Technological advances in biological experimentation are now enabling researchers to investigate living systems on an unprecedented scale by studying genomes, proteomes, or molecular networks in their entirety. Genomics technologies have led to a paradigm shift in biological experimentation because they measure (profile) most or even all components of one class (e.g. transcripts, proteins, etc.) in a highly parallel way. Whether gene expression analysis using microarrays, proteome and metabolome analysis using mass spectrometry, or large-scale screens for genetic interactions, high-throughput profiling technologies provide a rich source of quantitative biological information that allows researchers to move beyond a reductionist approach by both integrating and understanding interactions between multiple components in cells and organisms (Fig. 1; for a recent update of bioinformatics tools, see Pitzschke and Hirt, 2010). Currently, most genomics experiments involve profiling transcripts, proteins, or metabolites. Increasing efforts to complement molecular data with phenotypic information will further advance our understanding of the quantitative relationships between molecules in directing systems behavior and function. In the following Update we will briefly review recent advances in the field and highlight advantages and limitations of current approaches to develop models of genetic and molecular networks that aim to describe emergent properties of plant systems.
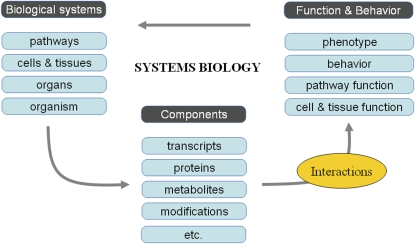
Figure 1.
Relationships between supracellular components (biological systems), intracellular components, and the function and behavior of these components are revealed by the interaction of individual components. Systems biological approaches aim at modeling these interactions to find primary relationships and to distinguish causality and effect. The understanding of how these interactions are regulated allows making predictions on function, behavior, and survival.
GENOMICS TECHNOLOGIES: THE POWER OF GENOME-SCALE QUANTITATIVE DATA RESOLUTION PROFILING TRANSCRIPTOMES
Transcript profiling offers the largest coverage and a wide dynamic range of gene expression information and can often be performed genome wide. Microarrays are currently most popular for transcript profiling and can be readily afforded by many laboratories. Various commercial and academic microarray platforms exist that vary in genome coverage, availability, specificity, and sensitivity (Table I). Microarrays manufactured by Affymetrix are probably most commonly used in plant biology (Redman et al., 2004; Rehrauer et al., 2010), but commercial arrays from Agilent or arrays from the academic Complete Arabidopsis Transcriptome MicroArray (CATMA) consortium are often used as well (for review, see Busch and Lohmann, 2007). Serial analysis of gene expression (SAGE) and massively parallel signature sequencing (MPSS) are well-established alternatives to microarrays. Both techniques can be superior to microarrays because they do not depend on prior probe selection. More recently, direct sequencing of transcripts by high-throughput sequencing technologies (RNA-Seq) has become an additional alternative to microarrays and is superseding SAGE and MPSS (Busch and Lohmann, 2007). Like SAGE and MPPS, RNA-Seq does not depend on genome annotation for prior probe selection and avoids biases introduced during hybridization of microarrays. On the other hand, RNA-Seq poses novel algorithmic and logistic challenges, and current wet-lab RNA-Seq strategies require lengthy library preparation procedures. Therefore, RNA-Seq is the method of choice in projects using nonmodel organisms and for transcript discovery and genome annotation. Because of their robust sample processing and analysis pipelines, often microarrays are still a preferable choice for projects that involve large numbers of samples for profiling transcripts in model organisms with well-annotated genomes. Tools such as Genevestigator (Hruz et al., 2008) and MapMan (Usadel et al., 2009) allow researchers to organize large gene expression datasets and analyze them for relational networks within a single experiment or across many experiments (contextual meta-analysis).
Table I.
Advantages and disadvantages of various technologies for the measurement of transcript and protein abundance
A systematic performance assessment for the different protein quantification techniques was recently conducted (Turck et al., 2007) and a detailed description of the different quantification techniques along with examples for application in the plant field is available (Baginsky, 2009).
