Abstract
Casein kinase II (CK2) is a protein kinase with an evolutionarily conserved function as a circadian clock component in several organisms, including the long-day plant Arabidopsis (Arabidopsis thaliana). The circadian clock component CIRCADIAN CLOCK ASSOCIATED1 (CCA1) is a CK2 target in Arabidopsis, where it influences photoperiodic flowering. In rice (Oryza sativa), a short-day plant, Heading date6 (Hd6) encodes a CK2α subunit that delays flowering time under long-day conditions. Here, we demonstrate that control of flowering time in rice by the Hd6 CK2α subunit requires a functional Hd1 gene (an Arabidopsis CONSTANS ortholog) and is independent of the circadian clock mechanism. Our findings from overexpressing the dominant-negative CK2 allele in rice support the independence of CK2 function from the circadian clock. This lack of control of the circadian clock by Hd6 CK2α might be due to the presence of glutamate in OsLHY (a CCA1 ortholog in rice) instead of the serine at the corresponding CK2 target site in CCA1. However, this glutamate is critical for the control of the OsPRR1 gene (a rice ortholog of the Arabidopsis TOC1/PRR1 gene) by OsLHY for regulation of the circadian clock. We also demonstrated that the other conserved CK2 target sites in OsLHY conferred robust rhythmic expression of OsLHY-LUC under diurnal conditions. These findings imply that the role of CK2 in flowering-time regulation in higher plants has diversified during evolution.
Plants flower at seasons appropriate for the survival of their progeny in various niches (Izawa, 2007a). This anticipation of the appropriate season is based on the interaction between the circadian clock and acute light signaling (Hayama and Coupland, 2004; Imaizumi and Kay, 2006; Izawa, 2007b). The integrated season signal has been introduced into the transcriptional regulation of florigen genes, including FT and Heading date3a (Hd3a; Hayama and Coupland, 2004). Recent studies in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) have revealed plant circadian clock and photoperiodic mechanisms. LATE ELONGATED HYPOCOTYL/CIRCADIAN CLOCK ASSOCIATED1 (LHY/CCA1) and TIMING OF CAB EXPRESSION1 (TOC1) are major components of the circadian oscillator in Arabidopsis (Schaffer et al., 1998; Wang and Tobin, 1998; Alabadi et al., 2001; Mizoguchi et al., 2002; Yanovsky and Kay, 2002). LHY and CCA1 are Myb-related transcription factors that are closely related to each other. These genes negatively regulate TOC1expression, which in turn positively regulates LHY and CCA1 expression. This feedback loop is considered to provide the basis of circadian oscillations (Alabadi et al., 2001). TOC1 encodes a pseudoresponse regulator (PRR) that shows circadian expression in antiphase to LHY and CCA1. Casein kinase II (CK2) is an evolutionarily well-conserved protein kinase found in various organisms, including yeast, mammals, and plants (Meggio and Pinna, 2003; Olsten and Litchfield, 2004). CK2 forms a tetrameric complex consisting of two α-subunits and two β-subunits; the α-subunit is a catalytic subunit and can function as a protein kinase in vitro (Pinna, 1994; Pagano et al., 2006). In Arabidopsis, CK2 can interact with, and phosphorylate, CCA1 (Sugano et al., 1998), and overexpression of a CK2β subunit, CKB3, leads to changes in the period of the circadian clock and early flowering under short-day (SD) conditions (Sugano et al., 1999). Degradation of another CK2β subunit in Arabidopsis, CKB4, is regulated by the circadian clock (Perales et al., 2006). Molecular genetics studies have further revealed that CK2 is a critical component of the circadian clock systems of various organisms (Allada and Meissner, 2005; Mizoguchi et al., 2006; Portoles and Mas, 2007). In addition, CK2 is involved in various biological phenomena: it phosphorylates a range of different proteins, including transcription factors, and can modulate DNA-binding ability, intracellular localization, and protein stability and by interacting with partner proteins in insects and animals (Miyata and Nishida, 2004; Riera et al., 2004; Krick et al., 2006; Pagano et al., 2006; Taghli-Lamallem et al., 2008). Consistent with this, a study in antisense CK2α lines has suggested the pleiotropic functions of CK2 in Arabidopsis (Lee et al., 1999). Recently, a dominant-negative form of CK2α was introduced into Arabidopsis to further confirm the pleiotropic functions of CK2 (Moreno-Romero et al., 2008).
In rice (Oryza sativa), a SD plant, we previously used natural variations among rice cultivars to identify the Hd6 gene as encoding an α-subunit of CK2 (Takahashi et al., 2001). Hd6 functional lines show late flowering under natural and long-day (LD) conditions. However, the molecular mechanism behind the promotion of late flowering by Hd6 has not been elucidated. In this paper, we analyzed the function of Hd6 in the rice circadian clock and in the photoperiodic flowering pathway. We revealed that the circadian clock has been disconnected from the function of CK2 in the photoperiodic control of flowering in rice, partly because of the presence of an evolutionarily conserved Glu site in OsLHY in rice, of which the corresponding site is Ser (a CK2 target site) in CCA1.
RESULTS
Hd6 Requires Functional Hd1 for Flowering Repression under LD Conditions
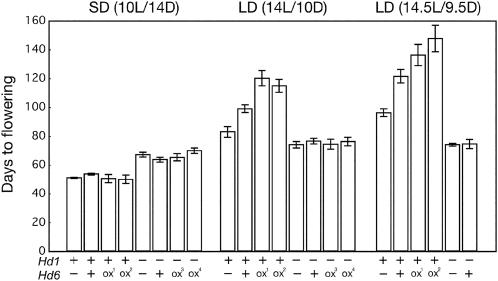
To elucidate the molecular function of Hd6, we examined flowering time in rice with functional, nonfunctional, or overexpressed Hd6 alleles (including nearly isogenic lines and transgenic lines) under LD and SD conditions (Fig. 1; Supplemental Fig. S1). Under LD conditions, Hd6 delayed flowering time in a dose-dependent manner (Fig. 1; Supplemental Fig. S2). This Hd6-induced delay required the presence of a functional Hd1 allele. A 30-min extension of the LD photoperiod caused a synergistic delay in flowering through the interaction of Hd6 with Hd1; therefore, Hd6 can enhance Hd1 floral repression activity under LD conditions. The time of flowering under SD conditions was also examined in plants with these Hd6-related alleles; the results suggested that Hd6 plays a critical role in Hd1 activity (Fig. 1; Supplemental Fig. S2).
Figure 1.
