Abstract
To investigate sepal/petal/lip formation in Oncidium Gower Ramsey, three paleoAPETALA3 genes, O. Gower Ramsey MADS box gene5 (OMADS5; clade 1), OMADS3 (clade 2), and OMADS9 (clade 3), and one PISTILLATA gene, OMADS8, were characterized. The OMADS8 and OMADS3 mRNAs were expressed in all four floral organs as well as in vegetative leaves. The OMADS9 mRNA was only strongly detected in petals and lips. The mRNA for OMADS5 was only strongly detected in sepals and petals and was significantly down-regulated in lip-like petals and lip-like sepals of peloric mutant flowers. This result revealed a possible negative role for OMADS5 in regulating lip formation. Yeast two-hybrid analysis indicated that OMADS5 formed homodimers and heterodimers with OMADS3 and OMADS9. OMADS8 only formed heterodimers with OMADS3, whereas OMADS3 and OMADS9 formed homodimers and heterodimers with each other. We proposed that sepal/petal/lip formation needs the presence of OMADS3/8 and/or OMADS9. The determination of the final organ identity for the sepal/petal/lip likely depended on the presence or absence of OMADS5. The presence of OMADS5 caused short sepal/petal formation. When OMADS5 was absent, cells could proliferate, resulting in the possible formation of large lips and the conversion of the sepal/petal into lips in peloric mutants. Further analysis indicated that only ectopic expression of OMADS8 but not OMADS5/9 caused the conversion of the sepal into an expanded petal-like structure in transgenic Arabidopsis (Arabidopsis thaliana) plants.
The ABCDE model predicts the formation of any flower organ by the interaction of five classes of homeotic genes in plants (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Jofuku et al., 1994; Pelaz et al., 2000, 2001; Theißen and Saedler, 2001; Pinyopich et al., 2003; Ditta et al., 2004; Jack, 2004). The A class genes control sepal formation. The A, B, and E class genes work together to regulate petal formation. The B, C, and E class genes control stamen formation. The C and E class genes work to regulate carpel formation, whereas the D class gene is involved in ovule development. MADS box genes seem to have a central role in flower development, because most ABCDE genes encode MADS box proteins (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Purugganan et al., 1995; Rounsley et al., 1995; Theißen and Saedler, 1995; Theißen et al., 2000; Theißen, 2001).
The function of B group genes, such as APETALA3 (AP3) and PISTILLATA (PI), has been thought to have a major role in specifying petal and stamen development (Jack et al., 1992; Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996; Kramer et al., 1998; Hernandez-Hernandez et al., 2007; Kanno et al., 2007; Whipple et al., 2007; Irish, 2009). In Arabidopsis (Arabidopsis thaliana), mutation in AP3 or PI caused identical phenotypes of second whorl petal conversion into a sepal structure and third flower whorl stamen into a carpel structure (Bowman et al., 1989; Jack et al., 1992; Goto and Meyerowitz, 1994). Similar homeotic conversions for petal and stamen were observed in the mutants of the AP3 and PI orthologs from a number of core eudicots such as Antirrhinum majus, Petunia hybrida, Gerbera hybrida, Solanum lycopersicum, and Nicotiana benthamiana (Sommer et al., 1990; Tröbner et al., 1992; Angenent et al., 1993; van der Krol et al., 1993; Yu et al., 1999; Liu et al., 2004; Vandenbussche et al., 2004; de Martino et al., 2006), from basal eudicot species such as Papaver somniferum and Aquilegia vulgaris (Drea et al., 2007; Kramer et al., 2007), as well as from monocot species such as Zea mays and Oryza sativa (Ambrose et al., 2000; Nagasawa et al., 2003; Prasad and Vijayraghavan, 2003; Yadav et al., 2007; Yao et al., 2008). This indicated that the function of the B class genes AP3 and PI is highly conserved during evolution.
It has been thought that B group genes may have arisen from an ancestral gene through multiple gene duplication events (Doyle, 1994; Theißen et al., 1996, 2000; Purugganan, 1997; Kramer et al., 1998; Kramer and Irish, 1999; Lamb and Irish, 2003; Kim et al., 2004; Stellari et al., 2004; Zahn et al., 2005; Hernandez-Hernandez et al., 2007). In the gymnosperms, there was a single putative B class lineage that duplicated to generate the paleoAP3 and PI lineages in angiosperms (Kramer et al., 1998; Theißen et al., 2000; Irish, 2009). The paleoAP3 lineage is composed of AP3 orthologs identified in lower eudicots, magnolid dicots, and monocots (Kramer et al., 1998). Genes in this lineage contain the conserved paleoAP3- and PI-derived motifs in the C-terminal end of the proteins, which have been thought to be characteristics of the B class ancestral gene (Kramer et al., 1998; Tzeng and Yang, 2001; Hsu and Yang, 2002). The PI lineage is composed of PI orthologs that contain a highly conserved PI motif identified in most plant species (Kramer et al., 1998). Subsequently, there was a second duplication at the base of the core eudicots that produced the euAP3 and TM6 lineages, which have been subject to substantial sequence changes in eudicots during evolution (Kramer et al., 1998; Kramer and Irish, 1999). The paleoAP3 motif in the C-terminal end of the proteins was retained in the TM6 lineage and replaced by a conserved euAP3 motif in the euAP3 lineage of most eudicot species (Kramer et al., 1998). In addition, many lineage-specific duplications for paleoAP3 lineage have occurred in plants such as orchids (Hsu and Yang, 2002; Tsai et al., 2004; Kim et al., 2007; Mondragón-Palomino and Theißen, 2008, 2009; Mondragón-Palomino et al., 2009), Ranunculaceae, and Ranunculales (Kramer et al., 2003; Di Stilio et al., 2005; Shan et al., 2006; Kramer, 2009).
