Abstract
Nodulation is the result of a mutualistic interaction between legumes and symbiotic soil bacteria (e.g. soybean [Glycine max] and Bradyrhizobium japonicum) initiated by the infection of plant root hair cells by the symbiont. Fewer than 20 plant genes involved in the nodulation process have been functionally characterized. Considering the complexity of the symbiosis, significantly more genes are likely involved. To identify genes involved in root hair cell infection, we performed a large-scale transcriptome analysis of B. japonicum-inoculated and mock-inoculated soybean root hairs using three different technologies: microarray hybridization, Illumina sequencing, and quantitative real-time reverse transcription-polymerase chain reaction. Together, a total of 1,973 soybean genes were differentially expressed with high significance during root hair infection, including orthologs of previously characterized root hair infection-related genes such as NFR5 and NIN. The regulation of 60 genes was confirmed by quantitative real-time reverse transcription-polymerase chain reaction. Our analysis also highlighted changes in the expression pattern of some homeologous and tandemly duplicated soybean genes, supporting their rapid specialization.
Nitrogen sources such as nitrate and ammonia are directly assimilated by plants. However, their low availability in soil is often a limitation to plant growth. Legume plants overcome this limitation through development of a symbiotic interaction with soil bacteria (rhizobia), which fix atmospheric dinitrogen into ammonia. The specificity of this interaction depends upon chemical recognition by both partners. Flavonoids synthesized and secreted by the plant lead to the synthesis and secretion of a bacterial chitooligosaccharide signal, termed Nod factor (NF; Oldroyd and Downie, 2008). NF is recognized by plant root hair cells, leading to rapid morphological changes (e.g. root hair cell curling followed by cortical cell division, leading to the formation of a nodule primordium) and biochemical changes (induction of NF-responsive plant genes, calcium spiking in the root hair cells, etc). The nodule primordium is the origin of the nodule, a new plant organ, where the bacteria differentiate into bacteroids and reduce atmospheric dinitrogen. In exchange, the plant provides carbon to the bacteroids.
Although not completely resolved, some genes involved in the NF signaling cascade have been identified, primarily in Lotus japonicus and Medicago truncatula, by screening for mutants affected in the root hair infection process (Oldroyd and Downie, 2008). Among them, LjNFR1, LjNFR5, and MtNFP, encoding LysM Ser/Thr receptor kinases, are considered to be NF receptors (Amor et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). Downstream of these receptors, LjCASTOR, LjPOLLUX (Kistner et al., 2005), and MtDMI1 (Catoira et al., 2000) encode ion channel proteins likely involved in intracellular calcium fluctuations, crucial to the nodulation process (Imaizumi-Anraku et al., 2005). This cascade ends with the regulation of several transcription factors, such as MtNSP1-2 and LjNSP1-2 encoding GRAS transcription factors (Catoira et al., 2000; Oldroyd and Long, 2003; Kalo et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006), LjASTRAY encoding a bZIP transcription factor (Nishimura et al., 2002), MtHAP2-1 encoding a CCAAT-binding domain transcription factor (Combier et al., 2006), and LjNIN encoding a bZIP transcription factor (Schauser et al., 1999; Marsh et al., 2007). To date, screening of L. japonicus and M. truncatula mutant libraries has resulted in the identification of approximately 20 genes critical for root hair infection and nodulation.
To further characterize genes involved in the nodulation process, gene expression studies were conducted during nodule development in L. japonicus (Colebatch et al., 2002, 2004; Kouchi et al., 2004; Asamizu et al., 2005; Høgslund et al., 2009), M. truncatula (Barnett et al., 2004; El Yahyaoui et al., 2004; Starker et al., 2006; Benedito et al., 2008), and soybean (Glycine max; Lee et al., 2004; Brechenmacher et al., 2008; Libault et al., 2009). However, the understanding of gene regulation during the earliest steps of the plant-symbiont interaction, especially in root hair cells, is lacking. To our knowledge, Lohar et al. (2006) and Høgslund et al. (2009) performed the only large-scale transcriptomic analysis focusing on the first hours of rhizobial infection of M. truncatula and L. japonicus roots. This analysis identified genes involved in root hair swelling and branching as well as root hair curling, cortical cell division, and nodule primordial development. A significant limitation of this analysis was the use of whole roots, leading to a dilution of root hair tissues, where the primary response to infection occurs.
To improve our knowledge of genetic regulation during root hair infection, we analyzed gene expression in isolated soybean root hairs mock inoculated or inoculated with B. japonicum. The recent sequencing of the soybean genome (http://www.phytozome.net/soybean; Schmutz et al., 2010) and the development of methodology to isolate soybean root hair cells (Wan et al., 2005) were essential developments that enabled this research. Using a combination of large-scale microarray and sequencing strategies, we analyzed the soybean root hair cell transcriptome in response to B. japonicum inoculation and then confirmed these analyses for selected genes using quantitative real-time reverse transcription (qRT)-PCR. The results identify genes specifically regulated during the root hair cell infection process. In addition, the results provide interesting insight into the evolution of genes specifically involved in the nodulation process.
RESULTS AND DISCUSSION
DNA Microarray and Sequence-Based Transcriptomic Analysis of Soybean Root Hair Infection by B. japonicum: an Overview
At the time of this study, the available Affymetrix soybean DNA microarray consisted of 37,641 features derived from over 350,000 EST sequences that were available at the time of the design. The recent release of the soybean genome sequence and prediction of associated genes enabled reexamination of the content and updated curation of this array. Accordingly, we blasted each probe set against the predicted soybean cDNA sequences (http://www.phytozome.net/soybean). A total of 34,015 probe sets (90.5%) matched against soybean cDNA sequences, while 3,578 (9.5%) did not (Supplemental Fig. S1; Supplemental Table S1). These 34,015 probe sets were divided into three categories, associated with one (13,949, 37.1%), two (16,509, 43.9%), and three or more (3,557, 9.5%) predicted soybean genes (Supplemental Fig. S1; Supplemental Table S1). The first category of probe sets targets 11,426 soybean genes (i.e. several probe sets match with the same gene). The second and third categories of probe sets target 22,568 and 10,915 soybean genes, respectively. In retrospect, given the duplicated nature of the soybean genome (Schlueter et al., 2004, 2007), this finding was not surprising. We looked at the chromosome location of pairs of genes matching the same probe set and identified blocks of homeologous genes based on the gene content, position, and orientation on different chromosome fragments (Supplemental Table S2). Furthermore, we documented the existence of 282 tandemly duplicated genes (Supplemental Table S3). The 3,557 probes matching three or more annotated soybean genes most likely represent conserved regions of large gene families. For example, the Gma.10755.1.S1_at probe set is predicted to cross-hybridize to 36 different predicted soybean genes, all encoding an anaphase-promoting complex subunit 10 (Supplemental Table S1).