Hd6 required functional Hd1 to delay flowering under LD conditions. Flowering-time phenotypes of Hd6 lines in rice are shown. Days to flowering after sowing were measured for each plant. Hd6+, Hd6−, and Hd6ox indicate the genotypes of each line (+, functional allele; −, nonfunctional allele; ox, overexpresser driven by the 35S promoter). Each line was grown under the indicated photoperiod conditions. Each bar represents the mean ± sd of flowering time from 10 to 20 plants. Representative results of two to four independent experiments are shown. Hd1+/hd6−, Nipponbare (an hd6-defective cultivar); Hd1+/ Hd6+, NIL(Hd6) (NIL in a Nipponbare genetic background with a functional Hd6 allele from a Kasalath indica cultivar; Supplemental Fig. S2); Hd1+/Hd6ox1 and Hd1+/Hd6ox2, two independent transgenic rice plants (T3) overexpressing Hd6 cDNA in Nipponbare; hd1−/hd6−, NIL(Hd1) (NIL in a Nipponbare genetic background with a nonfunctional Hd1 allele from the Kasalath cultivar); hd1−/Hd6+, NIL(Hd1,6) [progeny line from NIL(Hd1) and NIL(Hd6)]; hd1−/Hd6ox3 and hd1−/Hd6ox4, two independent transgenic rice plants (T3) overexpressing Hd6 cDNA in NIL(Hd1).
Hd6 Does Not Affect Circadian Rhythms in Rice
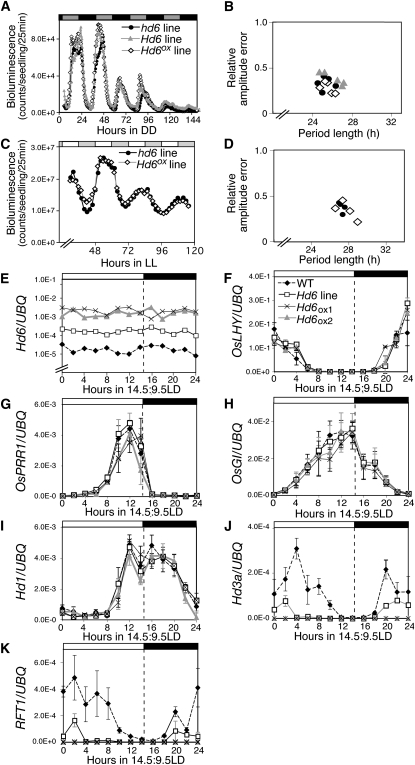
Because we found genes orthologous to the circadian clock genes of Arabidopsis in the rice genome (Izawa, 2007b), we used Hd6 alleles to examine the expression of circadian clock-controlled genes in rice. Under constant light or darkness, cab1R:LUCIFERASE (LUC) expression (Sugiyama et al., 2001) did not change in plants with these Hd6 alleles (Fig. 2, A–D). Expression of OsLHY and OsPRR1 (a TOC1 ortholog; Murakami et al., 2003, 2007) was also not affected under constant light (Supplemental Fig. S3, A and B). Furthermore, diurnal expression of OsLHY, OsPRR1, -95, -73, and -37, and OsGI (a GIGANTEA ortholog) did not change under LD conditions (Fig. 2, F–H; Supplemental Fig. S3C). These results clearly indicated that Hd6 is not a circadian clock component in rice. Therefore, Hd6 regulation of flowering time is not mediated by changes in circadian clock action.
Figure 2.
Hd6 repressed FT-like genes and acted downstream of Hd1 under LD conditions but did not control circadian clocks. A to D, Bioluminescence analysis of cab1R:LUC expression in constant darkness (DD; A) and constant light (LL; C) after 5 d of light/dark entrainment. Period lengths and relative amplitude errors in DD (B) and in LL (D) were calculated from luminescence data by a fast Fourier transform-nonlinear least squares method. Five to 10 seedlings of each line were assessed. The experiments were repeated three times. Hd6 and hd6 lines were T3 (or T4) progeny lines from a cross between a japonica cultivar with the Hd6 allele and another japonica cultivar with the hd6 allele carrying the cab1R:LUC gene. Hd6ox lines were from a cross between Hd6ox lines and cab1R:LUC lines. E to K, Expression levels of Hd6 (E), OsLHY (F), OsPRR1 (G), OsGI (H), Hd1 (I), Hd3a (J), and RFT1 (K) in Nipponbare. NIL(Hd6) and two Hd6ox lines were measured by real-time quantitative RT-PCR. Data were normalized against ubiquitin expression. Means and sd were plotted from three biological replicates. Each plant was grown under LD (14.5 h of light/9.5 h of dark) conditions for 60 d. Young leaves from three plants were collected every 2 h after dawn. WT, Wild type.
Hd6 Delays Flowering by Repressing FT-Like Genes under LD Conditions
We next analyzed the expression patterns of flowering-time genes in Nipponbare, a nearly isogenic line of Hd6 [NIL(Hd6)]; Supplemental Fig. S1], and in lines overexpressing Hd6 under LD conditions. Consistent with the delayed flowering phenotype, the overexpression of Hd6 repressed expression of Hd3a and RFT1 (another FT ortholog; Komiya et al., 2008) but had no effect on the expression profile of Hd1 (Fig. 2, I–K). To further confirm this genetic interaction, we analyzed the expression of Hd1, Hd3a, and RFT1 in an Hd1 nonfunctional background [NIL(Hd1) and NIL(Hd1,6)]. Hd3a and RFT1 were not dramatically repressed by Hd6 in the Hd1 nonfunctional background under LD conditions, although there seemed to be some Hd3a repression by Hd6 in the hd1 background (Supplemental Fig. S4). These results suggested that Hd6 modulates Hd1 repressor activity mainly by posttranslational modification.
Hd6 Has Protein Kinase Activity But Does Not Phosphorylate Hd1 in Vitro
To confirm the protein kinase activity of Hd6, we measured the CK2 activity of the Hd6 recombinant protein (rHd6; Sugano et al., 1998). We expressed Hd6 as a glutathione S-transferase (GST) fusion protein and isolated it from GST by thrombin treatment of the purified fusion protein bound to glutathione agarose beads (Supplemental Fig. S5A). We then measured the CK2 activity of the isolated rHd6. Purified rHd6 had nearly the same CK2 activity as a control human CK2α (Supplemental Fig. S5B).
To investigate Hd1 phosphorylation, GST fusion Hd1 protein was purified as above and its potential as an Hd6 substrate was characterized in vitro. Hd1 was not phosphorylated by rHd6 in vitro (Supplemental Fig. S6). The observation that flowering regulation by Hd6 required Hd1 and that Hd1 was not a direct Hd6 target suggests that Hd6 protein phosphorylates an unknown flowering repressor target that works together with Hd1.