Unlike the A or C class MADS box proteins, which form homodimers that regulate flower development, the ability of B class proteins to form homodimers has only been reported in gymnosperms and in the paleoAP3 and PI lineages of some monocots. For example, LMADS1 of the lily Lilium longiflorum (Tzeng and Yang, 2001), OMADS3 of the orchid Oncidium Gower Ramsey (Hsu and Yang, 2002), and PeMADS4 of the orchid Phalaenopsis equestris (Tsai et al., 2004) in the paleoAP3 lineage, LRGLOA and LRGLOB of the lily Lilium regale (Winter et al., 2002), TGGLO of the tulip Tulipa gesneriana (Kanno et al., 2003), and PeMADS6 of the orchid P. equestris (Tsai et al., 2005) in the PI lineage, and GGM2 of the gymnosperm Gnetum gnemon (Winter et al., 1999) were able to form homodimers that regulate flower development. Proteins in the euAP3 lineage and in most paleoAP3 lineages were not able to form homodimers and had to interact with PI to form heterodimers in order to regulate petal and stamen development in various plant species (Schwarz-Sommer et al., 1992; Tröbner et al., 1992; Riechmann et al., 1996; Moon et al., 1999; Winter et al., 2002; Kanno et al., 2003; Vandenbussche et al., 2004; Yao et al., 2008). In addition to forming dimers, AP3 and PI were able to interact with other MADS box proteins, such as SEPALLATA1 (SEP1), SEP2, and SEP3, to regulate petal and stamen development (Pelaz et al., 2000; Honma and Goto, 2001; Theißen and Saedler, 2001; Castillejo et al., 2005).
Orchids are among the most important plants in the flower market around the world, and research on MADS box genes has been reported for several species of orchids during the past few years (Lu et al., 1993, 2007; Yu and Goh, 2000; Hsu and Yang, 2002; Yu et al., 2002; Hsu et al., 2003; Tsai et al., 2004, 2008; Xu et al., 2006; Guo et al., 2007; Kim et al., 2007; Chang et al., 2009). Unlike the flowers in eudicots, the nearly identical shape of the sepals and petals as well as the production of a unique lip in orchid flowers make them a very special plant species for the study of flower development. Four clades (1–4) of genes in the paleoAP3 lineage have been identified in several orchids (Hsu and Yang, 2002; Tsai et al., 2004; Kim et al., 2007; Mondragón-Palomino and Theißen, 2008, 2009; Mondragón-Palomino et al., 2009). Several works have described the possible interactions among these four clades of paleoAP3 genes and one PI gene that are involved in regulating the differentiation and formation of the sepal/petal/lip of orchids (Tsai et al., 2004; Kim et al., 2007; Mondragón-Palomino and Theißen, 2008, 2009). However, the exact mechanism that involves the orchid B class genes remains unclear and needs to be clarified by more experimental investigations.
O. Gower Ramsey is a popular orchid with important economic value in cut flower markets. Only a few studies have been reported on the role of MADS box genes in regulating flower formation in this plant species (Hsu and Yang, 2002; Hsu et al., 2003; Chang et al., 2009). An AP3-like MADS gene that regulates both floral formation and initiation in transgenic Arabidopsis has been reported (Hsu and Yang, 2002). In addition, four AP1/AGAMOUS-LIKE9 (AGL9)-like MADS box genes have been characterized that show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis (Hsu et al., 2003; Chang et al., 2009). Compared with other orchids, the production of a large and well-expanded lip and five small identical sepals/petals makes O. Gower Ramsey a special case for the study of the diverse functions of B class MADS box genes during evolution. Therefore, the isolation of more B class MADS box genes and further study of their roles in the regulation of perianth (sepal/petal/lip) formation during O. Gower Ramsey flower development are necessary. In addition to the clade 2 paleoAP3 gene OMADS3, which was previously characterized in our laboratory (Hsu and Yang, 2002), three more B class MADS box genes, OMADS5, OMADS8, and OMADS9, were characterized from O. Gower Ramsey in this study. Based on the different expression patterns and the protein interactions among these four orchid B class genes, we propose that the presence of OMADS3/8 and/or OMADS9 is required for sepal/petal/lip formation. Further sepal and petal formation at least requires the additional presence of OMADS5, whereas large lip formation was seen when OMADS5 expression was absent. Our results provide a new finding and information pertaining to the roles for orchid B class MADS box genes in the regulation of sepal/petal/lip formation.
RESULTS
Isolation of OMADS5, OMADS8, and OMADS9 cDNAs from O. Gower Ramsey
To isolate MADS box genes from O. Gower Ramsey, a strategy that combined reverse transcription (RT)-PCR and 5′ and 3′ RACE was used. A DNA fragment was amplified by RT-PCR using total RNA from young floral buds as a template. Sequence comparison led to the identification of partial sequences for several MADS box genes. The full-length cDNA sequences for two AP3-like B class genes, OMADS5 and OMADS9, and one PI-like B class gene, OMADS8, were isolated.
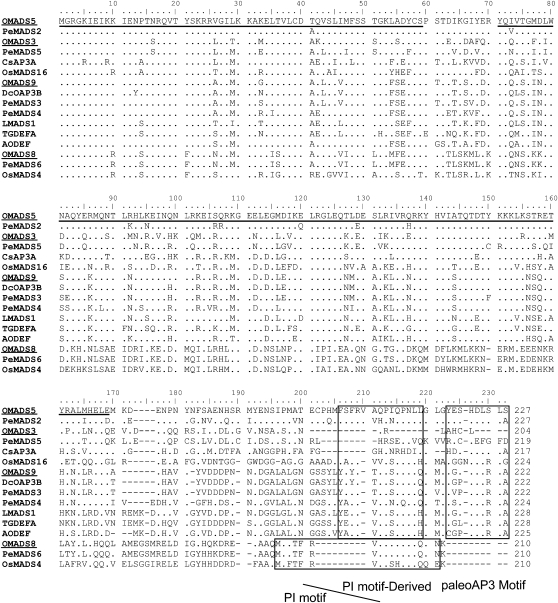
OMADS5 and OMADS9 encoded 227- and 222-amino acid proteins, respectively, that showed high sequence identity to the paleoAP3 lineage of B group MADS box genes. OMADS5 is closely related to clade 1 paleoAP3 genes of orchids, such as PeMADS2 of Phalaenopsis (Tsai et al., 2004) and DcOAP3A of Dendrobium (Xu et al., 2006), and also showed high sequence identity to another O. Gower Ramsey clade 2 paleoAP3 gene, OMADS3, which was previously reported by our laboratory (Hsu and Yang, 2002; Figs. 1 and 2). OMADS9 is closely related to clade 3 paleoAP3 genes, such as DcOAP3B of Dendrobium (Xu et al., 2006) and PeMADS3 of Phalaenopsis (Tsai et al., 2004; Figs. 1 and 2). Within their K boxes, a sequence (QYQRM) matching a conserved motif (Q/HYExM) in AP3 homologs (Kramer et al., 1998) was found (Fig. 1). In their C-terminal regions, two completely consensus PI-derived (FxFRLOPSQPNLH) and paleoAP3 (D[L/I]ITTFALLE) motifs that are unique to B class genes of monocots (Kramer et al., 1998; Moon et al., 1999; Tzeng and Yang, 2001) were identified (Fig. 1). Sequence identity between OMADS5 and OMADS9 and paleoAP3 orthologs indicates that OMADS5 and OMADS9 are O. Gower Ramsey B group MADS box paleoAP3 genes.