The remaining 3,578 probe sets (9.5%), which do not match predicted soybean cDNA sequences, call into question the nature of the EST sequences used to design the probe sets. Initially, we hypothesized that some of these probe sets targeted transcripts not predicted in the soybean genome annotation (e.g. splicing variants of known genes or transcripts of unannotated genes). To verify this hypothesis, we blasted the 3,578 probe sets against the soybean genome sequence (Supplemental Fig. S1; Supplemental Table S4). Interestingly, 2,662 probe sets returned matches from the soybean genome, while 916 did not. Among the 2,662 probe sets, 1,098 probe sets matched regions of the genome where no genes are predicted. The remaining 1,564 probes matched intron regions from 2,902 genes, supporting the existence of additional transcript variants for these genes. Among them, 1,050 probe sets matched only intron sequences of predicted genes (648 probe sets match the intronic region of one gene only [allowing quantification of the expression of 604 genes], and 402 match the intronic region of two or more genes). The remaining 514 probe sets matched both introns of predicted genes and genomic regions with no predicted genes. Altogether, we identified 1,612 Affymetrix probe sets (1,098 plus 514 probe sets) targeting the soybean genome where no genes are currently predicted (Supplemental Fig. S1). The absence of soybean sequences targeted by the remaining 916 probe sets (2.4% of the total soybean probe sets) remained problematic. Our analysis suggested that these EST sequences might not be soybean ESTs but were isolated from other organisms that were contaminating the soybean samples originally used for mRNA isolation. To verify this hypothesis, we blasted each probe of the 916 probe sets against the nucleotide and EST databases available on the National Center for Biotechnology Information. As expected, most of these probe sequences matched against plant virus, fungal, and bacterial sequences (data not shown). Altogether, although the Affymetrix array enables the quantification of a large number of soybean genes (36,486 out of the 66,153 predicted [55%]), the expression of only a subset of these (11,807, 17.8%) can be accurately quantified. The remainder of the array features map to two or more soybean genes.
Given the limitation of the current soybean Affymetrix DNA microarray platform, we decided to also employ Illumina transcriptome sequencing to obtain better coverage of the transcriptome by pooling total RNA isolated from three independent biological replications. In this analysis, we focused on total RNA isolated from mock-inoculated and B. japonicum-infected root hair cells harvested at 12, 24, and 48 h after inoculation (HAI). Similarly, a pool of total RNA extracted from stripped roots (i.e. roots devoid of root hairs) harvested at 48 HAI with B. japonicum were also sequenced. For each condition tested, 4.88 to 6.84 million 36-bp reads were generated, of which 63.2% to 75.6% aligned with the soybean genome (Table I). These reads were divided into three different groups: highly repetitive reads (i.e. matching against more than five loci), nonunique reads (i.e. matching against two to five loci), and unique reads (i.e. matching against only one soybean locus). The highly repetitive and nonunique read categories represented, on average, less than 0.62% and 20.5% of the total soybean reads. Interestingly, the nonunique read category mapped preferentially to homeologous or recently duplicated genes. The third category of reads represented 63.6% of the total reads. Since these reads reflect expression from a single, unique soybean gene, they were used to accurately and independently quantify the expression of each soybean gene. The expression of 69,145 soybean genes, including 2,982 transposable elements, was quantified by Illumina transcriptome sequencing. Transcripts of 48,281 predicted genes (69.8%) were detected in at least one out of the seven conditions tested. Transcripts of the remaining 20,864 genes (30.2%) were not detected, suggesting that these genes were not expressed or were expressed at levels too low to be detected (Supplemental Table S5). Compared with the 11,807 predicted genes uniquely quantifiable by Affymetrix array hybridization, Illumina transcriptome technology allowed a more thorough assessment of the soybean root hair cell transcriptome. In addition, when Affymetrix hybridization was not able to clearly establish the absence of transcripts due to the residual background hybridization, deep transcriptome sequencing allowed a clear distinction between low-abundance and undetectable transcripts (Supplemental Table S5).
Table I.
