. The resin was washed three times with Tris-buffered saline (50 mm Tris-HCl, 150 mm NaCl, pH 7.4). Proteins were eluted with 150 mg mL−1 3× Flag peptide according the manufacturer's instructions (Sigma). Eluted proteins were subjected to SDS-PAGE and the interesting bands were analyzed by MALDI-TOF-MS.
For the kinase assay, 2 μg of purified DSM1 protein was added into 20 μL of kinase buffer (50 mm Tris-HCl, 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, 2 μg of MBP, 0.2 mm ATP, 1 μCi γ-32P-ATP). The reaction was incubated at room temperature for 30 min and terminated by the addition of 5 μL of sample buffer and heating at 100°C for 5 min. After separation on a 15% SDS-PAGE gel, the gel was dried at 60°C for 2 h. The 32P-labeled bands were detected using Kodak x-ray film.
Microarray
Two independent biological replicates of 6-week-old dsm1-1 and wild-type plants under both normal and drought-stress conditions were used for microarray experiments. For microarray analysis, the process for experiment followed the standard protocol of Affymetrix GeneChip service (CapitalBio). To find differentially expressed genes, the signal ratio of each gene between the wild type and the mutant was calculated. Genes exhibiting more than 2-fold enhanced or 2.5-fold reduced transcription level in two biological replicates are listed in Supplemental Tables S2 to S5.
Western-Blot Analysis
Total leaf proteins were extracted using extraction buffer (100 mm HEPES, 5 mm EDTA, 5 mm EGTA, 10 mm DTT, 10 mm Na3VO4, 10 mm NaF, 50 mm P-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, 5 pg mL−1 aprotinin, 5 pg mL−1 leupeptin, 10% [v/v] glycerol). The total protein was separated on 12% SDS-PAGE gel and then transferred to polyvinylidene difluoride membrane. The membrane was incubated with anti-FLAG antibody and western blot was conducted according to Hou et al. (2009).
Measurement of POX Activity, Electrolyte Leakage and Chlorophyll Content, and DAB Staining
To measure POX activity, enzymes were extracted using 0.05 m potassium phosphate buffer (pH 7.0). The homogenate extract was centrifuged at 12,000g for 15 min at 4°C. The supernatant was used as the enzyme extract. Peroxidase activity was performed according to the method of Kwak et al. (1995) with minor modifications (Kwak et al., 1995). The reaction mixture (in a total volume of 5 mL) contained the enzyme extract (0.1 mL), 0.05 m potassium phosphate buffer (pH 5.5, 2.9 mL), 2% (v/v) H2O2 (1.0 mL), and 0.05 m guaiacol (1.0 mL) as substrates. The oxidation of guaiacol was monitored and absorbance measured at 470 nm every 30 s. For electrolyte leakage, six leaves from six plants were placed in 10 mL tubes containing 6 mL of 4 μm MV. The conductivity of the MV solution was determined with an ion leakage meter (Lei ci) at the designated time. Total electrolyte leakage was measured after boiling the sample for 15 min. The percentage of electrolyte leakage caused by MV treatment was determined by the ratio of ion leakage at a given time and total ion leakage. Chlorophyll content was carried out according to the method described by Lichtenthaler (1987). For DAB staining, leaves were vacuum infiltrated with 0.1 mg mL−1 3,3′-diaminobenzidine in 50 mm Tris-acetate buffer, pH 5.0. Samples were incubated for 24 h at room temperature in the light. To remove chlorophylls, the stained samples were transferred to 100% ethanol and incubated at 100°C for 15 min.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK100319.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The dsm1-1 mutants showed increased sensitivity to salt stress (three-leaf stage plants were treated with 100 mm NaCl).
Supplemental Figure S2. Alignment of DSM1 with plants known function MAPKKKs.
Supplemental Figure S3. Identification of DSM1 KD using MALDI-TOF-MS.