Figure 1.
Sequence comparison of OMADS3, OMADS5, OMADS8, and OMADS9 and the related B class MADS domain proteins. The functional MADS box proteins include PeMADS2, PeMADS3, PeMADS4, PeMADS5, and PeMADS6 (P. equestris), LMADS1 (L. longiflorum), OsMADS16 and OsMADS4 (O. sativa), CsAP3A (Crocus sativus), DcOPA3B (Dendrobium crumenatum), TGDEFA (T. gesneriana), and AODEF (Asparagus officinalis). The first and second underlined regions represent the MADS domain and the K domain, respectively. The three blocks in the C-terminal region represent the three motifs conserved among B class MADS box proteins. The paleoAP3 and PI-derived motifs are the two highly conserved motifs for paleoAP3 proteins of monocots. The PI motif is a highly conserved motif for PI orthologs. Amino acid residues identical to OMADS5 are indicated as dots. To improve the alignment, dashes were introduced into the sequence. The names of the OMADS3, OMADS5, OMADS8, and OMADS9 proteins are underlined. This sequence alignment was generated by the ClustalW Multiple Sequence Alignment Program at the DNA Data Bank of Japan (http://clustalw.ddbj.nig.ac.jp/top-e.html).
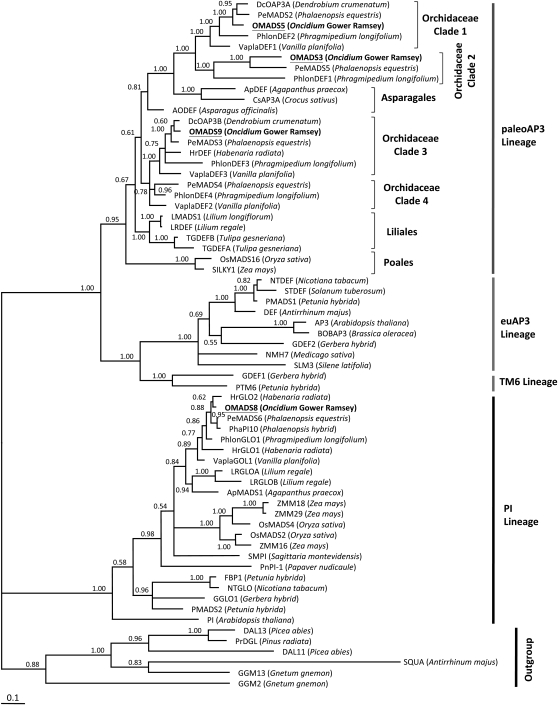
Figure 2.
Phylogenetic analysis of B class MADS domain proteins. Based on the amino acid sequence of the full-length protein, OMADS8 was closely related to PeMADS6 and OsMADS4 in the PI group of MADS box genes in monocots. OMADS3, OMADS5, and OMADS9 were closely related to genes in the paleoAP3 lineage of monocots. OMADS5 belongs to clade 1, OMADS9 belongs to clade 3, and OMADS3 belongs to clade 2 of paleoAP3 genes of orchids. The names of the OMADS3, OMADS5, OMADS8, and OMADS9 proteins are shown in boldface and underlined. The names of the plant species for each MADS box gene are listed behind the protein names. Amino acid sequences of B class MADS box genes were retrieved via the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/). The phylogenetic tree was generated using Bayesian analysis as described in “Materials and Methods.”
The OMADS8 cDNA encoded a 210-amino acid protein that showed high sequence identity to the B class gene PI of monocots (Figs. 1 and 2). In its C-terminal region, a consensus PI motif (MPFxFRVQPxQPNLQE), which is unique to B class PI genes (Kramer et al., 1998; Moon et al., 1999), was identified. Sequence identity between OMADS8 and PI homologs indicates that OMADS8 is an O. Gower Ramsey B group MADS box PI gene.
The alignment of amino acid sequences shown in Figure 1 and sequences for several other MADS box genes were used to construct a phylogenetic tree for B class genes (Fig. 2). Based on this analysis, OMADS8 was in the PI lineage, whereas OMADS5 and OMADS9 were in the paleoAP3 lineage.
OMADS8 Is Expressed in All Four Floral Organs as Well as in Vegetative Leaves
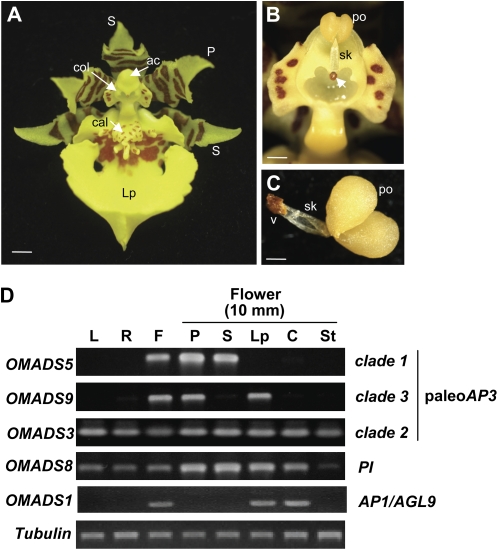
To explore the relationships between sequence identity and expression pattern for OMADS8, OMADS8 expression was detected by RT-PCR analysis. As shown in Figure 3D, OMADS8 mRNA was detected in vegetative leaves, roots, and flowers, indicating that the expression of OMADS8 was not flower specific. When floral organs from 8- to 10-mm mature floral buds (Fig. 3, A–C) were examined, OMADS8 was expressed in all floral organs, with relatively higher expression in the sepal, petal, and lips than in the stamen and carpel (Fig. 3D). When the expression of OMADS8 in sepal/petal/lip of flower buds in different early developmental stages (2, 3, and 5 mm in length; Fig. 4, A–C) was further analyzed, OMADS8 mRNA was consistently detected in sepals, petals, and lips of all three early flower buds (Fig. 4D), similar to that observed in mature flower buds (Fig. 3D). This expression pattern was different from that observed for PI orthologs of Arabidopsis and other orchids, which was floral specific and was only expressed in stamens and petals (Jack et al., 1992; Rounsley et al., 1995; Moon et al., 1999; Tsai et al., 2005; Xu et al., 2006; Guo et al., 2007; Kim et al., 2007). Interestingly, the expression pattern for OMADS8 was very similar to that observed for the clade 2 paleoAP3 gene OMADS3 (Fig. 3D; Hsu and Yang, 2002). OMADS3 mRNA was also consistently detected in sepals, petals, and lips of all three early flower buds (Fig. 4E), although it showed a relatively lower expression in lips than in sepal/petal (Fig. 4E). This result indicated that OMADS8 and OMADS3 possibly play similar roles in regulating the formation of the sepal, petal, and lip in O. Gower Ramsey.