Overexpression of a Dominant-Negative Form of Hd6 Causes Early Flowering under LD Conditions
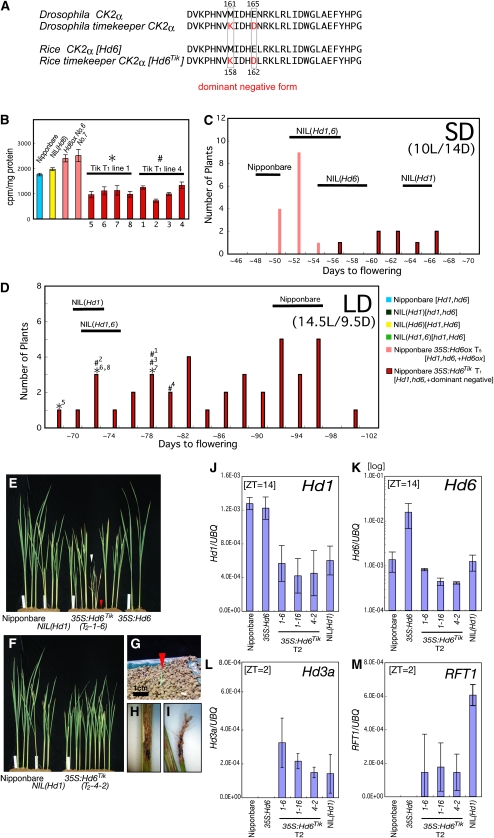
By a genome-wide homology search in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/), we found four genes with very high levels of identity to CK2α subunit genes in the rice genome (Supplemental Fig. S7A). Expression analysis revealed that, of the four CK2α genes, functional Hd6 (OsCKA2-1) and OsCKA2-2 were the two main ones expressed in various tissues of rice (Supplemental Fig. S7, B and C). Therefore, with deficiency of the Hd6 gene alone it was not easy to draw conclusions about the overall role of phosphorylation by CK2 in rice biology. Therefore, we used Hd6Tik, the Timekeeper (Tik) mutant form of Hd6, a dominant-negative CK2α allele that was originally identified in Drosophila (Akten et al., 2003; Moreno-Romero et al., 2008), to elucidate the role of CK2 in rice (Fig. 3A). We measured the CK2 activity of recombinant Hd6Tik and confirmed that the mutant protein had greatly reduced CK2 activity (Supplemental Fig. S5B). Next, we used the cauliflower mosaic virus (CaMV) 35S promoter to generate transgenic rice overexpressing Hd6Tik. With these Hd6Tik lines, we were able to reduce the total in vitro CK2 phosphorylation activity in cell extracts to about half of that observed with the nonfunctional Hd6 allele (Fig. 3B).
Figure 3.
Phenotypes of rice overexpressing a dominant-negative CK2α allele. A, The sequence on top shows the Tik mutation in a dominant-negative form of CK2α, with mutations of M158K and E162D sites in Hd6, referred from the Tik allele of Drosophila melanogaster (in red). B, CK2 activity in plant extracts. Data are means of three experiments for each line using distinct tillers after flowering. C and D, Flowering times in Nipponbare, NIL(Hd1), NIL(Hd6), NIL(Hd1,6), Hd6ox, and Hd6Tikox were examined. T1 plants were grown under SD (10 h of light/14 h of dark) and LD (14.5 h of light/9.5 h of dark) conditions. Histograms for days to flowering are shown. Rice plants of NIL(Hd6) and Hd6ox did not flower within 100 d after sowing under LD conditions. Black bars indicate ranges of flowering time for the tested lines (i.e. that all tested plants flowered within these ranges). E to I, Nipponbare, NIL(Hd1), Hd6Tikox, and Hd6ox plants under LD conditions 70 d after sowing. All tillers were removed. Most of the surviving Hd6Tikox T1 plants exhibited normal growth (E and F), but a few plants showed abnormal phenotypes (e.g. arrowhead in E). G, Hd6Tikox plants (red arrowhead) 30 d after sowing. H and I, Retarded panicles in Hd6Tikox plants. J to M, Gene expression in Hd6Tik lines.
Overexpression of the dominant-negative form (Allada and Meissner, 2005; Moreno-Romero et al., 2008; Smith et al., 2008) of Hd6 resulted in early flowering equivalent to that observed in plants with Hd1-defective alleles under LD conditions (Fig. 3, C and D). Some of the Hd6Tik lines failed to thrive in the T1 generation, especially under SD conditions, suggesting that CK2 has pleiotropic functions in rice, as recently found in Arabidopsis by the use of transgenic lines carrying a dominant-negative mutation in a CK2α allele and an antisense construct (Lee et al., 1999; Smith et al., 2008). Pleiotropic effects such as retarded growth were indeed observed in Hd6Tik lines (Fig. 3, E–I). We also observed slightly later flowering phenotypes in Hd6Tik lines than in hd1-deficient NIL plants under SD conditions (Fig. 3C), suggesting that there is extra floral control by CK2 that is independent of Hd1. In addition, Hd1 mRNA expression was reduced under LD conditions in Hd6Tik lines (Fig. 3, J–M), suggesting the involvement of CK2 in transcriptional control of Hd1. However, the occurrence of pleiotropic effects in the Hd6Tik lines (Fig. 3, E–I) does not support this possibility. To fully understand the mechanisms underlying the functions of CK2, further investigations of the biological roles of CK2, other than its role in photoperiodic flowering in rice, are required.
Overexpression of Hd6Tik Does Not Affect the Circadian Clock
To determine the effects of CK2 loss of function on molecular clock rhythms in rice, we next tested whether overexpression of a dominant-negative form of Hd6 altered the rhythmic expression of OsPRR1. The expression pattern of OsPRR1 in rice cells was monitored by LUC activity (Supplemental Fig. S8). Hd6Tik did not affect OsPRR1:LUC expression under light/dark and constant dark conditions (Fig. 4). By comparison, CK2 activity was essential for Hd1 activity in both promotion of flowering under SD conditions and repression of flowering under LD conditions (Fig. 3, C and D). These results indicated that the role of CK2 in photoperiodic flowering in rice is distinct from the role of CK2 reported in Arabidopsis (Sugano et al., 1999; Daniel et al., 2004) and that CK2 does not contribute markedly to regulation of the circadian clock in rice (Figs. 2–4).
Figure 4.
Hd6Tik did not markedly affect circadian rhythmic expression of OsPRR1:LUC. Independent transformed 35S:Hd6Tik calli carrying the same OsPRR1:LUC reporter gene were measured for OsPRR1:LUC in constant darkness (DD). Data from the original OsPRR:LUC line (control) and transformed 35S:Hd6 calli are presented together. Data for the period 48 to 60 h were missed accidentally.