| Technologies | Advantages | Disadvantages |
|---|---|---|
| Transcripts | ||
| MPSS | Sequences do not need to be known in advance | Relatively expensive, laborious |
| Microarrays | Genome wide, relatively cheap, streamlined handling, oligos | Sequences must be known in advance; limited sensitivity due to hybridization |
| Quantitative reverse transcription-PCR | High precision and high sensitivity Increasingly multiplexed | Not genome wide; data normalization sensitive to method/choice of reference genes |
| High-throughput sequencing | Sequences do not need to be known in advance; possibility to sequence very short sequences | Expensive at the moment, few solutions for downstream analysis; direct read out |
| Proteins | ||
| Relative quantification via iTRAQ | Established labeling protocol with stable isotopes, good reproducibility, relevant regulation factor can be determined from the data, multiplexing to up to eight samples, produces good quality tandem mass spectrometry spectra | Cost and effort, the analysis software is still not optimal, fluctuations between different softwares possible |
| Relative quantification via stable isotope labeling with amino acids in cell culture | Established protocol for the labeling of cell culture proteins, reliable quantification possible | Restricted to cell culture |
| Relative quantification via extracted ion chromatograms | Comes at no additional costs, software tools for alignment and normalization are available (e.g. SuperHirn; Mueller et al., 2007) | Only applicable to very similar samples and very similar liquid chromatography-mass spectrometry runs, done within a small time window, baseline normalization is sometimes a problem |
| Absolute quantification via AQUA peptides | Highly sensitive absolute quantification on the basis of isotopically labeled PTPs, targeted analyses possible via specific scan methods (e.g. SRM) | Finding suitable PTPs and characteristic parent to daughter ion transitions not straightforward, selectivity of the PTP transitions not always unambiguous |
| Absolute quantification via QconCAT | Excellent for the quantification of protein complex stoichiometry, lower cost compared to AQUA, PTPs are synthesized in a biological system | Unsuitable for the quantification of posttranslational modifications, optimization necessary, exact quantification of the standard is vital, incompatible with sample fractionation |
| Absolute quantification via protein standard absolute quantification | Excellent for the quantification of individual, low abundance proteins, compatible with fractionation | Restricted to few proteins, up scaling difficult, quantifications of posttranslational modifications not possible |
| Absolute quantification via normalized spectral counting (APEX; Lu et al., 2007) | No additional costs, produces reliable results with large-scale datasets | Quantification of individual proteins must be validated by additional tools, unreliable for small datasets |
PROFILING EPIGENOMES AND TRANSCRIPTION FACTOR BINDING
Much control of gene expression occurs at the level of transcription, and information on genome-wide chromatin profiles (epigenomes) and transcription factor binding to promoters is needed to decipher the inherent logic of transcriptional regulation. Chromatin immunoprecipitation (ChIP) coupled to microarray analysis (ChIP-chip) or high-throughput sequencing (ChIP-Seq) can generate such data. In plants, DNA methylation, repressive and activating chromatin marks, as well as histone variants have been mapped onto the genome (for review, see Zhang, 2008), but because such marks are expected to differ between cell types and developmental stages, more targeted epigenome profiling is needed in the future. Targeted analysis of DNA methylation during seed development, for instance, revealed unexpected genome-wide demethylation (Gehring et al., 2009; Hsieh et al., 2009). ChIP-chip was also used for global mapping of binding sites of transcription factors such as TGA2 and SEPALLATA3 and to refine definitions of binding motifs that were previously determined by in vitro experiments (Thibaud-Nissen et al., 2006; Kaufmann et al., 2009). It was found that SEPALLATA3 is a key component in the regulatory transcriptional network underlying the formation of floral organs. In a comparative experiment ChIP-chip and ChIP-Seq gave very similar results (Kaufmann et al., 2009). This is encouraging because bias introduced by the profiling technology seems not to severely confound studies on global protein-binding profiles. Currently, work is going on in several laboratories to establish a compendium of transcription factor binding sites in Arabidopsis (Arabidopsis thaliana). Thus, more genome-wide data sets are in reach that could provide causal explanations for transcriptional profiles.
PROFILING PROTEOMES
Gene expression is a highly regulated, multistep process, and it is impossible to predict the exact protein concentration or activity from the measurement of mRNA levels. Proteomics has therefore become a key tool in systems biology because it provides quantitative and structural information about proteins, which are the major functional determinants of cells. Phenotypic alterations associated with genetic perturbations often result from changes in protein accumulation or stability, or changes in protein posttranslational modifications, which can disrupt protein-protein interactions and network connectivity (Gstaiger and Aebersold, 2009). Quantitative protein information complements data from transcriptional profiling and metabolomics. It represents a key link between different levels of gene expression regulation and provides insights into their causal relationships. Unlike transcriptional profiling, however, comprehensive proteome analysis remains challenging, and information about proteome complexity and dynamics is far from complete (Cox and Mann, 2007). Moreover, the rate of metabolite synthesis is often controlled by regulatory posttranslational modifications of enzymes and not only by their abundance. Information about quantitative relationships between RNA and protein accumulation, posttranslational protein modifications, and metabolite levels is therefore required to fully understand regulatory circuits that control systems behavior and function.