Figure 3.
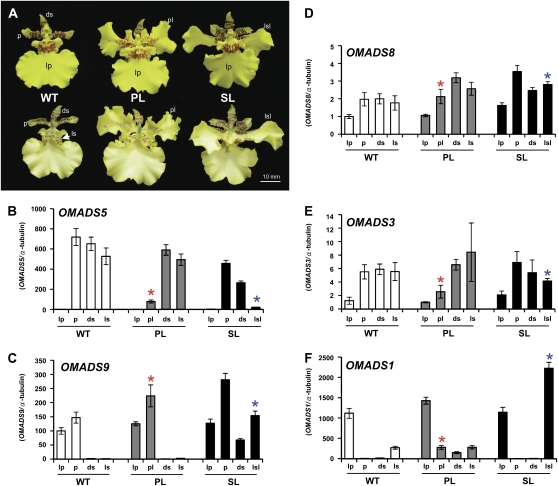
Detection of expression of OMADS3, OMADS5, OMADS8, OMADS9, and OMADS1 in O. Gower Ramsey. A, An O. Gower Ramsey mature flower bud (10 mm) consisting of three sepals (S), two petals (P), a lip (Lp) with red-brown around the callus (cal), and a reproductive organs column (col). Sepals and petals are yellow with red-brown bars and blotches toward the base of the segment. ac, Anther cap. Bar = 2 mm. B, Closeup of the column. The anther cap, which covers the reproductive organs column (col in A), was removed, revealing the pollinarium (male reproductive organ), which consists of two pollinia (po), a stalk (sk), and the brown viscidium (arrowed). Bar = 0.5 mm. C, Closeup of the pollinarium. po, Pollinia; sk, stalk; v, viscidium. Bar = 0.2 mm. D, Total RNAs isolated from leaves (L), roots (R), and the flower organs sepal (S), petal (P), lip (Lp), stamen (St), and carpel (C) of 10-mm-long floral buds (F) were used as templates to detect the expression of OMADS3, OMADS5, OMADS8, and OMADS9 by RT-PCR. In this study, two pollinia, a stalk of pollinarium, and the viscidium from the column were isolated as male reproductive organs (indicated as stamen). The remaining tissues of the column were used as female reproductive organs (indicated as carpel). The results indicated that OMADS8 and OMADS3 were expressed in all four floral organs as well as in vegetative leaves and roots. The mRNA for OMADS5 was only strongly detected in sepals and petals, whereas OMADS9 was only strongly detected in petals and lips. The AGL6-like gene OMADS1 was only expressed in lips and carpels. Each experiment was repeated twice with similar results. A fragment of the α-tubulin gene was amplified as an internal control.
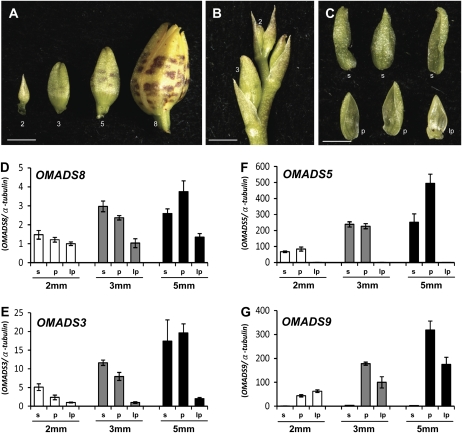
Figure 4.
Detection of expression of OMADS3, OMADS5, OMADS8, and OMADS9 in sepal/petal/lip during early flower development in O. Gower Ramsey. A, Flower buds of O. Gower Ramsey at different developmental stages (2, 3, 5, and 8 mm long). Bar = 2 mm. B, Closeup of the young flower buds on the top of the inflorescence in O. Gower Ramsey. The positions of the 2- and 3-mm-long flower buds are indicated by the numbers. Bar = 2 mm. C, A 2-mm-long O. Gower Ramsey flower bud consisting of three sepals (s), two petals (p), and a lip (lp). In this stage, the lip is small and morphologically similar to the petals. Bar = 1 mm. D to G, Total RNAs isolated from the sepal (s), petal (p), and lip (lp) of 2-, 3-, and 5-mm-long young flower buds from A were used as templates to detect the expression of OMADS8 (D), OMADS3 (E), OMADS5 (F), and OMADS9 (G) by real-time PCR. The results indicated that OMADS8 mRNA was consistently detected in sepals, petals, and lips of all three early flower buds. OMADS3 mRNA was also consistently detected in sepals, petals, and lips of all three early flower buds with a relatively lower expression in lips than in sepal/petal. OMADS5 mRNA was consistently expressed only in sepals and petals and was absent in lips, whereas OMADS9 mRNA was only expressed in petals and lips and was not detected in the sepals of all three early flower buds. In quantitative real-time PCR, the columns represent the relative expression of these genes. Transcript levels of these genes were determined using two to three replicates and were normalized using α-tubulin. Error bars represent sd. Each experiment was repeated three times with similar results.