CK2 Phosphorylation Sites in OsLHY Are Evolutionarily Conserved, Except One Found in CCA1
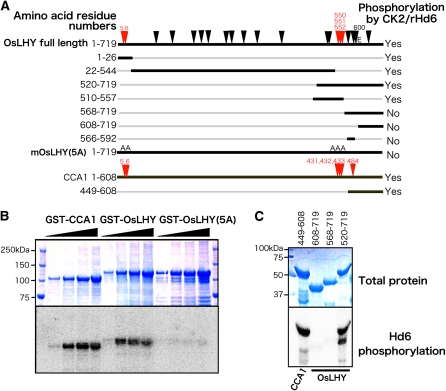
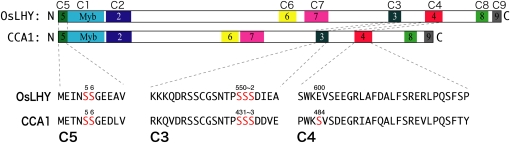
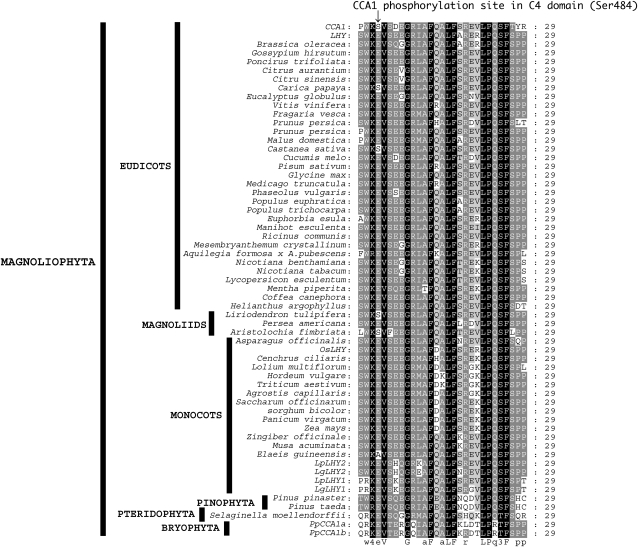
In Arabidopsis, CCA1 clock protein phosphorylation by CK2 is necessary for proper action of the circadian clock (Daniel et al., 2004). We started to investigate the mechanism underlying our finding by examining the phosphorylation sites in OsLHY proteins. First, GST-OsLHY protein was phosphorylated in vitro by rHd6 (Fig. 5). Deletion and amino acid exchange analysis revealed that OsLHY contained five phosphorylated Ser sites in two OsLHY domains, C5 and C3 (Okada et al., 2009; Figs. 5 and 6; Supplemental Fig. S9). We compared the corresponding positions of Ser sites between CCA1 and OsLHY and found that Ser-484 of CCA1 corresponded to Glu-600 of OsLHY, although five Ser sites (Ser-5, Ser-6, Ser-550, Ser-551, and Ser-552) in OsLHY were conserved as the corresponding sites in CCA1 (Fig. 6). We further compared the public database amino acid sequences of 22 CCA1 orthologs from 16 genera and found several motifs conserved in angiosperms (Fig. 6; Supplemental Figs. S9 and S10; Supplemental Tables S1 and S2; Miwa et al., 2006; Liu et al., 2009; Okada et al., 2009; Takata et al., 2009). Structurally, the LHY/CCA1-like genes are typified by the presence of nine highly conserved domains (C1–C9) in higher plants. Four domains (C1–C4) are conserved in moss (Okada et al., 2009). All the Ser sites in CCA1 were located in conserved domains (C5, C3, and C4). The Ser site in the C4 domain was changed to Glu in rice OsLHY (Fig. 6). Compared with other domains related to basic helix-loop-helix, the C4 domain was found only in CCA1, LHY, and OsLHY among all the annotations in the Arabidopsis and rice genomes, suggesting that the C4 domain is evolutionarily associated with the plant circadian clock (Fig. 7; Supplemental Fig. S9).
Figure 5.
CK2(rHd6) phosphorylated two regions in OsLHY. A, Schematic images of OsLHY and OsLHY fragments and OsLHY with mutations at all five possible phosphorylation sites from Ser to Ala, termed OsLHY(5A). Arrowheads indicate putative CK2 consensus sites (S/TXXD/E) found in OsLHY. Red arrowheads indicate five Ser residues at the phosphorylation sites corresponding to those in CCA1. Deleted OsLHY fragments are designated by the corresponding amino acid residue numbers in full-length OsLHY. The CK2 target assay results are summarized at the right. B, SDS-PAGE and autoradiograph of SDS-PAGE analyses of GST-CCA1, GST-OsLHY, and GST-mOsLHY(5A) after in vitro CK2(rHd6) phosphorylation assays. C, SDS-PAGE and autoradiographs of SDS-PAGE analyses of the C-terminal segments of OsLHY fused to GST after in vitro CK2(rHd6) phosphorylation assays. A fragment containing the C4 domain region (568–719) of OsLHY was not phosphorylated, whereas the corresponding CCA1 fragment (449–608) was phosphorylated.
Figure 6.
Distribution of phosphorylated regions in OsLHY and CCA1 relative to conserved domains. Diagrams of conserved domains in OsLHY and CCA1 are shown. C1 to C4 domains are conserved in land plant LHY/CCA1-like genes (Okada et al., 2009). C5 to C9 regions conserved among angiosperm LHY/CCA1-like genes are shown as colored boxes (Supplemental Fig. S9). The C1 domain is the Myb domain. The functions of the other domains are unknown (Supplemental Table S3). The three regions of phosphorylated residues (C5, C3, and C4) of OsLHY established in this study are indicated in red.
Figure 7.
Multiple alignment of C4 domain sequences in plants. This plant-specific motif is found only in LHY/CCA1 and OsLHY in the Arabidopsis and rice genomes, respectively. Likewise, this C4 domain is found only in predicted LHY-like gene(s) regions in the Sorghum, Populus, Vitis vinifera, and Physcomitrella genomes, respectively (Supplemental Fig. S9). Glu (E) is the amino acid site found in this domain, and one that corresponds to a CK2 phosphorylation site in CCA1. This amino acid is found in all C4 domain-like sequences in various plant species EST databases (Supplemental Table S4), with the exception of three species of eudicots, two of magnolids, and one of monocots (arrow).
Biological Roles of CK2 Phosphorylation Sites in OsLHY
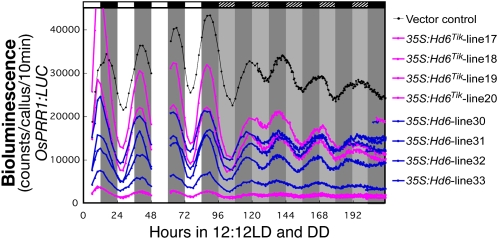
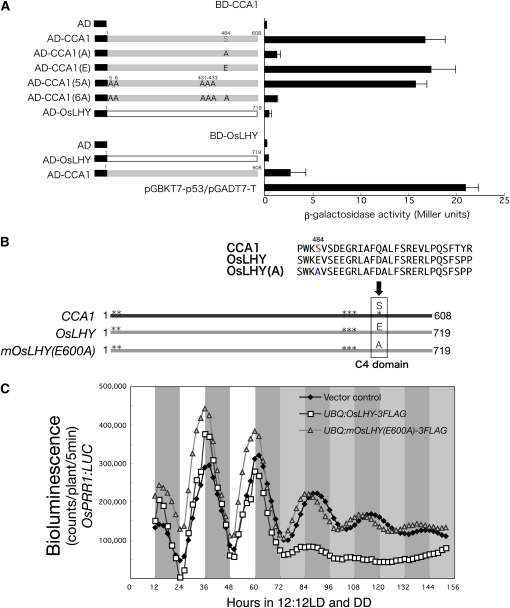
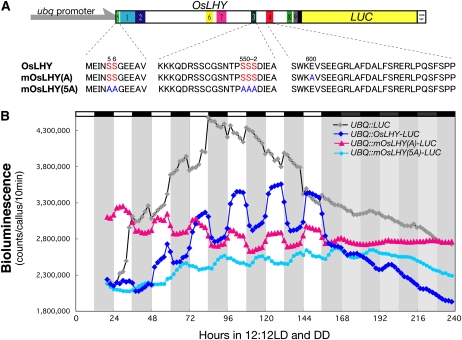
As described above, when the C4 domain was subjected to a homology search, we found that Glu (in Glu-600) was a major amino acid in the C4 domain and that Ser (in Ser-484) in CCA1 was an exception (Figs. 6 and 7). This conserved C4 domain fragment in OsLHY was not phosphorylated but that in CCA1 was, indicating that there was no Ser site for compensation for Ser-484 of CCA1 in the C4 domain of OsLHY (Fig. 5, A and C). A yeast two-hybrid (Y2H) assay further revealed that only the Ser-484 site, not the other five Ser sites, was critical for CCA1 homodimer formation (Fig. 8A). In contrast, OsLHY did not form a homodimer in the Y2H assay, suggesting that OsLHY and CCA1 had distinctly different biochemical properties. Although overexpression of tagged OsLHY was able to repress OsPRR1:LUC rhythmic expression in rice calli, that of tagged mOsLHY(A) (an E600A mutant of OsLHY) was not able to repress it (Fig. 8, B and C). These results suggested that OsLHY Glu-600 is essential for circadian clock function. In addition, to try to evaluate OsLHY stability in rice, we made transgenic rice calli overexpressing an OsLHY-LUC fusion protein driven by a maize (Zea mays) ubiquitin promoter (Fig. 9). The LUC activity of the OsLHY-LUC protein exhibited diurnal rhythms under light/dark conditions but no rhythms (and clear damping) under constant dark conditions. This strongly suggested that OsLHY protein becomes stable under light conditions and unstable under dark conditions. In contrast, OsLHY-LUC(5A), with five Ser-to-Ala mutations at the five conserved Ser sites in OsLHY, completely lost the light stabilization of OsLHY-LUC, suggesting that OsLHY stabilization by light signals was controlled through CK2 phosphorylation. On the other hand, the E600A mutation in OsLHY did not affect the light-induced stabilization of OsLHY-LUC, suggesting that Glu-600 was not involved in the control of OsLHY stabilization by light. It is also possible that this rhythm of LUC activity came from translational regulation and protein modification.