Supplemental Table S1. General information about MAPK cascades T-DNA mutants.
Supplemental Table S2. Genes down-regulated in dsm1-1 mutant lines compared to wild type (DSM1/DSM1) in normal condition.
Supplemental Table S3. Genes up-regulated in dsm1-1 mutant lines compared to wild type (DSM1/DSM1) in normal condition.
Supplemental Table S4. Genes down-regulated in dsm1-1 mutant lines compared to wild type (DSM1/DSM1) in drought stress.
Supplemental Table S5. Genes up-regulated in dsm1-1 mutant lines compared to wild type (DSM1/DSM1) in drought stress.
Supplemental Table S6. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Arabi dopsis Biological Resource Center (Columbus, Ohio) for providing plasmid HBT-SGFP (Sheen et al., 1995). We also thank Lei Wang (National Center of Plant Gene Research, Huazhong Agricultural University, Wuhan) for providing plasmid 35:CFP-GHD7. We also thank Rui Li and Yihua Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for help with the rice protoplast.
This work was supported by grants from the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the National Program on the Development of Basic Research, the National Natural Science Foundation of China, and the Project from the Ministry of Agriculture of China for Transgenic Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lizhong Xiong (lizhongx@mail.hzau.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, et al (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 373–399 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983 [DOI] [PubMed] [Google Scholar]
- Banno H, Hirano K, Nakamura T, Irie K, Nomoto S, Matsumoto K, Machida Y (1993) NPK1, a tobacco gene that encodes a protein with a domain homologous to yeast BCK1, STE11, and Byr2 protein kinases. Mol Cell Biol 13 4745–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304 1494–1497 [DOI] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218 443–448 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu JK, Gong Z (2006) Mutations in ABO1/ELO2, a subunit of holo-elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol 26 6902–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 132 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittoor JM, Leach JE, White FF (1997) Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 10 861–871 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413 217–226 [DOI] [PubMed] [Google Scholar]
- Covic L, Silva NF, Lew RR (1999) Functional characterization of ARAKIN (ATMEKK1): a possible mediator in an osmotic stress response pathway in higher plants. Biochim Biophys Acta 1451 242–254 [DOI] [PubMed] [Google Scholar]
- Eichberg J, Iyer S (1996) Phosphorylation of myelin protein: recent advances. Neurochem Res 21 527–535 [DOI] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM (1991) Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol 200 38–62 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42 462–468 [DOI] [PubMed] [Google Scholar]
- Hou X, Xie K, Yao J, Qi Z, Xiong L (2009) A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc Natl Acad Sci USA 106 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33 221–233 [DOI] [PubMed] [Google Scholar]
- Ichimura K (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7 301–308 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281 36969–36976 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim J, et al (2006. a) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45 123–132 [DOI] [PubMed] [Google Scholar]
- Jeong MJ, Lee SK, Kim BG, Kwon TR, Cho WS, Park YT, Lee JO, Kwon HB, Byun MO, Park SC (2006. b) A rice (Oryza sativa L.) MAP kinase gene, OsMAPK44, is involved in response to abiotic stresses. Plant Cell Tissue Organ Cult 85 151–160 [Google Scholar]
- Jonak C, Okresz L, Bogre L, Hirt H (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5 415–424 [DOI] [PubMed] [Google Scholar]
- Jouannic S, Hamal A, Leprince AS, Tregear JW, Kreis M, Henry Y (1999) Plant MAP kinase kinase kinases structure, classification and evolution. Gene 233 1–11 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10 103–105 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441 [DOI] [PubMed] [Google Scholar]
- Kim YH, Kim CY, Song WK, Park DS, Kwon SY, Lee HS, Bang JW, Kwak SS (2008) Overexpression of sweet potato swpa4 peroxidase results in increased hydrogen peroxide production and enhances stress tolerance in tobacco. Planta 227 867–881 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Zeng W, Sheen J (1998) Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395 716–720 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Jester PJ, Gottwald JR, Sussman MR (2002) An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 14 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak KSK, Lee MS, Jung KH, Park IH, Liu JR (1995) Acidic peroxidases from suspension-cultures of sweet potato. Phytochemistry 39 981–984 [Google Scholar]