OMADS5 and OMADS9 Showed Different Expression Patterns in Floral Organs
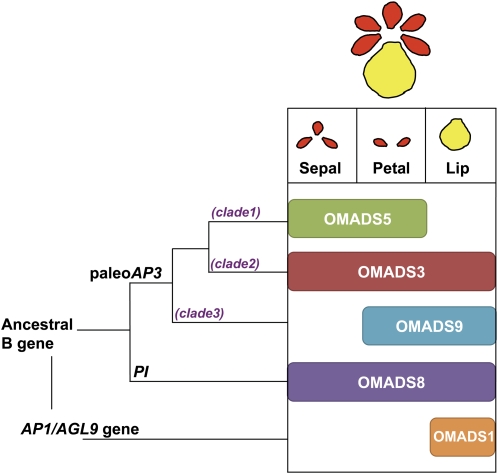
To explore the relationships between sequence identity and expression pattern for OMADS5 and OMADS9, expression of OMADS5 and OMADS9 was detected by RT-PCR. As shown in Figure 3D, the OMADS5 mRNA was flower specific and absent in vegetative leaves and roots. When floral organs from 8- to 10-mm mature floral buds (Fig. 3, A–C) were examined, OMADS5 was only expressed in sepals and petals (Fig. 3D). Similar to OMADS5, OMADS9 was also strongly detected in flowers and absent in vegetative leaves and roots (Fig. 3D). In flowers, the OMADS9 mRNA was also highly expressed in petals (Fig. 3D). However, in contrast to OMADS5, OMADS9 was highly expressed in the lip and was not detected in the sepal (Fig. 3D). To examine the consistency of the OMADS5 and OMADS9 expression, the expression of OMADS5 and OMADS9 in sepal/petal/lip of flower buds in different early developmental stages (2, 3, and 5 mm in length; Fig. 4, A–C) was further analyzed. The result indicated that OMADS5 mRNA was consistently only expressed in sepals and petals and was absent in lips of all three early flower buds (Fig. 4F). Similar to that observed in mature flower buds, OMADS9 mRNA was also only expressed in petals and lips and was not detected in the sepals of all three early flower buds (Fig. 4G). This result indicated that OMADS5 and OMADS9 may play different roles in controlling the formation of the sepal, petal, and lip in O. Gower Ramsey. The formation of the sepal and petal at least requires the presence of OMADS5, whereas the formation of lips requires no less than the presence of OMADS9 and the absence of OMADS5. The expression patterns in the sepal/petal/lips and the phylogenetic relationship between the four B class genes OMADS3, OMADS5, OMADS9, and OMADS8 and one AGL6-like gene, OMADS1, of O. Gower Ramsey are illustrated in Figure 5.
Figure 5.
Gene duplications and expression patterns of the B class genes in O. Gower Ramsey. A major duplication event from an ancestral B gene generated the paleoAP3 and PI lineages. In orchid O. Gower Ramsey, there is only one PI gene, OMADS8, and two more duplications occurred in the paleoAP3 gene that generated at least three paleoAP3-like genes, OMADS5 (clade 1), OMADS3 (clade 2), and OMADS9 (clade 3). OMADS8 and OMADS3 were expressed in the sepal/petal/lip, OMADS5 was expressed in the sepal/petal, while OMADS9 was expressed in the petal/lip. The AGL6-like gene OMADS1 was expressed in lip and carpel. [See online article for color version of this figure.]
OMADS5 Was Significantly Down-Regulated in Lip-Like Petals/Sepals in the Peloric Mutants
To further explore the roles of OMADS5, OMADS9, OMADS8, and OMADS3 in regulating sepal, petal, and lip formation in O. Gower Ramsey, the expression of these four genes in peloric mutants of O. Gower Ramsey with conversions of either petal to lip (Fig. 6A, middle) or the lateral sepal to lip (Fig. 6A, right) was examined by real-time PCR. As shown in Figure 6B, the OMADS5 mRNA was undetectable in lips and was highly expressed in normal sepals and petals of wild-type and both peloric mutant flowers. The expression of OMADS5 was significantly reduced (Fig. 6B) in lip-like petals of peloric mutant flowers (Fig. 6A, middle). A similar reduction of the OMADS5 mRNA was also observed (Fig. 6B) in lip-like lateral sepals of peloric mutant flowers (Fig. 6A, right). This result clearly indicated a strong correlation between lip formation and reduction in OMADS5 expression. Since the sizes of both lip-like petals and lip-like lateral sepals in peloric mutant flowers were relatively smaller than normal lips (Fig. 6A), lower OMADS5 expression also associated with the larger size of the lips produced.
Figure 6.
Detection of expression of OMADS1, OMADS3, OMADS5, OMADS8, and OMADS9 in peloric mutants of O. Gower Ramsey. A, Flower phenotypes of wild-type plants (WT), peloric mutants with two petals converted into lips (PL), and peloric mutants with two lateral sepals converted into lips (SL) of O. Gower Ramsey. The top row represents the front side and the bottom row represents the back side of the flower. ds, Dorsal sepal; ls, lateral sepal; p, petal; lp, lip; pl, lip-like petal; lsl, lip-like lateral sepal. Bar = 10 mm. B to F, Total RNAs isolated from the dorsal sepal (ds), lateral sepal (ls), petal (p), and lip (lp) of mature flowers from A, the wild type (WT), peloric mutants with petals converted into lips (PL), and peloric mutants with lateral sepals converted into lips (SL), were used as templates to detect the expression of OMADS5 (B), OMADS9 (C), OMADS8 (D), OMADS3 (E), and OMADS1 (F) by real-time PCR. The level of gene expression in lip-like petals is marked with red stars, and expression in the lip-like lateral sepal (lsl) is marked with blue stars. The results indicated that OMADS5 was significantly down-regulated in both lip-like petals and lip-like lateral sepals in peloric mutants. OMADS9 was significantly up-regulated in both lip-like lateral sepals and normal dorsal sepals in peloric mutants (A, right). OMADS1 was clearly up-regulated in both lip-like petals and lip-like lateral sepals in peloric mutants. In quantitative real-time PCR, the columns represent the relative expression of these genes. Transcript levels of these genes were determined using two to three replicates and were normalized using α-tubulin. Error bars represent sd. Each experiment was repeated three times with similar results.
When the expression of OMADS9 was examined, its mRNA was not detected (Fig. 6C) in normal sepals of either wild-type or peloric mutant flowers with lip-like petals (Fig. 6A, middle). In contrast, the OMADS9 mRNA was increased (Fig. 6C) in the lip-like lateral sepals of peloric mutant flowers (Fig. 6A, right). Interestingly, the increase in OMADS9 expression was also observed in the normal dorsal sepal (Fig. 6C) of peloric mutant flowers (Fig. 6A, right). This result revealed that the increase in OMADS9 expression is not absolutely linked to the formation of lips.