Figure 8.
Biochemical and biological roles of the Glu-600 and Ser-484 sites in OsLHY and CCA1. A, Y2H assays. Mutations in CCA1 altered the interactions with CCA1 in yeast. The Ser-484 site in CCA1 was critical for homodimer formation of CCA1 in yeast. OsLHY did not interact with OsLHY itself in yeast. All experiments were done in accordance with the manufacturer's instructions for the Matchmaker kit (BD Clontech). Average values of β-galactosidase activities from five individual colonies from a representative experiment are shown ± se at right. B, Schematic view of tested constructs with site-specific mutations. The arrow indicates the position of Glu/Ser in the C4 domain. The identified CK2 target sites are shown as asterisks for CCA1 and OsLHY. C, Bioluminescence analysis of OsPRR1:LUC expression in rice calli transformed with UBQ:mOsLHY(A)-3FLAG or UBQ:OsLHY-3FLAG in light/dark (LD) followed by constant darkness (DD) cycles. About 10 independent transformed T0 cell lines carrying the same OsPRR1:LUC reporter gene were measured. Average counts are shown.
Figure 9.
Light-dependent OsLHY protein stability in transgenic rice calli. A, Structure of OsLHY-LUC used for this experiment. B, Four distinct transgenic cell lines carrying UBQ:LUC, UBQ:OsLHY-LUC, UBQ:OsLHY(A)-LUC, and UBQ:OsLHY(5A)-LUC were grown under 12-h-light/12-h-dark conditions (12:12LD), and their LUC bioluminescence activity was monitored for 1 week under 12:12LD conditions and 3 d under constant darkness (DD) conditions. Light dependency of LUC stability was observed in UBQ:OsLHY-LUC lines. The quintuple mutations in the phosphorylation sites of OsLHY in the mOsLHY(5A) transgenic line may have resulted in loss of the light dependency of fusion protein stability in rice cells. By comparison, the Ala mutation at the Glu site in mOsLHY(A) did not greatly affect LUC activity. Similar bioluminescence profiles were reproducibly obtained. A representative result is shown.
DISCUSSION
Using rice NILs and lines overexpressing Hd6, we demonstrated here that flowering-time control by the previously identified quantitative trait locus, Hd6, in rice requires a functional Hd1 gene (an Arabidopsis CONSTANS ortholog) but does not occur through circadian clock action (Figs. 1 and 2; Supplemental Figs. S2 and S4). Because the Hd6 gene is a member of the CK2α subfamily and is one of two major expressed CK2α subunits (Supplemental Fig. S7), transgenic lines overexpressing a dominant-negative CK2α were further examined to evaluate the role of CK2 in the circadian clock in the photoperiodic control of flowering in rice. Because CK2 activity was not completely absent in these dominant-negative lines (Fig. 3), we were able to conclude that circadian clock activity was disconnected from the control of flowering time by Hd6 but not from all CK2 activity. However, we confirmed that the circadian clock in rice was less affected by a reduction in CK2 activity than was flowering-time control (Figs. 3 and 4). We did not identify the target protein that could be phosphorylated by Hd6 CK2α to control flowering time. Our genetic analysis, however, strongly suggested that any protein that could work together with Hd1 might be a target. Recently, the Ghd7 gene was reported to be a strong repressor of rice flowering (Xue et al., 2008). We found that Hd6 did not phosphorylate either Hd1 or Ghd7 protein in vitro (Supplemental Fig. S6; data not shown). In addition, our preliminary data revealed that some protein associated with Hd1 in vitro could be an Hd6 target for the control of Hd1 repressor activity (data not shown). We are now attempting to identify this Hd6 target.
Although there is no genetic evidence of a role for OsLHY in the circadian clock in rice, we demonstrated here that overexpression of tagged OsLHY repressed the rhythmic expression of OsPRR1:LUC in rice cells (Fig. 8C). This result strongly suggested that OsLHY is a core component of the circadian clock in rice and was consistent with the concept of OsLHY as a sole ortholog of Arabidopsis CCA1/LHY in the rice genome. Notably, we tried a simple way of making transgenic rice plants overexpressing OsLHY cDNA (without the tag) driven by both CaMV 35S and a maize ubiquitin promoter, but we were not able to obtain any rice plants overexpressing OsLHY mRNA (data not shown). In addition, we succeeded in overexpressing the OsLHY-LUC gene in rice cells (Fig. 9), but no associated phenotypes were observed. We obtained circadian clock-related phenotypes associated with the repression of OsPRR1:LUC rhythmic expression, but only when we overexpressed the tagged OsLHY. Unlike with LHY/CCA1, the simple overexpression of which disturbs the circadian clock in Arabidopsis (Schaffer et al., 1998; Wang and Tobin, 1998), in the case of OsLHY its function may be too critical in rice to obtain a simple overexpresser.