Distribution of Illumina-Solexa 36-bp reads according to their alignment against soybean and B. japonicum genomes
| Sample | Soybean Unique | Soybean Nonunique | Soybean Repetitive | Unaligned | Total Reads |
|---|---|---|---|---|---|
| 12-HAI inoculated root hair | 2,397,650 | 662,769 | 23,727 | 1,798,189 | 4,882,335 |
| 12-HAI mock-inoculated root hair | 2,828,246 | 719,626 | 38,901 | 2,024,736 | 5,611,509 |
| 24-HAI inoculated root hair | 3,527,919 | 855,962 | 22,741 | 1,787,847 | 6,194,469 |
| 24-HAI mock-inoculated root hair | 3,612,912 | 988,558 | 25,973 | 2,199,529 | 6,826,972 |
| 48-HAI inoculated root hair | 3,219,116 | 821,617 | 14,848 | 1,788,904 | 5,844,485 |
| 48-HAI mock-inoculated root hair | 4,086,965 | 1,052,457 | 28,314 | 1,670,473 | 6,838,209 |
| 48-HAI stripped root | 3,855,751 | 1,006,813 | 22,786 | 1,767,904 | 6,653,254 |
During our investigation, we detected expression from coalescent genes (i.e. pairs of genes predicted on both the Watson and Crick DNA strands at the same chromosome location), leading to an increase in the complexity of our analysis (Supplemental Table S5). For example, among the 69,145 predicted genes, 198 genes, forming 97 groups, were coalescent, preventing a clear assignment of the direction of gene transcription (Supplemental Table S6). Mapping of Illumina reads highlighted a fact previously suggested by the Affymetrix probe set analysis: that is, a significant number of Illumina reads matched 6,565 soybean genome loci where no genes were predicted, strongly suggesting the existence of additional transcripts representing unannotated genes (Supplemental Table S7). We also compared the locations on the genome of the 6,565 transcribed loci identified by sequencing to the 1,612 Affymetrix probe sets matching the soybean genome but not an annotated soybean gene. Among these 1,612 probe sets, 492 matched loci where no genes were predicted but where transcripts were detected based on sequencing data. The existence of EST sequences used to design these probe sets and the detection of transcripts based on RNA sequence analysis strongly support the existence of currently unannotated genes in the soybean genome. As an initial effort to validate this analysis, we identified a portion of the 5′ untranslated region of the Glyma01g03890 gene (Supplemental Fig. S2). The Gm01:3359551 to Gm01:3359812 Illumina coverage region was localized 4 bp upstream of the start codon of Glyma01g03890 (Glyma01g03890 ATG: Gm01-3359816). By PCR and sequencing, we showed that the Gm01:3359551 to Gm01:3359812 fragments were part of the Glyma01g0389 transcript, extending the Glyma01g03890 5′ untranslated region by 261 bp. These results show the value of the Illumina sequencing to improve and update the soybean genome annotation.
B. japonicum Infection of Soybean Root Hair Cells Strongly Affects the Soybean Root Hair Cell Transcriptome
Affymetrix arrays were used to identify soybean genes regulated during root hair cell infection by comparing the transcriptome of B. japonicum-inoculated and mock-inoculated root hairs 6, 12, 18, 24, 36, and 48 HAI. Three independent biological replications were produced for each time point and condition. The differential hybridization for each of the 37,641 probe sets is reported in Supplemental Table S8. Performing a stringent analysis of these data (log2[fold change] > 1.6 or < −1.6 and false discovery rate [FDR] P < 0.01), 173 Affymetrix probe sets showed significant differential hybridization for at least one time point of the time course (Supplemental Table S9). Based on these results, the transcriptional response of soybean root hair cells to B. japonicum inoculation was maximal at 12 and 18 HAI (Supplemental Table S9). Of the 173 Affymetrix probe sets differentially hybridized during root hair infection, 171 targeted the soybean genome (98.9%), while two probe sets showing differential hybridization did not match the soybean genome. Only 81 probe sets targeted a single soybean gene, allowing the accurate quantification of the expression of 73 genes (Supplemental Table S10). Of these 73 genes, 40 were significantly regulated at several time points, reflecting an important role for these genes during infection.
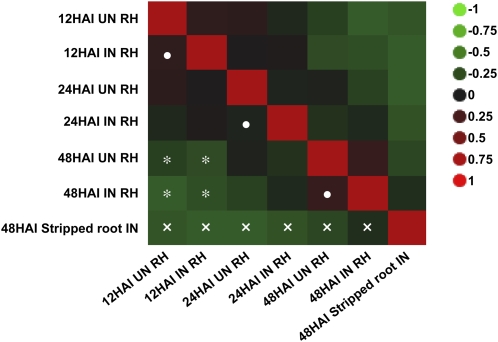
To better investigate changes in the soybean root hair cell transcriptome occurring after B. japonicum inoculation, we compared the expression of soybean genes between inoculated and mock-inoculated root hairs isolated 12, 24, and 48 HAI using Illumina transcriptome sequencing. In addition, we also included 48-HAI stripped root. Gene expression patterns were significantly different between root hair and stripped root samples (Fig. 1, crosses). In addition, changes in soybean gene expression patterns were more dependent on root hair developmental stage (e.g. low correlation between 48- and 12-HAI root hair transcriptome independent of inoculation by B. japonicum; Fig. 1, asterisks) than to the response to B. japonicum inoculation (Fig. 1, dots indicating correlation of developmental stage independent of inoculation state).
Figure 1.
Comparison of the expression of soybean genes in root hairs (RH) and stripped roots inoculated (IN) and mock-inoculated (UN) with B. japonicum at 12, 24, and 48 HAI. Ward hierarchal clustering of log2-transformed gene distribution in inoculated and uninoculated root samples was based on Pearson correlation calculated between numeric variables, principal components of the correlation matrix, and outlier analysis. The color scale indicates the degree of correlation (green, low correlation; red, strong correlation). Dots, asterisks, and crosses highlighted changes in the transcriptome of root hair cells based on B. japonicum inoculation, root hair developmental stage, and tissues, respectively. The heat map was generated using JMP Genomics 4.0. [See online article for color version of this figure.]
To identify the genes regulated during root hair infection through transcriptome sequencing, we combined a binomial test (P < 0.05) and a 3-fold change cutoff of the gene expression levels between inoculated and mock-inoculated conditions (log2[fold change]IN/UN < −1.6 or > 1.6]. Considering all time points and taking into account the presence of coalescent genes, transcriptome sequence analysis allowed the identification of 1,949 predicted genes regulated during root hair infection (Supplemental Table S11). A total of 1,056 and 870 genes were up- and down-regulated, respectively, while the expression of 23 genes was up-regulated at one time point and then down-regulated at another time point of the time course. Interestingly, transcripts of 41 of the 1,949 up-regulated genes were absent in mock-inoculated root hair cells, indicating that these genes respond specifically to B. japonicum inoculation (Supplemental Table S12). Among them, some were predicted to be involved in the plant defense response, modification of cell wall composition, signal transduction, and basic metabolic processes. Conversely, none of the remaining 1,908 genes was expressed in mock-inoculated root hairs or in inoculated root hairs. By combining Affymetrix and Illumina transcriptome analyses, a total of 1,973 genes were identified that were significantly regulated (49 genes were found regulated by both platforms, 24 by Affymetrix array only, and 1,900 by Illumina sequencing only).