Furthermore, the expression of OMADS8 in lip-like petals (Fig. 6A, middle) or lip-like lateral sepals (Fig. 6A, right) of peloric mutant flowers was similar to that observed in normal sepals or petals, respectively (Fig. 6D). The expression of OMADS3 was also detected in lip-like petals (Fig. 6A, middle) or lip-like lateral sepals (Fig. 6A, right) of peloric mutant flowers and was slightly reduced to a level similar to that observed in normal lips (Fig. 6E). When the expression of OMADS1, an AGL6-like gene in O. Gower Ramsey that is specifically expressed in the lip and carpel of flowers (Fig. 3; Hsu et al., 2003), was examined, an increase in expression (Fig. 6F) was observed in both lip-like petals (Fig. 6A, middle) and lip-like sepals (Fig. 6A, right) of peloric mutant flowers.
The Formation of Larger Lips Is Due to an Increase in Cell Proliferation Rather Than Cell Expansion
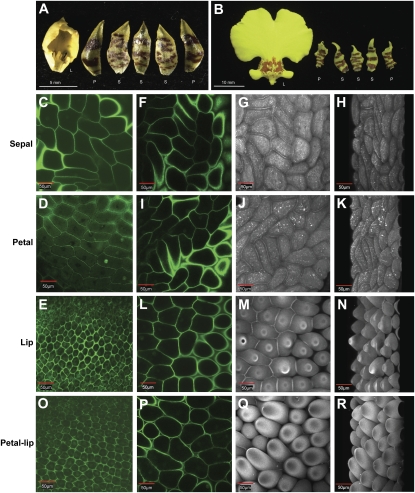
A major difference between the lip and sepal/petal in O. Gower Ramsey is size. The well-expanded large lip in a mature Oncidium flower is about 15- to 20-fold larger than the sepal and petal (Figs. 6A, left, and 7B). It is interesting to investigate whether the increased size of the lips, possibly caused by the absence of OMADS5 expression, was due to a difference in the total number of cells produced or the increased size of each individual cell. Therefore, the epidermal cells in these three organs were examined by confocal laser scanning microscopy. In flower buds (Fig. 7A), the epidermal cells in the sepal (Fig. 7C) and petal (Fig. 7D; 100 × 40 μm2) were about eight times larger than those observed in lips (25 × 20 μm2; Fig. 7E). In this stage, the size of the lips was only approximately twice that of the sepal and petal (Fig. 7A). This indicated that there were about 16 times more cells in the lips than in the sepal and petal during the floral bud stage. When the epidermal cells in the lips (Fig. 7, L and M) of a mature flower (Fig. 7B) were examined, they were almost the same size (100 × 40 μm2) as those of the sepal (Fig. 7, F and G) and petal (Fig. 7, I and J) cells, although these papillate cells showed a cobblestone-like appearance with a protuberance on the top (Fig. 7, M and N) that was morphologically different from the irregularly shaped flat sepal (Fig. 7, G and H) or petal (Fig. 7, J and K) cells. Interestingly, these papillate cells found in the lip epidermis are actually the characteristic of petals for plants such as Arabidopsis and Petunia. This result indicated that the total number of cells in the lips during early flower development was already about 16 times more than in the sepal and petal. These epidermal cells in the lips grew rapidly during maturation, reached the same size as sepal/petal cells, and caused the 15 to 20 times larger size of the lip than the sepal and petal. Therefore, it is clear that the increase in cell proliferation was the main reason for the larger size of the lips in Oncidium. When the epidermal cells in lip-like petals (Fig. 6A, middle) of peloric mutant flowers were examined, they were morphologically identical to the wild-type lip epidermis (Fig. 7, E and L–N) in either the floral bud (Fig. 7O) or the mature flower (Fig. 7, P–R) and distinct from the wild-type sepal/petal epidermis.
Figure 7.
Confocal laser scanning microscopy of various floral organs in O. Gower Ramsey. A, A lip (L), two petals (P), and three sepals (S) of a 10-mm O. Gower Ramsey floral bud. The sepals and petals look the same. Bar = 5 mm. B, A lip (L), two petals (P), and three sepals (S) of an O. Gower Ramsey mature flower. The sepals and petals look the same. Bar = 10 mm. C, Irregularly shaped cells in the epidermis of sepals in the wild-type floral bud in A. Bar = 50 μm. D, Irregularly shaped cells in the epidermis of petals in the wild-type floral bud in A. Bar = 50 μm. E, Small cobblestone-like cells in the epidermis of lips in the wild-type floral bud in A. Bar = 50 μm. F to H, Irregularly shaped cells in the epidermis of sepals in the wild-type mature flower in B. Bar = 50 μm. I to K, Irregularly shaped cells in the epidermis of petals in the wild-type mature flower in B. Bar = 50 μm. L to N, Cobblestone-like cells in the epidermis of lips in the wild-type mature flower in B. Bar = 50 μm. O, Small cobblestone-like cells in the epidermis of lip-like petals in a peloric mutant floral bud (Fig. 3A, middle). Bar = 50 μm. P to R, Cobblestone-like cells in the epidermis of lip-like petals in a peloric mutant mature flower. Bar = 50 μm. The cell wall was counterstained with propidium iodide. C to F, I, L, O, and P are single slices to indicate the cell outline; G, J, M, and Q are Z-stacks to present the cell morphology; and H, K, N, and R are three-dimensional rotated images from the Z-stack images using the Fluoview 1000 software.
Homodimer and Heterodimer Formation of the OMADS5, OMADS9, OMADS8, and OMADS3 Proteins
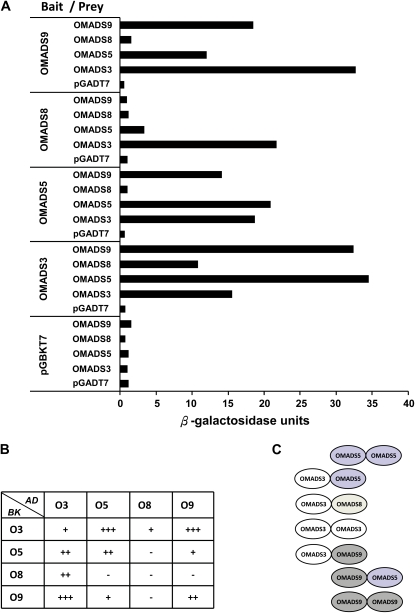
LMADS1, the AP3 homolog from the monocot lily, has been reported to be able to form homodimers (Tzeng and Yang, 2001; Tzeng et al., 2004). Since OMADS5 and OMADS9 showed high sequence identity to monocot AP3 homologs, it was interesting to study the possible interaction between OMADS5 and OMADS9. To achieve this, a yeast two-hybrid analysis was performed. Similar to that observed for LMADS1 (Tzeng and Yang, 2001), our result indicated that the OMADS5 and OMADS9 proteins were also able to form strong homodimers (Fig. 8, A and B). When the interaction between OMADS5 and OMADS9 was analyzed, a high level of β-galactosidase activity was also detected (Fig. 8, A and B). This indicated that OMADS5 and OMADS9 are not only able to form homodimers but also formed heterodimers with each other.