- Lagrimini LM, Bradford S, Rothstein S (1990) Peroxidase-induced wilting in transgenic tobacco plants. Plant Cell 2 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq J, Adams-Phillips LC, Zegzouti H, Jones B, Latche A, Giovannoni JJ, Pech JC, Bouzayen M (2002) LeCTR1, a tomato CTR1-like gene, demonstrates ethylene signaling ability in Arabidopsis and novel expression patterns in tomato. Plant Physiol 130 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148 350–383 [Google Scholar]
- Lin LL, Wartmann M, Lin AY, Knopf JL, Seth A, Davis RJ (1993) cPLA2 is phosphorylated and activated by MAP kinase. Cell 72 269–278 [DOI] [PubMed] [Google Scholar]
- Lin Z, Alexander L, Hackett R, Grierson D (2008) LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J 54 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kiegerl S, Hirt H (2004) OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J Biol Chem 279 26959–26966 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10 339–346 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Soukupova H, Schikora A, Zarsky V, Hirt H (2006) A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281 38697–38704 [DOI] [PubMed] [Google Scholar]
- Nishihama R, Ishikawa M, Araki S, Soyano T, Asada T, Machida Y (2001) The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev 15 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Soyano T, Ishikawa M, Araki S, Tanaka H, Asada T, Irie K, Ito M, Terada M, Banno H, et al (2002) Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109 87–99 [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) ASCORBATE AND GLUTATHIONE: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49 249–279 [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24 255–265 [DOI] [PubMed] [Google Scholar]
- Polle A (2001) Dissecting the superoxide dismutase-ascorbate-glutathione-pathway in chloroplasts by metabolic modeling: computer simulations as a step towards flux analysis. Plant Physiol 126 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga M, Guerrero C, Botella MA, Barcelo A, Amaya I, Medina MI, Alonso FJ, de Forchetti SM, Tigier H, Valpuesta V (2000) A tomato peroxidase involved in the synthesis of lignin and suberin. Plant Physiol 122 1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Overexpression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23 319–327 [DOI] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J 8 777–784 [DOI] [PubMed] [Google Scholar]
- Shou H, Bordallo P, Wang K (2004) Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J Exp Bot 55 1013–1019 [DOI] [PubMed] [Google Scholar]
- Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y (2003) NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev 17 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill TW, Ray LB (1986) Muscle proteins related to microtubule associated protein-2 are substrates for an insulin-stimulatable kinase. Biochem Biophys Res Commun 134 565–571 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105 13679–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Innes RW (2002) Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J 32 975–983 [DOI] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4 392–400 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Li X, Yuan W, Chen G, Kilian A, Li J, Xu C, Zhou DX, Wang S, Zhang Q (2003) Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35 418–427 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40 761–767 [DOI] [PubMed] [Google Scholar]
- Yang Z, Wu Y, Li Y, Ling HQ, Chu C (2009) OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol 70 219–229 [DOI] [PubMed] [Google Scholar]
- Young SA, Guo A, Guikema JA, White FF, Leach JE (1995) Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas oryzae pv oryzae. Plant Physiol 107 1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B, Xiong L, Xue W, Xing Y, Luo L, Xu C (2005) Genetic analysis for drought resistance of rice at reproductive stage in field with different types of soil. Theor Appl Genet 111 1127–1136 [DOI] [PubMed] [Google Scholar]
- Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q (2006) Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172 1213–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li S, Qian Q, Zeng D, Zhang M, Guo L, Liu X, Zhang B, Deng L, Luo G, et al (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57 446–462 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong XJ, Li DP, Gu LK, Li DQ, Liu LX, Hu XL (2009) Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 229 485–495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.