We next revealed that OsLHY could not form a homodimer in yeast, whereas CCA1 could do so through the C4 domain (Ser-484 site), one of the previously identified CK2 phosphorylation sites (Daniel et al., 2004; Fig. 8A; data not shown). This finding is consistent with that of a previous report in Arabidopsis (Daniel et al., 2004). By comparison, the corresponding site (Glu-600) in OsLHY in rice was critical for repression of the rhythmic expression of OsPRR1. In Arabidopsis, the findings that lines overexpressing CKB3 and CKB4 have circadian clocks with short free-running periods and show early flowering under SD conditions indicate that CK2 can control the circadian clock and flowering time in this species and that this control is mediated by CCA1 phosphorylation (Sugano et al., 1999; Daniel et al., 2004). CCA1 and LHY function as TOC1/PRR1 repressors (Mizuno and Nakamichi, 2005). These actions form a negative feedback loop in the circadian clock of Arabidopsis (Alabadi et al., 2001; Mizuno and Nakamichi, 2005). As expected, OsLHY expression is likely to be regulated by the circadian clock (Izawa et al., 2002). Because CCA1 phosphorylation is necessary for CCA1 homodimerization and normal clock function, OsLHY phosphorylation is likely to be important for rice circadian clock function. In fact, OsLHY was phosphorylated by Hd6, and the phosphorylation sites were conserved, with one exception (Figs. 6 and 7; Supplemental Table S3). However, NIL(Hd6) lines, Hd6-overexpressing lines, and dominant-negative lines had little effect on the rice circadian clock (Figs. 2 and 4). Only one unconserved site, OsLHY Glu-600, corresponding to CCA1 Ser-484, was present in a highly conserved motif, termed the C4 domain in land plants (Fig. 7). CCA1 Ser-484 is necessary for homodimerization (Fig. 8A). In addition, mOsLHY E600A overexpression did not repress OsPRR1:LUC in rice calli, whereas OsLHY overexpression did (Fig. 8, B and C; data not shown). These results indicate that this Glu site in the C4 domain was critical for OsLHY function related to the circadian clock. They also strongly suggest that the change from Ser to Glu at the Glu-600 site in OsLHY is a critical cause of the disconnection of Hd6 from rice circadian clock regulation. During plant evolution, CCA1 may have acquired the capacity for Ser-484 phosphorylation, which may then have led to dimer formation for fine regulation of the circadian clock in Arabidopsis. The inability of OsLHY to form homodimers (Fig. 8A) suggests that OsLHY has a role in regulation of the circadian clock system in rice and that this role is distinct from that of CCA1 found in Arabidopsis.
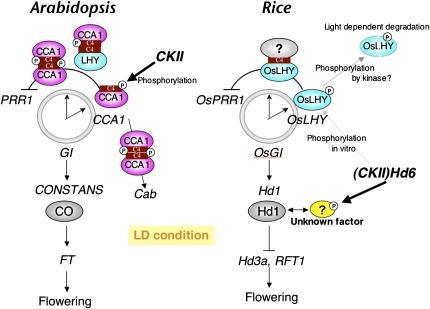
Interestingly, of the other five Ser sites phosphorylated in vitro by rHd6, those corresponding to CCA1 sites might be involved in the light-dependent protein stability of an OsLHY-LUC fusion protein (Fig. 9), although the molecular nature of the rhythms of OsLHY-LUC has not been confirmed. This result for OsLHY-LUC suggests that OsLHY activity under daily light/dark cycles is controlled by phosphorylation by CK2 (or other kinases) and that control of this nature, through the sites of phosphorylation of OsLHY, is clearly distinct from the control provided by the Glu-600 site of OsLHY for circadian clock oscillation in rice, which confers rhythmic gene expression under constant light or dark conditions. The molecular nature of light-regulated translation of LHY protein has also been reported in Arabidopsis (Kim et al., 2003). Therefore, the regulation of protein stability by light in LHY and OsLHY is likely to be conserved among plant species through phosphorylation of the five conserved amino acids by CK2. The control of light-dependent protein stability (Kim et al., 2003) by the phosphorylation Ser sites in OsLHY implies that the robustness of diurnal rhythmic expression of OsLHY is an evolutionarily conserved function. The contribution of CK2 to circadian clock regulation under constant light or dark conditions might be a recently developed function of CK2 observed in Arabidopsis. The finding that Glu was a major amino acid at Glu-600 in OsLHY (corresponding to Ser-484 in CCA1) strongly supports this idea. These findings might shed light on the molecular mechanisms underlying the development of circadian clock systems and photoperiodic flowering during plant evolution, since Arabidopsis and rice are believed to have diverged about 200 million years ago (Fig. 10).
Figure 10.
Model of CK2(Hd6) regulation of the circadian clock and photoperiodic control of flowering in Arabidopsis and rice. In Arabidopsis, CK2 may control the circadian clock through CCA1 dimer formation. In rice, it is likely that CK2 does not control the circadian clock, since OsLHY (an ortholog of CCA1) cannot form homodimers and the amino acid site corresponding to the CK2 target site of CCA1 has been changed to Glu. It is still possible that phosphorylation of OsLHY by CK2 controls protein stability. In addition, CK2 may control Hd1 activity through the phosphorylation of an unknown interacting factor to control flowering time in rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions for Measuring Flowering Time
One rice (Oryza sativa) cultivar, three NILs (Supplemental Fig. S1) that contained an Hd1 or Hd6 chromosomal region of the Kasalath genome, and two overexpression lines were used. The DNA markers for selection of the Kasalath fragments for the Hd1 and Hd6 regions were Arg-2171 to Arg-2654 and Arg-2311 to Cys-217, respectively. The constructs for the overexpression lines were built in the following way. Nipponbare (genotype Hd1/hd6), NIL(Hd6) (genotype Hd1/Hd6), NIL(Hd1) (genotype hd1/hd6), NIL(Hd1,6) (genotype hd1/hd6), and Hd6ox transgenic [genetic background: Nipponbare and NIL(Hd1)] plants (Supplemental Fig. S1) were grown under SD (10 h of light/14 h of dark) or LD (14 h of light/10 h of dark or 14.5 h of light/9.5 h of dark) conditions. All tillers after the fourth were removed to save space under LD conditions. A metal halide lamp was used as the light source in the growth chambers. Photosynthetic photon flux density ranged from 450 to 500 μmol m−2 s−1.
Establishment of Rice Cell Lines for Bioluminescence Analysis
Rice cells were transformed with the OsPRR1:LUC construct using Gly-418 as a selection marker, and the cell lines obtained were selected on the basis of stable rhythmic expression of OsPRR1:LUC in constant darkness. The selected OsPRR1:LUC cell line was then further transformed with plasmids [35S:OsLHY-3FLAG, 35S:OsLHY(A)-3FLAG, etc.]. The effects of OsPRR1:LUC expression were examined in about 10 independent cell lines.
Bioluminescence Assays of Circadian Rhythm
Transgenic lines containing cab:LUC together with Hd6 were selected in the F5-471 (the F5 progeny from a cross between a japonica rice with an hd6 allele, Norin8, carrying cab1R:LUC [Sugiyama et al., 2001] and another Japonica line with an Hd6 allele, Kinmaze). A few seedlings were grown with 3 mL of Murashige and Skoog medium in a 35- × 10-mm petri dish (Becton-Dickinson) and entrained for 5 d in 12-h-light/12-h-dark cycles at 30°C. The light source was a mixture of red and blue light-emitting diode lamps (Sanyo) with a photon flux density of 200 μmol m−2 s−1. The bioluminescence in transformed cell lines (callus) containing OsPRR1:LUC was measured in a 90- × 20-mm petri dish (Bio-Bik). An Aquacosmos photon-counting system (Hamamatsu Photonic Systems) was used for all bioluminescence imaging experiments. Periods and relative amplitude errors were estimated from data starting from 24 h after the last dark-to-light transition using fast Fourier transform-nonlinear least squares software, as described by Plautz et al. (1997).