Figure 8.
Protein interactions among B class proteins of O. Gower Ramsey in a yeast two-hybrid assay. A, β-Galactosidase activity in yeast cells transformed with combinations of truncated OMADS3, OMADS5, OMADS8, and OMADS9 in either the binding domain plasmid (Bait) or the activation domain plasmid (Prey) calculated according to Miller (1992). Yeast cells transformed with OMADS3, OMADS5, OMADS8, and OMADS9 in the binding domain plasmid or the activation domain plasmid alone were used as controls for background activity. B, A summary of the strength of the β-galactosidase activity obtained in A. The number of + signs indicates the relative strength of the activity detected in each reaction. The – sign indicates that no activity was detected. AD, Activation domain in plasmid pGADT7; BK, binding domain in plasmid pGBKT7. C, A summary of the homodimers and heterodimers among the four B class proteins of O. Gower Ramsey obtained in A. [See online article for color version of this figure.]
When OMADS8 was analyzed in the yeast two-hybrid analysis, it was not only unable to form homodimers itself but also unable to form heterodimers with either OMADS5 or OMADS9 (Fig. 8, A and B). Interestingly, OMADS8 was able to form strong heterodimers with OMADS3 (Fig. 8, A and B). When OMADS3 was further analyzed, it showed a completely different pattern from OMADS8, since it was able to form strong homodimers and heterodimers with OMADS5, OMADS9, and OMADS8 (Fig. 8, A and B). The interactions among the four orchid B class proteins are illustrated in Figure 8C.
Ectopic Expression of OMADS8 Converted Sepals into Petal-Like Structures in Transgenic Arabidopsis Plants
To further investigate the functions of OMADS5, OMADS9, and OMADS8, ectopic expression of these genes in transgenic plants was necessary. cDNAs for these three genes driven by the cauliflower mosaic virus 35S promoter were transformed into Arabidopsis plants for functional analysis.
Twenty independent 35S∷OMADS8 transgenic Arabidopsis plants were obtained. Nine plants were phenotypically indistinguishable from wild-type plants, whereas the other 11 plants showed identical novel phenotypes in both vegetative and reproductive development. These plants flowered earlier than wild-type plants by producing fewer rosette leaves. Flowers produced in the inflorescence of these plants (Fig. 9C) were different from those observed in wild-type plants (Fig. 9A). Unlike wild-type flowers (Fig. 9B), homeotic conversion of first whorl green sepals into white petal-like structures (Fig. 9, D and E) was observed in these 35S∷OMADS8 flowers. When the epidermal cells (Fig. 9F) in these first whorl petal-like structures were examined, they were morphologically similar to wild-type petal epidermal cells (Fig. 9G) and distinct from the wild-type sepal epidermis (Fig. 9H). To explore whether the severe phenotype correlated with OMADS8 expression in the transgenic plants, RT-PCR analysis was performed. As shown in Figure 9I, higher OMADS8 expression was observed in the transgenic plants with the severe phenotype compared with transgenic plants with less severe phenotypes or phenotypes indistinguishable from wild-type plants. This result clearly indicated that the phenotypes generated in the 35S∷OMADS8 transgenic Arabidopsis were due to the ectopic expression of the orchid OMADS8 gene.
Figure 9.
Phenotypic analysis of transgenic Arabidopsis plants ectopically expressing OMADS8 or OMADS8-ΔMADS. A, A wild-type inflorescence contained flower buds and mature flowers with normal sepals (s) and petals (p). B, A mature wild-type Arabidopsis flower consisted of four whorls of organs, including four sepals (s), four elongated petals (p), six stamens (arrow), and two fused carpels. The sepals had not opened in this stage. C, A 35S∷OMADS8 inflorescence contained flower buds (fb) with green/white sepals in the first whorl, fully opened mature flowers with white elongated petal-like sepals (ps) in the first whorl, and normal petals (p) in the second whorl of the flower. D and E, Closeup views of the 35S∷OMADS8 transgenic Arabidopsis flowers. White elongated petal-like sepals (ps) and normal petals (p) were produced in the first and second whorls of the flowers, respectively. F, Scanning electron micrograph of the surface cells of the epidermis of the first whorl petal-like sepal of a 35S∷OMADS8 flower was similar to the mature wild-type petal epidermis in G. Bar = 10 μm. G, Scanning electron micrograph of the surface cells of the epidermis of a mature wild-type petal. Bar = 10 μm. H, Scanning electron micrograph of the surface cells with irregular shapes in the epidermis of wild-type sepals. Bar = 10 μm. I, Total RNA isolated from two severe (T5 and T6) and four less severe (T1–T4) 14-d-old 35S∷OMADS8 transgenic Arabidopsis plants and from one untransformed wild-type plant (WT) used as a template. The results indicated that OMADS8 (OM8) was clearly expressed higher in the T5 and T6 than in the T1 to T4 transgenic plants. A fragment of the ACTIN (ACT) gene was amplified as an internal control.
Twenty and 29 independent 35S∷OMADS5 and 35S∷OMADS9 transgenic Arabidopsis plants were obtained, respectively. In contrast to the flowers in 35S∷OMADS8 described above, no homeotic conversion in the floral organs was observed in the flowers of these 35S∷OMADS5 and 35S∷OMADS9 plants.
DISCUSSION
To investigate the role of B class MADS box genes in the regulation of flower development in the orchid O. Gower Ramsey, three genes, OMADS5, OMADS8, and OMADS9, were identified and characterized in this study. Based on sequence alignment, the conserved motifs in the C-terminal regions of the proteins, and phylogenetic tree analysis, OMADS5 belongs to clade 1 whereas OMADS9 belongs to clade 3 of paleoAP3 genes of orchids. OMADS8 is a PI-like gene and is closely related to monocot PI orthologs (Figs. 1 and 2).