Protein quantification can be absolute or relative (Table I). While relative protein quantification mostly depends on stable isotopes, absolute quantification of comprehensive protein sets is much more difficult. Recent improvements in statistical data evaluation and increasing accuracy of mass spectrometry instruments allow quantifying large numbers of proteins in shotgun-type experiments on the basis of spectral counting (Lu et al., 2007). This method is reliable and comparable to most other quantification methods, including two-dimensional PAGE-based protein staining; however, the protein dataset must be very large. More accurate information about the exact in vivo concentration of individual proteins requires specialized targeted approaches.
Current methods for absolute protein quantification include isotope dilution strategies using isotopically labeled peptides as internal standards (for a comprehensive review, see Brun et al., 2009). Signature peptides for internal standardization are characteristic for a protein of interest, and are often referred to as proteotypic peptides (PTPs). In AQUA, PTPs are added to analytical protein samples in known concentrations. The protein samples are subsequently scanned for PTPs of interest. Using the extracted ion chromatograms the native peptide can then be quantified relative to the added PTP (Kuster et al., 2005). A modification of this strategy accounts for quantification errors derived from incomplete tryptic digest of the analytical sample. In QconCAT (for quantification concatamer), a synthetic protein with concatenated, isotopically labeled PTPs is expressed as recombinant protein in a biological system, added to the sample prior to Trypsin treatment and carried through the digestion procedure, such that losses from incomplete tryptic digestion will also affect the quantity of the PTPs. Both the AQUA and the QconCAT strategies are incompatible with upstream fractionation techniques, which is a potential problem in biomarker quantification. A way around this constraint is offered by the protein standard absolute quantification strategy, which uses isotopically labeled protein standards that are added to the sample prior to fractionation. Several prediction tools exist that help to define the most suitable PTPs for the detection and quantification of specific proteins. However, only experimental data provide the necessary reliability for PTP selection because in practice PTP prediction often deviates from experimental observations. Therefore, efforts are under way to catalogue PTPs for model organism proteomes. Proteome maps for Arabidopsis generated PTPs for 4,105 proteins, many of which may be optimal for the detection of proteins in different organs (Baerenfaller et al., 2008).
Similar quantitative approaches are also used for metabolites, because in addition to RNA and protein levels, understanding the function and behavior of metabolic networks requires global information about metabolite concentrations and fluxes as well. In recent years, much progress has been made in metabolic profiling, and the interested reader is referred to recent reviews (e.g. Issaq et al., 2009, and refs. therein).
TRANSCRIPTS AND MORE TRANSCRIPTS: WHAT CAN WE LEARN FROM GENE EXPRESSION ANALYSIS?
During the analysis of large gene expression datasets the researcher is often confronted with several questions. How do we interpret a mathematical relationship between genes or between genes and conditions? For example, does a high correlation between two genes mean that they are coregulated, or could one of them be the positive regulator of the other? Or can we assume that they are involved in the same pathway or biological process? Although it is not possible to answer these questions conclusively from gene expression data alone, a number of parallel approaches can be useful to distinguish between different scenarios. For example, Gene Ontology enrichment analysis can provide confidence that a given gene cluster is enriched in genes that are known to have a common function, cellular location, or biological process. Similarly, conserved cis-regulatory elements in the promoters of genes from the same cluster indicate that they are likely coregulated. Although these methods do not establish proof of the nature of the relationship between genes, they allow formulating hypotheses that can be tested in the laboratory. In summary, although gene expression analysis by itself is rather descriptive (i.e. describing how genes respond to various test conditions or tissues), it is a valuable validation tool and an excellent starting point to study novel cellular process and to formulate novel hypotheses.