Constructs
All primer sequence information is given in Supplemental Table S1. Plasmids for overexpressing the target gene product were constructed using either a pBI vector containing the CaMV 35S promoter or a related pSK2 vector (Izawa et al., 2002) containing a maize (Zea mays) ubiquitin promoter. Constructs for overexpressing the Hd6 cDNA fragment and the mutated Hd6Tik cDNA fragment were generated by means of the megaprimer method using a full-length Hd6 cDNA clone as template. The primer sets used were Hd6-1F and Hd6-R for Hd6 and Hd6-1F and Hd6-Tik-R2, Hd6-Tik-F2 and Hd6-R for Hd6Tik. Rice line Nipponbare was used for transformation in making these overexpressers.
The 35S-strepII-3xFLAG-Hd1 construct was generated from PCR fragments using designed oligonucleotides. For the 3xFLAG tag, the following primers were used: FlagNF1, FlagNF2, FlagN1R1, FlagN1R2, FlagN2R1, FlagN2R2, FlagN3R1, and FlagN3R2 (Supplemental Table S1). For the strepII tag, the following oligonucleotide linkers were used: XbaI-strepII-BamHI-F and XbaI-strepII-BamHI-R. Constructs with mutated OsLHY and CCA1 [(OsLHY(A), CCA1(484A), CCA1(484E), CCA1(5A), and CCA1(6A)] were generated by the megaprimer method (Sarkar and Sommer, 1990) and amplified using full-length OsLHY and CCA1 clones as templates. The primer sets used were as follows: OsLHY-5,6S>A-SalI-F and mOsLHY-SSS-R, mOsLHY-SSS-F and OsLHY-SpeI-R for mOsLHY(5S); OsLHY-SalI-F and mOsLHY-600E>A-R, mOsLHY-600E-A-F and OsLHY-SpeI-R for mOsLHY(E600A); CCA1-SalI-F and CCA1-R, CCA1-S>A-F and CCA1-S>A-R for CCA1(484A); CCA1-S>E-F and CCA1-S>E-R for CCA1 (484E); CCA1-56,7-S>A-F for CCA1(56,57A); and CCA1-S>AAA-F and CCA1-S>AAA-R for CCA1(5A) and CCA1(6A). All constructs containing OsLHY or CCA1, as well as the final mutated binary constructs, were verified by sequence analysis of inserts. For Hd6 phosphorylation tests using Escherichia coli-produced proteins, pGEX-OsLHY, mOsLHYs, and OsLHY, the deletion lines, and related CCA1 constructs (GST fusion proteins) were generated as above. Initially, full-length OsLHY and CCA1 cDNA subclones were digested with SalI and NotI and ligated into pGEX-4T vector (GE Healthcare Life Sciences). The OsPRR1:LUC construct was generated by PCR with primers KpnI-OsPRR1pro-F and HindIII-OsPRR1ex2-R. These fragments were fused to the modified firefly LUC+ gene on pSP-luc+ (Promega) at the KpnI and HindIII sites. The OsPRR1:LUC constructs were cloned onto the KpnI and SacI sites of an Agrobacterium tumefaciens binary vector, pBINPLUS. Nipponbare was transformed by a typical Agrobacterium-mediated method. We isolated several independent transgenic cell lines with OsPRR1:LUC. To generate the UBQ:OsLHY-3FLAG and UBQ:mOsLHY(A)-3FLAG constructs, non-stop-codon OsLHY was amplified by PCR with the primers OsLHY-SalI-F and OsLHY-stopX-XhoI-R. These isolated fragments were fused to TOPO vector (Invitrogen), and SalI/NotI fragments were fused to pBI221 vector. To generate 3xFLAG fusion OsLHY, 3xFALG oligonucleotide linker was ligated into the C terminus of OsLHY (XhoI/NotI) using XhoI-3FLAG-F and XhoI-3FLAG-R and NotI-3FLAG-F and NotI-3FLAG-R. The OsLHY-3FLAG-nos and mOsLHY(A)-3FLAG-nos constructs were cloned onto the SalI and EcoRI sites of an Agrobacterium binary vector, pPZP-UBQ. The calli of one line of OsPRR1:LUC were selected and transformed by the same Agrobacterium-mediated method with a different selectable antibiotic marker. We isolated several independent transgenic cell lines.
Real-Time Quantitative PCR
Total RNA was prepared by a conventional SDS-phenol method. The reverse transcription (RT) reaction was performed with SuperScript II reverse transcriptase (Invitrogen), oligo(dT)12–18 (Invitrogen), and 5 μg of total RNA in accordance with the manufacturer's instructions. Real-time quantitative RT-PCR analysis was performed as described previously. Briefly, quantitative RT-PCR was performed by the Taq-Man PCR method using an ABI PRISM 7900 Sequence Detection System in accordance with the manufacturer's instructions. Primer and probe sequence information for the quantitative RT-PCR is listed in Supplemental Table S4.
CK2 Activity Assay
One hundred milligrams of frozen seedlings was ground and extracted with 100 μL of extraction buffer (50 mm Tris-HCl [pH 7.6], 15 mm MgCl2, 0.1 m KCl, 0.25 m Suc, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture [Sigma], phosphatase inhibitor mixture [Sigma], and 14 mm 2-mercaptoethanol). After centrifugation of the sample at 15,000g for 10 min, the supernatant was sampled and the protein concentration was measured. CK2 activity in these extracts was measured with a CK2 kinase assay kit (Upstate) in accordance with the manufacturer's instructions.
Recombinant Protein Purification
GST-Hd6, GST-OsLHY, and GST-mOsLHY series and GST-CCA1 fusion proteins were overexpressed in E. coli (strain BL21) and purified. To remove the GST affinity tail from the GST-Hd6 fusion protein, protease cleavage by biotinylated thrombin protease combined with Glutathione-Sepharose 4B (Novagen) was used in accordance with the manufacturer's instructions. After the removal of thrombin by the addition of streptavidin agarose, the recombinant Hd6 protein was stored at 20°C until required for the in vitro assay to detect Hd6 target proteins.
Y2H Assay
The protein-coding sequences of OsLHY, mOsLHYs, CCA1, and mCCA1s were cloned into the plasmids provided in the Matchmaker GAL4 Two-Hybrid System 3 kit (BD Clontech). All assays were performed in accordance with the manufacturer's instructions. Expression of all proteins was confirmed by immunoblot analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Graphical genotypes of NILs.
Supplemental Figure S2. Functional Hd1 gene is necessary for the late flowering observed in Hd6-overexpressing lines under LD conditions.
Supplemental Figure S3. Hd6ox does not affect the rhythms of clock-controlled gene expression in continuous light (LL).