Several L. japonicus and M. truncatula genes have known roles in root hair invasion by symbiotic bacteria (Oldroyd and Downie, 2008). We hypothesized that soybean homologs to these genes might be regulated in response to B. japonicum infection. Recently, a total of 100 soybean genes were identified based on their homology to previously characterized L. japonicus and M. truncatula genes known to be involved in the nodulation process (Schmutz et al., 2010). Using this resource, we identified 10 and three genes that were up- and down-regulated during the root hair infection process (Supplemental Table S13). In addition, Glyma11g13390 (homolog of LjNIN2; Schauser et al., 2005) showed a switch in its induction during root hair infection from up- to down-regulated. The identification of these known nodulation genes responsive to B. japonicum inoculation in our data set validates our initial approach to utilize transcriptomic analysis to identify key soybean nodulation genes.
In addition, 415 soybean sequences not predicted to encode proteins were also regulated during root hair infection (Supplemental Table S11). Presumably, some of these sequences represent genes not predicted by the current soybean genome annotation. Among them, four sequences were repetitively regulated during root hair infection at the three time points of the time course. They were located on chromosomes Gm02(1820937–1821450), Gm05(12241696–12241991), Gm10(43271119–43271407), and Gm17(1320036–1320705). We annotated these genomic sequences using FGENESH on 20 kb surrounding each sequence. This method failed to predict an open reading frame around Gm10(43271119–43271407). The three remaining unannotated sequences were predicted to encode 628-, 538-, and 340-amino acid proteins, respectively. Using InterProScan, we predicted that these putative genes encode a WRKY transcription factor, an ankyrin repeat region protein, and a tRNA Δ(2)-isopentenylpyrophosphate transferase, respectively. Comparing these predictions with the currently annotated soybean genes, we identified Gm02(1820937–1821450), Gm05(12241696–12241991), and Gm17(1320036–1320705), which led to the extension of the predicted cDNA sequences of Glyma02g02430, Glyma05g12090-Glyma05g12100, and Glyma17g02080 genes. These results further support the ability to improve genome annotation via the use of global expression data.
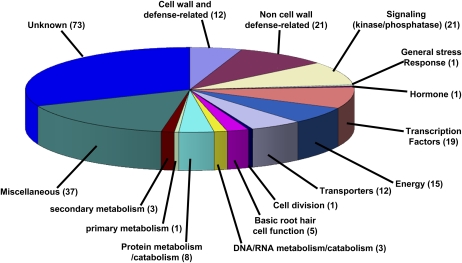
Among the 1,949 regulated soybean genes identified by Solexa sequencing, 219 were regulated at several time points of the time course (189 and 30 genes are up- and down-regulated, respectively). Combined with Affymetrix array analyses, a total of 233 genes were consistently regulated across time points during root hair infection (193 by Illumina, 14 by Affymetrix array hybridization alone, 26 by both platforms). Among them, 11 array analysis genes consistently regulated during root hair infection were also identified by Illumina sequencing as regulated during root hair infection but only at one time point. These 233 genes are promising candidates for further investigation of the root hair infection process in soybean (Supplemental Table S14). Among them, 26 were expressed only when root hair cells were inoculated by B. japonicum (Supplemental Table S12). Consequently, we decided to classify the 233 genes according to their putative functions (Fig. 2). First, known protein domains were not identified for 73 of these genes (32%). Thirty-three genes (14%) are predicted to play a role in the plant defense response (e.g. cell wall and plant defense-related genes). Interestingly, after a strong initial induction of their expression at 12 and 24 HAI, expression of most of these genes decreased by 48 HAI, suggesting that B. japonicum may have the ability to suppress an initial plant defense response that could be detrimental to the infection process. Our observation confirms previous studies in soybean, L. japonicus, and M. truncatula documenting the increased abundance of defense-related transcripts specifically during the early stages of nodulation (Colebatch et al., 2002, 2004; El Yahyaoui et al., 2004; Kouchi et al., 2004; Lohar et al., 2006; Brechenmacher et al., 2008). In addition, 42 genes (18%) were identified as part of signaling cascades (e.g. kinase/phosphatase/signaling-related genes, stress response-related genes, hormone-related genes, and transcription factor genes). Based on previous studies describing the role of kinase/phosphatase/signaling-related genes in root hair infection (e.g. CCaMK [Gleason et al., 2006; Tirichine et al., 2006]), we hypothesized that these genes might be important to trigger the plant cell responses to B. japonicum inoculation by activating the expression of several genes necessary for subsequent steps of root hair infection (e.g. calcium spiking, root hair deformation).
Figure 2.
Distribution of soybean genes regulated during root hair infection according to their function. A putative function was assigned to all soybean genes regulated during root hair infection by B. japonicum. Gene annotation was performed based on the identification of conserved amino acid domains (PFAM, KOG, and PANTHER domain predictions). Accordingly, these genes were classified into 16 different groups (Supplemental Table S14).
To further refine the root hair-specific transcriptome, we compared inoculated root hair cells (48-HAI inoculated root hair cells) and inoculated stripped roots (roots devoid of root hair cells isolated 48 HAI). A total of 7,236 predicted genes and 563 unannotated sequences were differentially regulated (binomial test P < 0.05 and log2[Δfold change] < −1.6 or > 1.6; Supplemental Table S15). These genes reflect both the specialization of root hair cells and also the differential responses of root hairs and root cells to the B. japonicum infection process (i.e. root hair cell curling and calcium spiking, induction of root cortex cell division). Among the 233 genes regulated at several stages of the root hair infection, 94 were also differentially expressed when comparing isolated root hairs with stripped roots, indicating that they play a key role specifically in the root hair cell infection. The remaining 139 genes were regulated in both root hair cells and other root cells, suggesting that these genes might have a dual role in response to B. japonicum inoculation; that is, during root hair infection and in further steps of the infection occurring in the root.