A major challenge of genome-scale transcription analysis is the very large number of predictors (genes) compared to a generally small number of measurements (microarrays). Without appropriate statistical measures to correct for multiple testing and including false discovery rates, almost any approach will yield significant genes, including many false positives. The creation of large databases in recent years has brought an additional layer of complexity and precautions to take (see Table II). For example, large databases such as Genevestigator (Hruz et al., 2008) not only profile a large number of genes, but also allow contextual meta-analysis of several hundred conditions, each of which is covered by only a small number of replicates (usually 3–5). While some genes will respond to a small number of conditions and therefore their expression is easier to contextualize and interpret, other genes will respond to dozens or hundreds of conditions. It is often very difficult to distinguish primary effects from secondary effects, because the intensity of the effect does not necessarily relate to the direct involvement of the corresponding condition in regulating a specific target gene. Breaking down these effects into local patterns (e.g. by using a biclustering algorithm; Prelic et al., 2006) helps in finding out conditions that are more directly linked to the gene of interest.
Table II.
Overview of some of the most popular plant gene expression microarray platforms and the number of available experiments in ArrayExpress
The Arabidopsis ATH1 array is the most frequently used microarray, followed by the CATMA 25k and 23k arrays. In all, approximately 750 Arabidopsis microarray experiments have been published so far. Rice (Oryza sativa) and barley (Hordeum vulgare) are the second and third plant species in terms of microarray experiments published. Soybean (Glycine max) also has a high number of arrays, but this is due to a single very large experiment containing 2,521 arrays. IPK, Leibniz Institute of Plant Genetics and Crop Plant Research; TIGR, The Institute for Genomic Research.
| Species | Provider | Array Format | Array Name | Experiments | Arrays |
|---|---|---|---|---|---|
| Arabidopsis | Affymetrix | 8K | AG | 41 | 352 |
| Affymetrix | 22K | ATH1 | 554 | 8,895 | |
| Agilent | 22K | Arabidopsis 2 | 34 | 253 | |
| Agilent | 44K | Arabidopsis 3 | 7 | 60 | |
| CATMA | 25K | CATMA2_URGV to CATMA2.3_URGV | 83 | 851 | |
| CATMA | 23K | CATMA Arabidopsis 23K array | 50 | 1,290 | |
| TIGR | 26K | TIGR Arabidopsis whole genome | 6 | 264 | |
| Rice | Affymetrix | 57K | GeneChip Rice Genome Array | 29 | 418 |
| Agilent | 21K | Agilent Rice Oligo Microarray | 22 | 164 | |
| Barley | Affymetrix | 22K | GeneChip Barley Genome Array | 35 | 1,165 |
| IPK | 6K + 4K | IPK barley PGRC1_A and B | 7 | 324 | |
| Medicago | Affymetrix | 61K | GeneChip Medicago Genome Array | 19 | 218 |
| Maize | Affymetrix | 17K | GeneChip Maize Genome Array | 22 | 370 |
| Soybean | Affymetrix | 61K | GeneChip Soybean Genome Array | 22 | 3,236 |
| Tomato (Solanum lycopersicum) | Affymetrix | 10K | GeneChip Tomato Genome Array | 6 | 127 |
| Grape (Vitis vinifera) | Affymetrix | 16K | GeneChip Vitis vinifera Genome Array | 6 | 239 |
| Wheat (Triticum aestivum) | Affymetrix | 61K | GeneChip Wheat Genome Array | 25 | 811 |
| Total | 968 | 19,037 |
APPROACHING THE TARGET: FROM ORGANS TO TISSUES AND CELLS
Most transcript and protein profiling experiments analyze mixtures of tissues containing different cell types and organelles. This approach reveals certain global patterns, but quantitative analyses and modeling is limited with such complex data. Therefore methods for organ (or better) cell-type-specific transcript and protein profiling as well as for organelle-specific proteomics are needed. Four types of approaches are now commonly used to sample RNA and/or proteins from selected cell types: (1) micropipetting, (2) laser capture microdissection (LCM), (3) protoplasting and sorting, and (4) polysome immunopurification (for review, see Zanetti et al., 2005; Hennig, 2007; Nelson et al., 2008).