Supplemental Figure S4. Hd6 does not repress Hd3a and RFT1 critically in an hd1 nonfunctional background.
Supplemental Figure S5. Activity of purified recombinant Hd6 and Hd6Tik proteins expressed in E. coli.
Supplemental Figure S6. CK2(Hd6) phosphorylation assay for Hd1.
Supplemental Figure S7. CK2α genes in rice.
Supplemental Figure S8. Circadian clock monitoring by using OsPRR1:LUC in rice cells.
Supplemental Figure S9. Multiple alignment of LHY, CCA1, OsLHY, and LHY-like proteins.
Supplemental Figure S10. Evolutionary relationships of 22 taxa in the angiosperm CCA1/LHY-like gene.
Supplemental Table S1. C1 to C9 domains and conserved CCA1 phosphorylation sites from LHY/CCA1-like proteins in angiosperms.
Supplemental Table S2. C4 domain-like sequences in database.
Supplemental Table S3. Primers used for constructs in this work.
Supplemental Table S4. Primers and Taq-Man probes for quantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Drs. Tokitaka Oyama (Kyoto University) and Masayuki Serikawa (Nagoya University) for their discussions and for their efforts in transient analysis using Lemna. We also thank Dr. Tsuyoshi Tanaka (National Institute of Agrobiological Sciences) for discussions and for construction of the molecular phylogenetic trees and Dr. Shoji Sugano (National Institute of Agrobiological Sciences) for biochemical technical advice.
This work was supported by the Integrated Research Project for Plant, Insect, and Animal using Genome Technology (grant no. IP1001 to T.I. and M.Y.), by Grants-in-Aid for Scientific Research in Priority Areas from the Program for the Promotion of Basic Research Activities for Innovative Biosciences to T.I., and by Genomics for Agricultural Innovation (grant no. GPN0001 to T.I. and M.Y.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Takeshi Izawa (tizawa@nias.affrc.go.jp) and Masahiro Yano (myano@nias.affrc.go.jp).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR (2003) A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6 251–257 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Allada R, Meissner RA (2005) Casein kinase 2, circadian clocks, and the flight from mutagenic light. Mol Cell Biochem 274 141–149 [DOI] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Coupland G (2004) The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol 135 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11 550–558 [DOI] [PubMed] [Google Scholar]
- Izawa T (2007. a) Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot 58 3091–3097 [DOI] [PubMed] [Google Scholar]
- Izawa T (2007. b) Daylength measurements by rice plants in photoperiodic short-day flowering. Int Rev Cytol 256 191–222 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carré IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135 767–774 [DOI] [PubMed] [Google Scholar]
- Krick R, Aschrafi A, Hasgun D, Arnemann J (2006) CK2-dependent C-terminal phosphorylation at T300 directs the nuclear transport of TSPY protein. Biochem Biophys Res Commun 341 343–350 [DOI] [PubMed] [Google Scholar]
- Lee Y, Lloyd AM, Roux SJ (1999) Antisense expression of the CK2 alpha-subunit gene in Arabidopsis: effects on light-regulated gene expression and plant growth. Plant Physiol 119 989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang H, Gao P, Xü J, Xü T, Wang J, Wang B, Lin C, Fu YF (2009) Analysis of clock gene homologs using unifoliolates as target organs in soybean (Glycine max). J Plant Physiol 166 278–289 [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17 349–368 [DOI] [PubMed] [Google Scholar]
- Miwa K, Serikawa M, Suzuki S, Kondo T, Oyama T (2006) Conserved expression profiles of circadian clock-related genes in two Lemna species showing long-day and short-day photoperiodic flowering responses. Plant Cell Physiol 47 601–612 [DOI] [PubMed] [Google Scholar]
- Miyata Y, Nishida E (2004) CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Mol Cell Biol 24 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Putterill J, Ohkoshi Y (2006) Kinase and phosphatase: the cog and spring of the circadian clock. Int Rev Cytol 250 47–72 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nakamichi N (2005) Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol 46 677–685 [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J, Carme Espunya M, Platara M, Arino J, Martinez MC (2008) A role for protein kinase CK2 in plant development: evidence obtained using a dominant-negative mutant. Plant J 55 118–130 [DOI] [PubMed] [Google Scholar]
- Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T (2003) The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol 44 1229–1236 [DOI] [PubMed] [Google Scholar]
- Murakami M, Tago Y, Yamashino T, Mizuno T (2007) Comparative overviews of clock-associated genes of Arabidopsis and Oryza sativa. Plant Cell Physiol 48 110–121 [DOI] [PubMed] [Google Scholar]
- Okada R, Kondo S, Satbhai SB, Yamaguchi N, Tsukuda M, Aoki S (2009) Functional characterization of CCA1/LHY homolog genes, PpCCA1a and PpCCA1b, in the moss Physcomitrella patens. Plant J 60 551–563 [DOI] [PubMed] [Google Scholar]
- Olsten ME, Litchfield DW (2004) Order or chaos? An evaluation of the regulation of protein kinase CK2. Biochem Cell Biol 82 681–693 [DOI] [PubMed] [Google Scholar]
- Pagano MA, Cesaro L, Meggio F, Pinna LA (2006) Protein kinase CK2: a newcomer in the ‘druggable kinome.’ Biochem Soc Trans 34 1303–1306 [DOI] [PubMed] [Google Scholar]
- Perales M, Portoles S, Mas P (2006) The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J 46 849–860 [DOI] [PubMed] [Google Scholar]
- Pinna LA (1994) A historical view of protein kinase CK2. Cell Mol Biol Res 40 383–390 [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12 204–217 [DOI] [PubMed] [Google Scholar]
- Portoles S, Mas P (2007) Altered oscillator function affects clock resonance and is responsible for the reduced day-length sensitivity of CKB4 overexpressing plants. Plant J 51 966–977 [DOI] [PubMed] [Google Scholar]
- Riera M, Figueras M, Lopez C, Goday A, Pages M (2004) Protein kinase CK2 modulates developmental functions of the abscisic acid responsive protein Rab17 from maize. Proc Natl Acad Sci USA 101 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G, Sommer SS (1990) The “megaprimer” method of site-directed mutagenesis. Biotechniques 8 404–407 [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Smith EM, Lin JM, Meissner RA, Allada R (2008) Dominant-negative CK2alpha induces potent effects on circadian rhythmicity. PLoS Genet 4 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA 95 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM (1999) The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci USA 96 12362–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Izawa T, Oikawa T, Shimamoto K (2001) Light regulation of circadian clock-controlled gene expression in rice. Plant J 26 607–615 [DOI] [PubMed] [Google Scholar]
- Taghli-Lamallem O, Hsia C, Ronshaugen M, McGinnis W (2008) Context-dependent regulation of Hox protein functions by CK2 phosphorylation sites. Dev Genes Evol 218 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc Natl Acad Sci USA 98 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N, Saito S, Tanaka Saito C, Nanjo T, Shinohara K, Uemura M (2009) Molecular phylogeny and expression of poplar circadian clock genes, LHY1 and LHY2. New Phytol 181 808–819 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weing X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40 761–767 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.