Differential Expression of Homeologous and Tandemly Duplicated Genes during Root Hair Infection Supports Functional Specialization of Duplicated Genes
The BLAST analysis of the Affymetrix probe sets allowed the identification of blocks of soybean homeologous genes based on gene content, order, and orientation. Although these analyses cannot identify all homeologies existing in soybean, this represents a first step in the analysis of soybean genome architecture. Utilizing this analysis and the extensive transcriptomic analysis, we compared the expression patterns of homeologous genes in inoculated root hair cells and stripped roots and in mock-inoculated root hair cells. This led to a specific focus on five homeologous regions representing 430 genes based on transcriptome sequencing (Supplemental Table S2).
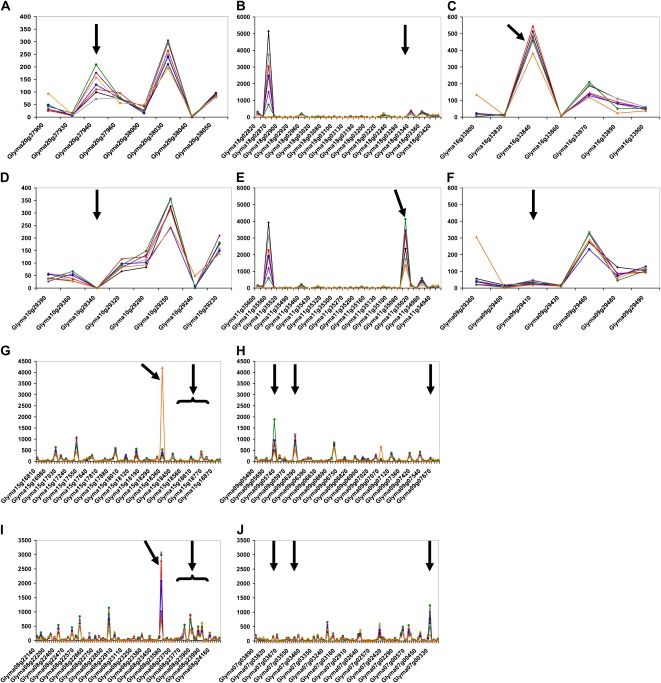
This analysis revealed a strong overall conservation of gene expression patterns in the homeologous regions after gene duplication in terms of gene expression level and tissue specificity (Fig. 3). Therefore, we can assume that key promoter elements were conserved during genome duplication. However, gene expression varied considerably when specific homeologous genes pairs were compared. These differences may reflect the silencing or activation of one of the two homeologous genes and, in some cases, might lead to the specialization of one of the two homeologous genes in soybean (Fig. 3, black arrows). Similarly, we also analyzed the expression of tandemly duplicated soybean genes in the seven conditions tested. Although gene expression is typically conserved, some duplicated genes showed dramatic changes in their expression, suggesting a rapid differentiation of their promoter sequences during evolution (Supplemental Table S16). Clearly, the soybean genome duplication not only increased the size of the gene pool but also facilitated the specialization of homeologs, resulting in greater transcriptional plasticity.
Figure 3.
Expression of soybean homeologs in B. japonicum-inoculated and uninoculated root hair cells and in inoculated stripped roots. Expression of homeologous genes was quantified by ultra-high-throughput sequencing after normalization of the read number by the total number of soybean reads (y axis) on five blocks of homeologous genes (x axis; A–D, B–E, C–F, G–I, and H–J). Expression was analyzed in seven different conditions: 12-HAI inoculated (gray squares) and uninoculated (black diamonds) root hair cells, 24-HAI inoculated (blue circles) and uninoculated (red triangles) root hair cells, 48-HAI inoculated (green circles) and uninoculated (purple diamonds) root hair cells, and 48-HAI inoculated stripped roots (orange triangles). Black arrows highlight homeologous genes differentially expressed.
Sixty Genes, Including Soybean NFR5, NSP1, NSP2, and NIN Homeologs, Are Regulated during Soybean Root Hair Infection
To confirm the regulation of some of the 1,973 regulated soybean genes and unannotated sequences identified during root hair infection, we analyzed their expression by qRT-PCR. Primer sets were carefully designed to ensure maximum specificity despite the soybean genome duplication. A second preparation of inoculated and mock-inoculated root hair cells was produced to confirm the reproducibility of the plant tissues harvested (0, 3, 6, 12, 18, 24, 36, 48, and 72 HAI). The expression of ENOD40, a gene described to be highly expressed during nodulation (Minami et al., 1996), was quantified at each time point and used as a positive control. Gene expression was normalized against Cons6 expression, one of the reference genes identified in soybean (Libault et al., 2008).
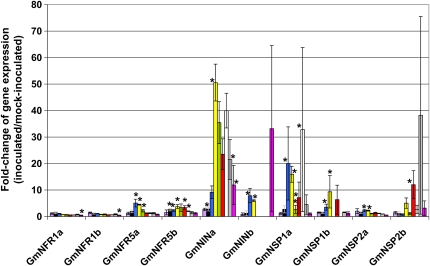
Using Illumina sequencing, we identified homologs to LjNIN-PsSYM35 (Glyma04g00210; GmNINa), LjNSP1 (Glyma16g01020; GmNSP1a), and LjNFR5-MtNFP-PsSYM1 (Glyma11g06740; GmNFR5a) up-regulated in response to B. japonicum inoculation. While induction of GmNINa and GmNSP1a was previously reported in inoculated L. japonicus and M. truncatula roots (Heckmann et al., 2006; Marsh et al., 2007), no induction of NFR5 genes was reported in other legume models. By qRT-PCR, we confirmed the induction of GmNINa, GmNSP1a, and GmNFR5a during root hair infection (Fig. 4; Supplemental Table S17). Moreover, GmNINb, GmNSP1b, GmNFR5b, and GmNSP2a and GmNSP2b were induced during root hair infection. In addition, we also quantified the expression of NFR1 and NSP2 homologous soybean genes (GmNFR1a and GmNFR1b, GmNSP2a and GmNSP2b) due to their key role in the cascade leading to the infection of root hairs by symbiotic bacteria (Radutoiu et al., 2003; Hirsch et al., 2009). GmNFR1a and GmNFR1b expression was slightly repressed only at 72 HAI, while GmNSP2a and GmNSP2b expression was slightly induced. These observations confirmed the lack of regulation of L. japonicus NFR1 gene during nodulation (Radutoiu et al., 2003).