Micropipetting using microcapillaries directly extracts the contents from selected cells. It has been successfully applied to various leaf cell types and for phloem but extraction is more difficult from internal cells. LCM involves sectioning of frozen or embedded tissue, and subsequent dissection of the region of interest using laser excision. Applications of LCM include studies of vascular tissue, epidermis, and pericycle in maize (Zea mays) and seed development in Arabidopsis. Micropipetting and LCM are usually very labor intensive and difficult for isolation of small cells such as in meristems. Because of the limited amount of material that can be captured, they work well for transcript profiling, which can use amplification steps, but provide only a very small coverage of the proteome. As an alternative, protoplasting and cell sorting offers rapid and accurate isolation of RNA from small cells. Specific tissues or cell types that are labeled by expression of GFP are isolated by protoplasting and sorted through a fluorescence-activated cell sorter. Millions of cells can be processed within 1 to 2 h, but care has to be taken to exclude changes in gene expression profiles by sample processing. This technique was successfully applied to measure genome-wide expression profiles in more than 15 root regions, establishing a compendium of digital in situ data (Birnbaum et al., 2003; Cartwright et al., 2009). It will be interesting to test whether this approach can also be used for protein profiling. Polysome immunopurification is based on the tissue-specific expression of the FLAG-tagged ribosomal protein L18 in transgenic plants (Zanetti et al., 2005). In contrast to micropipetting, LCM, and sorting of protoplasts, which all can be used to isolate total cellular RNA, polysome immunopurification can be used to isolate transcripts that are associated with ribosomes (translatome). Discrepancies between total RNA levels and representation translatome can reveal regulation at the level of translation (Mustroph et al., 2009). In the future, translatome datasets, which bridge transcriptomics and proteomics, can help to interpret unusual transcript-to-protein ratios (see below).
Alternatively, it is possible to identify cell-type-specific transcripts and proteins by comparing wild-type plants with mutants that lack specific cells or tissue types. In Arabidopsis, for instance, a series of homeotic mutants that lack various floral organs was used to identify several hundreds of floral organ-specific genes (Wellmer et al., 2004). If no appropriate mutants exist, specific cell types can be genetically ablated by expression of a cell-autonomous toxin, such as diphtheria toxin subunit A or RNase, under the control of cell-type-specific promoters. Again, these approaches have been proven to work for transcript profiling (Tung et al., 2005) but it remains to be tested whether they could be useful for protein profiling.
DECREASING COMPLEXITY BY ORGANIZING ORGAN AND SUBCELLULAR PROTEOMES
Systematic analysis of accurate protein localization is essential to understand cellular networks in the context of compartmentalization, which is a fundamental design principle of eukaryotic cells. Organelle proteomics has therefore become a very active research field. Until recently, the protein inventory of cell organelles was based on proteins from isolated organelles, such as mitochondria, chloroplast, and peroxisomes (Lilley and Dupree, 2007; Baginsky, 2009). This approach has limitations because true low-abundant organelle proteins often cannot be distinguished from contaminating proteins. Two approaches have been used to deal with this problem. First, a recently reported isolation procedure for mitochondria used the electrostatic characteristics of the mitochondrial surface to separate mitochondria from other organelles in an electric field. This procedure results in mitochondria preparations with higher purity, but the yield is low (Eubel et al., 2007). Second, information about the quantitative distribution of proteins along density gradients has been used to determine if a protein was enriched by the organellar isolation procedure. In practice, the abundance distribution profile of unknown proteins is compared to known organelle marker proteins. This strategy is referred to as protein correlation profiling (Foster et al., 2006) or LOPIT (Dunkley et al., 2006).
Both procedures, however, are of limited use for the analysis of proteome dynamics in response to a stimulus because the long time that is needed to isolate and purify organelles affects their proteome properties. This is especially critical for transient posttranslational protein modifications. Thus, proteome dynamics is best analyzed at the cell or tissue level, followed by sorting of proteins into their respective organelle a posteriori. This strategy is now possible because substantial information about the protein complement of different cell organelles has accumulated (a comprehensive collection of proteome databases is for example available in Lu and Last, 2009). The SUBA database is most suitable for this purpose, because it is frequently updated and well maintained. SUBA generates lists of organelle proteins using reliability criteria, for example evidence from several different proteomics studies, targeting prediction, or GFP-localization assays, or a combination of this information (Heazlewood et al., 2007). For the chloroplast, two proteome reference tables have been established (Yu et al., 2008; Reiland et al., 2009). The overlap between these two proteome reference tables has generated a list of 1,156 proteins that can be considered high-confidence chloroplast proteins.
Although the number of organelle proteins is constantly increasing, it is not clear when an organelle proteome can be considered complete. Organelle proteomes are dynamic and functional organelle proteomes differ significantly during development, in different cell types or tissues, and in different conditions. This problem can be addressed by considering organelles as cellular subnetworks and applying flux-balance modeling to assess network consistency. Initial modeling approaches with mitochondria and chloroplasts focused on a limited number of reactions, such as those of the Calvin cycle, amino acid biosynthesis, or the tricarboxylic acid cycle. Also, mitochondrial network reconstructions based on proteomics data are available and the existing models allow prediction of metabolite accumulation for a limited number of metabolites (Vo and Palsson, 2007). A recent flux-balance model of the primary metabolism in Chlamydomonas reinhardtii localized reactions into chloroplasts, mitochondria, and the cytosol and assessed systematically the contribution of different organelles to biomass production (Boyle and Morgan, 2009). The above examples illustrate the excellent suitability of metabolic network reconstruction to identify gaps in existing knowledge.
THE CHALLENGE OF DATA INTEGRATION: GENOME-SCALE ANALYSIS OF RNA-PROTEIN CORRELATIONS
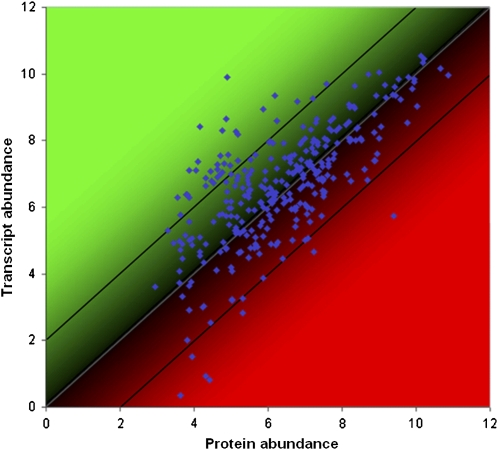
Quantitative information about protein accumulation at genome scale offers entirely new insights into network function and the behavior of organs, tissues, and cells. Because gene expression is regulated at different levels, a comparison between transcript and protein accumulation can provide information about the rate of protein translation and the degree of posttranscriptional regulation. We have recently analyzed the correlation between protein and transcript abundance in representative samples from different plant organs and found mostly positive correlations in the range from 0.5 to 0.68 (Baerenfaller et al., 2008). The lowest correlation was observed for seeds, which accumulate stable storage proteins whose abundance is largely uncoupled from transcription. The highest correlation was obtained in leaves, suggesting that the most abundant photosynthetic proteins are predominantly regulated at the transcriptional level. It is clear that such a genome-scale analysis only offers a global view of regulatory events and does not allow a systematic assessment of individual enzyme regulation. A more refined comparison of protein and transcript levels showed that the correlation between transcript and protein abundance can vary significantly between different pathways (Kleffmann et al., 2004) and most likely also between different enzymes in the same pathway. Figure 2 shows an example of a correlation analysis of a representative leaf transcriptome and proteome for a selection of 345 genes/proteins from primary and secondary metabolism pathways. Although the data was collected from various sources and summarized (see also Baerenfaller et al., 2008), the protein-to-transcript ratio was similar for most proteins, indicating that this analysis is robust. The majority of proteins in most metabolic pathways were found within the typical protein-to-transcript range. Only a small fraction of proteins deviated significantly from these ratios, both up or down and to a similar extent. This preliminary genome-scale correlation analysis revealed combined and consistent effects for particular pathways that are worth considering in further experimentation, for example by taking into account the effects of circadian rhythm, light, and nutrient status. The generation of protein and transcript data from the exact same samples is urgently needed to precisely address these questions and to understand how and under which conditions the protein-to-transcript ratio varies.
Figure 2.
Correlation analysis of transcript and protein abundance in Arabidopsis leaves based on 345 genes from various primary and secondary metabolism pathways. Transcript abundance was calculated as a representative expression vector derived from multiple Affymetrix ATH1 array measurements from leaf samples (data from Genevestigator, Hruz et al., 2008). The proteome data was obtained from distinct leaf samples. Approximately 20% of these genes/proteins had ratios of protein to transcript abundance deviating strongly from 1.
FROM GENES AND PROTEINS TO NETWORK DISCOVERY
Network discovery is a generic term describing the effort of elucidating the nature of relationships between molecules and associated properties emerging from of a biological network. Multiple types of networks have been described with respect to the types of molecules involved and the dimension of the molecular network (genome scale or small scale). While genome-/proteome-/metabolome-scale analyses aim at identifying novel properties of the global network, smaller scale networks usually incorporate additional data types that cannot be obtained on a global scale and use models that allow a more precise prediction of network behavior. A more recent development is the integration of various networks into an evolutionary ecology of networks, in which networks are considered as strategies that interact and possibly compete for resources (Weitz et al., 2007).