Figure 4.
Regulation of the expression of soybean homologs of M. truncatula and L. japonicus nodulation-related genes. After qRT-PCR, the fold change of the expression of GmNFR1a, GmNFR1b, GmNFR5a, GmNFR5b, GmNINa, GmNINb, GmNSP1a, GmNSP1b, GmNSP2a, and GmNSP2b between inoculated and mock-inoculated root hair cells was calculated (y axis). These comparisons were performed on root hair cells isolated 0 (purple), 3 (black), 6 (blue), 12 (yellow), 18 (green), 24 (red), 36 (white), 48 (gray), and 72 (pink) HAI. Significant differences across three independent replicates based on a Student's t test (P < 0.05) are highlighted with asterisks. The largest error bars are due to the very low expression of some genes in the mock-inoculated samples, leading to a lack of detection of a qRT-PCR product in some replicates.
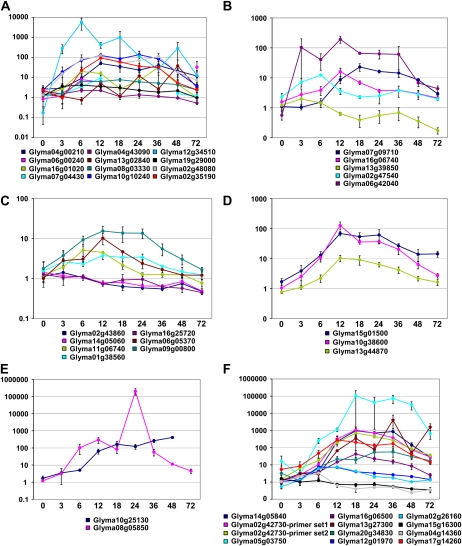
We also confirmed the regulation of 46 other genes, leading to a total of 56 predicted soybean genes during root hair infection (Supplemental Table S17). Overall, a strong regulation of their expression was observed mainly between 12 and 24 HAI, followed by a decrease of this induction at 48 to 72 HAI (Fig. 5). After classifying these genes according to their putative functions, it appeared that each gene group was induced at a specific time after B. japonicum inoculation. For example, genes encoding putative transcription factors and transporters were induced as soon as 3 to 12 HAI (e.g. Glyma12g34510 and Glyma10g10240 encoding two CCAAT-binding transcription factors [Glyma10g10240 is ortholog to MtHAP2-1; Libault et al., 2009] and Glyma02g47540 encoding a calcium transporting ATPase [Fig. 5]). Later, maximum induction of genes involved in signal cascades, gibberellin biosynthesis, and root hair restructuring was observed from 6 to 24 HAI. Gibberellin was described as an important phytohormone in controlling root hair infection and nodulation in Pisum sativum, Sesbania rostrata, and L. japonicus (Ferguson et al., 2005; Lievens et al., 2005; Maekawa et al., 2009). In legumes, cell wall composition is modified during nodulation (Brewin, 2004), and an extensin was also described to play a role in root hair cell deformation. The identification of one soybean extensin (Glyma10g25130) strongly induced during the root hair infection time course is consistent with the changes in root hair morphology during the infection process (i.e. curling, branching, and swelling). Finally, as mentioned above, plant defense system gene expression was induced between 12 and 48 HAI, followed by a decrease in the expression of these genes by 48 and 72 HAI. In addition, we confirmed the regulation of the currently unannotated soybean sequences [e.g. Gm02(1820937–1821450), Gm05(12241696–12241991), Gm10(43271119–43271407), and Gm17(1320036–1320705)] by qRT-PCR.
Figure 5.
Classes of soybean genes differentially regulated at different times following B. japonicum inoculation. Gene expression fold change (y axis) in response to B. japonicum inoculation was investigated in six groups of genes (A, transcription factors; B, transporters; C, signal transduction; D, gibberellin biosynthesis; E, control of root hair shape/elongation; F, plant defense system) across a time course of 0 to 72 HAI (x axis). Details are available in Supplemental Table S17.
In summary, our data support a rapid transcriptional response by soybean root hair cells upon inoculation with B. japonicum, suggesting that the signal cascade following NF recognition leads to a rapid, global change in the root hair cell transcriptome. Interestingly, soybean gene expression is modified chronologically. Initially, transcription factors are up-regulated, followed by downstream targets of signal cascades such as gibberellin biosynthesis-related and root hair deformation-related genes involved in the infection process.
CONCLUSION
In this study, we highlighted the limits and usefulness of the soybean Affymetrix array and took advantage of the Illumina Solexa sequencing platform to dissect the perturbation of the soybean root hair transcriptome in response to mutualistic bacterial infection. Our analysis allowed us to measure changes in the transcriptome of a single cell in response to microbial infection. Our data indicated that roughly 2,000 soybean root hair cell genes respond to B. japonicum inoculation. These genes now provide important targets for further investigation of the initial steps in B. japonicum infection of soybean leading to symbiotic nitrogen fixation.
MATERIALS AND METHODS
Bacterial Culture
Bradyrhizobium japonicum USDA110 was grown at 30°C in HM medium (Cole and Elkan, 1973) supplemented with yeast extract (0.025%), d-Ara (0.1%), and chloramphenicol (0.004%). After 3 d, B. japonicum cells were pelleted by centrifugation (4,000 rpm, 10 min), washed with sterile water three times, and diluted in water to an optical density at 600 nm = 0.8.
Plant Treatment
Soybean (Glycine max) seeds were surface sterilized according to Wan et al. (2005) and then placed on nitrogen-free B&D agar medium (Broughton and Dilworth, 1971). After 3 d of germination in growth chambers (dark conditions, 80% humidity, 27°C), a B. japonicum cell suspension or water (mock inoculation) was sprayed on approximately 2,000 seedlings growing on B&D agar medium. Treated seedlings were returned to the original growth chamber for incubation. Root hair cells were isolated according to Wan et al. (2005) and stored at −80°C.