Metabolic network reconstructions have received increasing attention in the last few years and several genome-scale models are now available for microorganisms and human tissues. However, none of the existing models can currently provide a full view of all reactions in a cell organelle. For example, even the most advanced genome-scale models for Saccharomyces cerevisiae only contain approximately 1,200 of the predicted approximately 2,200 metabolic genes and only approximately 70% of the modeled reactions are functional (Feist et al., 2009). The situation is worse for higher organisms, and for plants no genome-scale model exists to date although efforts to build knowledge databases are under way (Tsesmetzis et al., 2008). Progress in metabolic network reconstruction critically depends on the functional annotation of the genes that encode proteins with unknown functions, which is still the case for about 30% of the Arabidopsis genome. Accordingly, current plant metabolic network reconstruction focuses mostly on specific pathways such as fatty acid synthesis or Asp metabolism (Chen et al., 2009; Curien et al., 2009). Systematic efforts to improve genome annotation are under way, and the community is well connected via The Arabidopsis Information Resource (www.arabidopsis.org), which supports and facilitates the exchange of material and information. One example of such a functional characterization pipeline is the Chloroplast 2010 project that was launched at Michigan State University (Lu et al., 2008). Here, homozygous knockout lines for all known chloroplast proteins are generated and phenotypically characterized, also at the metabolite level. In brief, a dramatic improvement in the functional annotation of genes is key to networks with improved quality and consistency.
Similar efforts are under way to construct plant transcriptional regulatory networks, for example those that control flower and root development (Grieneisen et al., 2007), photomorphogenesis (Jiao et al., 2007; Nemhauser, 2008), or the circadian clock (Zeilinger et al., 2006). On this smaller scale, graphical models, in particular Bayesian networks, are increasingly used in reverse engineering of genetic regulatory networks. Graphical models, such as sparse graphical Gaussian modeling, are powerful for a small number of genes and have been used to model the isoprene biosynthesis pathway network from temporal transcriptome data to discover new genes associated with this network (Wille et al., 2004). A similar graphical Gaussian modeling was carried out on a larger set of genes in Arabidopsis and allowed to discover novel components in various networks (Ma et al., 2007). In the future, analysis of transcriptional regulatory networks needs to incorporate also epigenome and transcription factor binding data from ChIP-chip and ChIP-Seq experiments.
Regulatory network construction is also increasingly being used in plant breeding. For example, Keurentjes et al. (2007) showed that for many genes variation in expression could be explained by expression quantitative trait loci using recombinant inbred lines of Arabidopsis. By combining expression quantitative trait loci mapping and regulator candidate gene selection, gene regulatory networks for flowering time could be built that were in agreement with published data. The combination of omics data with quantitative genetics data is expected to facilitate the understanding of complex regulatory networks governing important phenotypic traits such as yield, pathogen resistance, and nutrient acquisition and utilization. A variety of models exist for mapping complex traits and linking phenotypic outputs to changes in genomic regions (Hammer et al., 2006).
Acknowledgments
We apologize to all colleagues whose work could not be cited due to space constraints.
This work was supported by the European Union (EU Framework Program 6, AGRON-OMICS; grant no. LSHG–CT–2006–037704), the Swiss National Science Foundation, CTI (Swiss Innovation Promotion Agency), ETH Zurich, and the Functional Genomics Center Zurich for our profiling experiments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wilhelm Gruissem (wgruissem@ethz.ch).
References
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320 938–941 [DOI] [PubMed] [Google Scholar]
- Baginsky S (2009) Plant proteomics: concepts, applications, and novel strategies for data interpretation. Mass Spectrom Rev 28 93–120 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Morgan JA (2009) Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst Biol 3 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun V, Masselon C, Garin J, Dupuis A (2009) Isotope dilution strategies for absolute quantitative proteomics. J Proteomics 72 740–749 [DOI] [PubMed] [Google Scholar]
- Busch W, Lohmann JU (2007) Profiling a plant: expression analysis in Arabidopsis. Curr Opin Plant Biol 10 136–141 [DOI] [PubMed] [Google Scholar]
- Cartwright DA, Brady SM, Orlando DA, Sturmfels B, Benfey PN (2009) Reconstructing spatiotemporal gene expression data from partial observations. Bioinformatics 25 2581–2587 [DOI] [PubMed] [Google Scholar]
- Chen M, Mooney BP, Hajduch M, Joshi T, Zhou M, Xu D, Thelen JJ (2009) System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol 150 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2007) Is proteomics the new genomics? Cell 130 395–398 [DOI] [PubMed] [Google Scholar]
- Curien G, Bastien O, Robert-Genthon M, Cornish-Bowden A, Cárdenas ML, Dumas R (2009) Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol Syst Biol 5 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Lee CP, Kuo J, Meyer EH, Taylor NL, Millar AH (2007) Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J 52 583–594 [DOI] [PubMed] [Google Scholar]
- Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO (2009) Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol 7 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125 187–199 [DOI] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AFM, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449 1008–1013 [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Aebersold R (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet 10 617–627 [DOI] [PubMed] [Google Scholar]
- Hammer G, Cooper M, Tardieu F, Welch S, Walsh B, van Eeuwijk F, Chapman S, Podlich D (2006) Models for navigating biological complexity in breeding improved crop plants. Trends Plant Sci 11 587–593 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35 D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L (2007) Patterns of beauty—omics meets plant development. Trends Plant Sci 12 287–293 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendrop F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD (2009) Analytical and statistical approaches to metabolomics research. J Sep Sci 32 2183–2199 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8 217–230 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7 e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJ, Vreugdenhil D, Koornneef M, Jansen RC (2007) Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA 104 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14 354–362 [DOI] [PubMed] [Google Scholar]
- Kuster B, Schirle M, Mallick P, Aebersold R (2005) Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6 577–583 [DOI] [PubMed] [Google Scholar]
- Lilley KS, Dupree P (2007) Plant organelle proteomics. Curr Opin Plant Biol 10 594–599 [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25 117–124 [DOI] [PubMed] [Google Scholar]
- Lu Y, Last RL (2009) Web-based Arabidopsis functional and structural genomics resources. In The Arabidopsis Book. The American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0118, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, Dellapenna D, Ohlrogge JB, Osteryoung KW, Weber AP, et al (2008) New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol 146 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ (2007) An Arabidopsis gene network based on the graphical Gaussian model. Genome Res 17 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Muller M (2007) SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7 3470–3480 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Gandotra N, Tausta SL (2008) Plant cell types: reporting and sampling with new technologies. Curr Opin Plant Biol 11 567–573 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL (2008) Dawning of a new era: photomorphogenesis as an integrated molecular network. Curr Opin Plant Biol 11 4–8 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H (2010) Bioinformatic and systems biology tools to generate testable models of signaling pathways and their targets. Plant Physiol 152 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelic A, Bleuler S, Zimmermann P, Wille A, Bühlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22 1122–1129 [DOI] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38 545–561 [DOI] [PubMed] [Google Scholar]
- Rehrauer H, Aquino C, Gruissem W, Henz S, Hilson P, Laubinger S, Naouar N, Patrignani A, Rombauts S, Shu H, et al (2010) AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol 152 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD (2006) Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J 47 152–162 [DOI] [PubMed] [Google Scholar]
- Tung CW, Dwyer KG, Nasrallah ME, Nasrallah JB (2005) Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol 138 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck CW, Falick AM, Kowalak JA, Lane WS, Lilley KS, Phinney BS, Weintraub ST, Witkowska HE, Yates NA (2007) The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 study: relative protein quantitation. Mol Cell Proteomics 6 1291–1298 [DOI] [PubMed] [Google Scholar]
- Tsesmetzis N, Couchman M, Higgins J, Smith A, Doonan JH, Seifert GJ, Schmidt EE, Vastrik I, Birney E, Wu G, et al (2008) Arabidopsis reactome: a foundation knowledgebase for plant systems biology. Plant Cell 20 1426–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ 32 1211–1229 [DOI] [PubMed] [Google Scholar]
- Vo TD, Palsson BO (2007) Building the power house: recent advances in mitochondrial studies through proteomics and systems biology. Am J Physiol Cell Physiol 292 C164–C177 [DOI] [PubMed] [Google Scholar]
- Weitz JS, Benfey PN, Wingreen NS (2007) Evolution, interactions, and biological networks. PLoS Biol 5 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille A, Zimmermann P, Vranová E, Bleuler S, Fürholz A, Hennig L, Laule O, Prelíc A, von Rohr P, Thiele L, et al (2004) Sparse graphical gaussian modeling for genetic regulatory network inference. Genome Biology 5 R92.1–R92.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Li G, Wang G, Sun JC, Wang PC, Wang C, Mi HL, Ma WM, Cui J, Cui YL, et al (2008) Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana. Cell Res 18 1007–1019 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X (2008) The epigenetic landscape of plants. Science 320 489–492 [DOI] [PubMed] [Google Scholar]