Three sets of root hairs were isolated (two for Affymetrix/Illumina analyses [i.e. the first set was group 6, 12, and 18 HAI and the second set was group 24, 36, and 48 HAI] and one for qRT-PCR analyses [0–72 HAI]). For each set, at least three independent biological replicates were produced to ensure the reproducibility of the plant tissues analyzed. A single time point and biological replicate were produced per sowing.
Affymetrix Array Hybridization and Analysis
Total RNA was used for Affymetrix Soybean Genome GeneChip analysis following the manufacturer's recommended protocols (Affymetrix). Fragmentation of biotin-labeled copy RNA was confirmed before hybridization by running samples on the Agilent 2100 Bioanalyzer using an RNA Nano Chip. Microarray hybridizations were performed at the Keck Center for Comparative and Functional Genomics following the manufacturer's recommended protocols. The microarrays were scanned with a GeneChip model 3000 7G Plus high-resolution scanner. Individual scans were analyzed with the MAS5 algorithm in Expression Console version 1.1 of the Affymetrix GeneChip Command Console suite and quality checked for the presence of housekeeping and control genes, background, and other quality-control indicator values.
Because the 6-, 12-, and 18-HAI time points were done in one experiment and the 24-, 36-, and 48-HAI time points were done in a separate experiment, these array sets were normalized and analyzed separately. Comparisons were made only between the infected and mock-infected samples at each time point. No direct statistical analysis was made between samples of the different experiments. The 23,300 probe sets on the array that were designed for two common soybean pathogens were removed prior to all data processing using R (R Development Core Team, 2009) and Bioconductor (Gentleman et al., 2004). Data preprocessing was done separately for each experiment using the GCRMA algorithm (Wu et al., 2004, Wu and Irizarry, 2005). Each experiment was fit with an ANOVA model with effects for infection treatment and time using the Limma package (Smyth, 2005) that uses an empirical Bayes correction (Smyth, 2004) to improve power by borrowing information across probe sets. Contrasts between the infected and mock-infected treatments at each time point were pulled from the models, and the FDR method (Benjamini and Hochberg, 1995) was used to correct the raw P values for multiple hypothesis testing. The experiment with the 6-, 12-, and 18-HAI time points had an additional infection group (data not shown), but only the results of the B. japonicum infection versus mock-infected samples are reported here.
Illumina Sample Preparation, Read Alignment, and Statistical Analysis
mRNAs isolated from 12-, 24-, and 48-HAI inoculated and mock-inoculated soybean root hair cells were reverse transcribed using standard protocols. After first- and second-strand cDNA synthesis, the cDNAs were end repaired preliminary to the ligation of Illumina adaptors. The products were sequenced on an Illumina platform.
Illumina Genome Analyzer II image data were base called and quality filtered using the default filtering parameters of the Illumina GA Pipeline GERALD stage (Illumina). Alignments of passing 36-nucleotide reads to the Glyma1 8× Soybean Genome assembly (Soybean Genome Project, Department of Energy Joint Genome Institute) were performed using GSNAP (T. Wu, personal communication), an alignment program derived from GMAP (Wu and Watanabe, 2005) with optimizations for aligning short transcript reads to genomic reference sequences. Alignments were processed using the Alpheus pipeline (Miller et al., 2008), keeping only alignments that had at least 34 out of 36 identities and had no more than five equivalent best hits. Read counts used in expression analyses were based on the subset of uniquely aligned reads that also overlapped the genomic spans of the Glyma1 and MAGE gene predictions in the case of soybean and B. japonicum, respectively.
Illumina statistical analysis was performed as described previously (Miller et al., 2008) using JMP Genomics 3.2 (SAS). Briefly, soybean and B. japonicum data were normalized by multiplying each read number in a treatment by the ratio of the whole-experiment average to the treatment average. The data were exported into SAS JMP Genomics format. Expression data (reads) were log2 transformed and filtered for 2-fold differences in expression at the various time points (12, 24, and 48 h) for inoculated versus mock-inoculated samples. Venn diagrams were made comparing differently expressed genes at the various time points. The selected 2-fold difference genes were selected, and log2 expression data were used to create hierarchical cluster heat maps.
qRT-PCR
Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer's instructions and subsequently purified using chloroform extraction. Purified RNA was treated with TURBO DNase (Ambion) to remove any contaminating genomic DNA according to the manufacturer's instruction. One to 2.5 μg of DNA-free RNA was used with oligo(dT) for first-strand cDNA synthesis using the Moloney murine leukemia virus for RT-PCR (Promega) according to the manufacturer's instruction. The lack of genomic DNA contamination was verified by qRT-PCR using primers able to amplify genomic DNA.
Primers used for qRT-PCR were designed using primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) with the following criteria: melting temperature of 60°C, PCR amplicon lengths of 80 to 120 bp, and yielding primer sequences with lengths of 19 to 23 nucleotides with an optimum at 21 nucleotides and guanine-cytosine contents of 40% to 60%. The list of the primers used in this study is available in Supplemental Table S17.
qRT-PCR was performed as described by Libault et al. (2008) using Cons6 to normalize the expression levels of GmFWL genes (i.e. the cycle threshold [Ct] value of the reference genes was subtracted from the Ct values of the genes analyzed [ΔCt]). Expression levels (E) of each gene were calculated according to the equation E = Peff(−ΔCt), where Peff is the primer set efficiency calculated using LinRegPCR (Ramakers et al., 2003). To calculate fold changes, the ratios of the expression levels between inoculated and mock-inoculated plants were calculated at the different time points of the time course and for the three different biological replicates. The average ratio between the three different replicates is represented, and a Student's t test between the three inoculated expression levels on the one side and the three mock-inoculated expression levels on the other side was also calculated at each time point of the time course to statistically validate the differences.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Affymetrix probe set distribution according to specificity for soybean predicted cDNA and genomic DNA sequences.
Supplemental Figure S2. Confirmation of the identification of the 5′ untranslated region of Glyma01g03890.
Supplemental Table S1. Soybean genes represented on the soybean Affymetrix array.
Supplemental Table S2. Gene expression of predicted soybean genes found in five homeologous regions.
Supplemental Table S3. Identification of tandemly duplicated genes hybridizing with the same Affymetrix probe sets.
Supplemental Table S4. Soybean genomic DNA sequences hybridizing against the Affymetrix probe sets that do not match against a predicted gene sequence.
Supplemental Table S5. Expression levels (i.e. normalized Illumina reads) of soybean genes in B. japonicum-inoculated and mock-inoculated root hair cells and stripped roots.
Supplemental Table S6. Expression levels (i.e. normalized Illumina reads) of soybean coalescent genes in B. japonicum-inoculated and mock-inoculated root hair cells and stripped roots.
Supplemental Table S7. Expression levels (i.e. normalized Illumina reads) of unannotated soybean sequences in B. japonicum-inoculated and mock-inoculated root hair cells and stripped roots.
Supplemental Table S8. Soybean Affymetrix array hybridization analysis using inoculated and mock-inoculated root hair cells isolated 6 to 48 HAI.
Supplemental Table S9. Affymetrix probe sets showing a significant differential hybridization (log2[fold change] > 1.585 or < −1.585 and FDR P < 0.01).
Supplemental Table S10. Soybean genes quantified accurately by Affymetrix array hybridization and showing a significant differential hybridization (log2[fold change] > 1.585 or < −1.585 and FDR P < 0.01).
Supplemental Table S11. Soybean annotated and unannotated sequences significantly and differentially expressed in root hair cells in response to B. japonicum inoculation.
Supplemental Table S12. Soybean predicted genes expressed exclusively when root hair cells were inoculated with B. japonicum.
Supplemental Table S13. Regulation of gene expression during root hair infection for soybean genes known to be involved in nodulation or homologous to L. japonicus and M. truncatula nodulation-related genes.
Supplemental Table S14. Soybean predicted genes consistently regulated across the root hair-B. japonicum inoculation time course based on Illumina sequencing and Affymetrix array hybridization.
Supplemental Table S15. Soybean annotated and unannotated sequences significantly and differentially expressed between inoculated root hairs and stripped roots.
Supplemental Table S16. Expression pattern of soybean tandemly duplicated genes in B. japonicum-inoculated and mock- inoculated root hair cells and stripped roots.
Supplemental Table S17. Gene expression levels of 56 soybean predicted genes and four unannotated sequences by qRT-PCR.
Supplementary Material
Acknowledgments
We thank William Franck for critical reading of the manuscript and Chia Riou Yeo for technical assistance in the root hair cell isolation.
This work was supported by the National Science Foundation, Plant Genome Research Program (grant no. DBI–0421620).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gary Stacey (staceyg@missouri.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Denarie J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34 495–506 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2005) Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol Plant Microbe Interact 18 487–498 [DOI] [PubMed] [Google Scholar]
- Barnett MJ, Toman CJ, Fisher RF, Long SR (2004) A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci USA 101 16636–16641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55 504–513 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57 289–300 [Google Scholar]
- Brechenmacher L, Kim MY, Benitez M, Li M, Joshi T, Calla B, Lee MP, Libault M, Vodkin LO, Xu D, et al (2008) Transcription profiling of soybean nodulation by Bradyrhizobium japonicum. Mol Plant Microbe Interact 21 631–645 [DOI] [PubMed] [Google Scholar]
- Brewin NJ (2004) Plant cell wall remodelling in the Rhizobium-legume symbiosis. Crit Rev Plant Sci 23 293–316 [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MA, Elkan GH (1973) Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother 4 248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15 411–420 [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, et al (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Ross JJ, Reid JB (2005) Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol 138 2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Munoz A, Poovaiah BW, Oldroyd GE (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441 1149–1152 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Munoz A, Heckmann AB, Downie JA, Oldroyd GE (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS One 4 e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531 [DOI] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Webb KJ, et al (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11 263–274 [DOI] [PubMed] [Google Scholar]
- Lee H, Hur CG, Oh CJ, Kim HB, Pakr SY, An CS (2004) Analysis of the root nodule-enhanced transcriptome in soybean. Mol Cells 18 53–62 [PubMed] [Google Scholar]
- Libault M, Joshi T, Takahashi K, Hurley-Sommer A, Puricelli K, Blake S, Xu D, Nguyen H, Stacey G (2009) Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol 151 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1 44–54 [Google Scholar]
- Lievens S, Goormachtig S, Den Herder J, Capoen W, Mathis R, Hedden P, Holsters M (2005) Gibberellins are involved in nodulation of Sesbania rostrata. Plant Physiol 139 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Maekawa-Yoshikawa M, Takeda N, Imaizumi-Anraku H, Murooka Y, Hayashi M (2009) Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J 58 183–194 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NA, Kingsmore SF, Farmer AD, Langley RJ, Mudge J, Crow JA, Gonzalez AJ, Schilkey FD, Kim RJ, van Velkinburgh J, et al (2008) Management of high-throughout DNA sequencing projects: Alpheus. J Comp Sci Syst Biol 1 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami E, Kouchi H, Cohn JR, Ogawa T, Stacey G (1996) Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules. Plant J 10 23–32 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S (2006) Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13 255–265 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Kawaguchi M (2002) The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol 43 853–859 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR (2003) Identification and characterization of Nodulation-Signaling Pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Schauser L, Wieloch W, Stougaard J (2005) Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol 60 229–237 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47 868–876 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Lin JY, Schlueter SD, Vasylenko-Sanders IF, Deshpande S, Yi J, O'Bleness M, Roe BA, Nelson RT, Scheffler BE, et al (2007) Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463 178–183 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed]
- Smyth GK (2005) Limma: linear models for microarray data. In R Gentleman, V Carey, S Dudoit, R Irizarry, W Huber, eds, Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, pp 397–420
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156 [DOI] [PubMed] [Google Scholar]
- Wan J, Torres M, Ganapathy A, Thelen J, DaGue BB, Mooney B, Xu D, Stacey G (2005) Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol Plant Microbe Interact 18 458–467 [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21 1859–1875 [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Martinez Murillo F, Spencer F (2004) A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99 909–917 [Google Scholar]
- Wu Z, Irizarry RA (2005) Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol 12 882–893 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.