Like the other clade 1 paleoAP3 genes, such as PeMADS2 of Phalaenopsis (Tsai et al., 2004), the mRNA for OMADS5 was only strongly detected in sepals and petals. The OMADS9 mRNA was only strongly detected in petals and lips, similar to the clade 3 paleoAP3 genes DcOAP3B, PeMADS3, and HrDEF (Tsai et al., 2004; Xu et al., 2006; Kim et al., 2007). The expression of both OMADS5 and OMADS9 was absent in stamens and leaves. Interestingly, the expression patterns of OMADS5 and OMADS9 were largely different from OMADS3, which was expressed in all flower organs and leaves (Fig. 3; Hsu and Yang, 2002). This indicated the possibility of functional and transcriptional diversification of OMADS5, OMADS9, and OMADS3. OMADS9 and OMADS5 may have experienced significant expression diversification from OMADS3 after the gene duplication in the orchid during evolution (Fig. 5).
The possible role for OMADS5 and OMADS9 in regulating perianth formation is interesting. The detection of only OMADS9 (clade 3) expression in lips suggests that OMADS9 may have a positive role in lip formation. However, it is unclear why the petals did not convert into lips, since OMADS9 was also expressed in petals. In addition, a high amount of OMADS9 expression was also observed in the normal dorsal sepal of peloric mutant flowers (Fig. 6C). This dorsal sepal could be converted into a lip-like structure. This result revealed that the presence of OMADS9 expression is not necessarily associated with the formation of lips. One possibility is that the increase of OMADS9 alone is necessary but not sufficient for the transformation of sepals or petals into lips. It has been proposed that a lip-specific clade 4 gene, such as PeMADS4 of Phalaenopsis, was responsible for lip formation (Tsai et al., 2004; Mondragón-Palomino and Theißen, 2008, 2009). OMADS9 may need to work together with a clade 4 gene that remains to be identified in O. Gower Ramsey to control lip formation.
Alternatively, the difference for petal and lip formation may be due to the expression of OMADS5 in the petal and the absence of OMADS5 expression in the lip. This suggested a possible negative role for OMADS5 in regulating lip formation. Based on this assumption, in the presence of OMADS5 expression, lip formation will be suppressed and the organs will be converted into sepals/petals, no matter whether OMADS9 expression is present or absent. Thus, a high amount of OMADS9 expression did not convert normal dorsal sepal of peloric mutant flowers into lips, since a certain high amount of OMADS5 expression was also detected (Fig. 6, B and C). In the absence of OMADS5 expression, the organs will be converted into lips in the presence of OMADS9 expression. This assumption was supported by the expression analysis of OMADS5 and OMADS9 in peloric mutants of orchid. OMADS5 mRNA was significantly reduced in both lip-like petals and lip-like sepals of peloric mutant flowers (Fig. 6B). Taken together, these results support that the loss of OMADS5 (clade 1) expression is likely the main reason for the lip specification and for the conversion of the sepals/petals into lips in O. Gower Ramsey and perhaps also in other orchids with lips and petals in different sizes. Interestingly, these peloric mutants of orchid are also likely affected in floral symmetry, which was regulated by two duplicated TCP transcription factors, CYCLOIDEA (CYC) and DICHOTOMA (DICH; Mondragón-Palomino and Theißen, 2009; for review, see Preston and Hileman, 2009). It has been reported that the expression of CYC depends on the transcription of the B class gene DEF in A. majus (Clark and Coen, 2002). Therefore, the change of expression for B class genes OMADS5/OMADS9 in orchid may cause the alteration for CYC/DICH expression and result in the changes in floral symmetry. However, it has been shown that there are no true orthologs of the floral symmetry genes CYC and DICH outside core eudicots (for review, see Preston and Hileman, 2009). This implies a possible different genetic mechanism underlying dorsoventral asymmetry in monocot orchids.
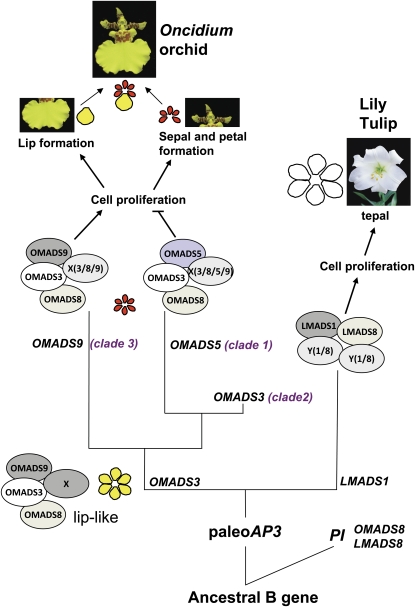
Similar to OMADS3, the mRNA for OMADS8 was detected in all four floral organ whorls as well as in vegetative leaves and roots (Fig. 3). The expression in organs other than flowers has also been reported in some B class genes such as GDEF1 (G. hybrida), TGGLO (T. gesneriana), ZMM16 (Z. mays), and EgGLO2 (Elaeis guineensis) that showed a similar expression pattern to OMADS8/3 by expressing in both flower organs and leaves (Yu et al., 1999; Münster et al., 2001; Kanno et al., 2003; Adam et al., 2007). This revealed that in addition to regulating flower organ formation, B class genes such as OMADS3/8, GDEF1, TGGLO, ZMM16, and EgGLO2 are also possibly involved in the regulation of leaf development. However, this assumption still requires further investigation. The expression of OMADS3 and OMADS8 in sepals, petals, and lips suggested that OMADS8 and OMADS3 may play similarly general and necessary roles in regulating the formation of these three flower organs in O. Gower Ramsey. Thus, we proposed that the presence of at least OMADS3/8/5 and/or OMADS9 is required for sepal and petal formation, whereas the presence of OMADS3/8/9 and the absence of OMADS5 are likely required for lip formation in O. Gower Ramsey (Figs. 5 and 10).
Figure 10.
Possible evolutionary relationships between B class genes in the regulation of sepal/petal/lip formation for O. Gower Ramsey. B group genes in monocots are thought to have been produced by a major duplication event from an ancestral gene that generated the paleoAP3 and PI lineages. In orchid O. Gower Ramsey, there is only one PI gene, OMADS8, and at least two more duplications occurred in the paleoAP3 gene OMADS3 that generated the paleoAP3-like genes OMADS5 and OMADS9. The ancestral complex containing OMADS3/8 and other A/E proteins (X) is sufficient to convert the first and second whorl organs into well-expanded lip-like sepals/petals (in yellow). After gene duplication, OMADS9 may retain the ancestral B gene function of possibly promoting cell proliferation, whereas OMADS5 might have been altered in relation to this function. This caused the possible suppression of cell proliferation for any complex that includes OMADS5 (e.g. OMADS3/8/5/9/X) and might result in the formation of six short sepals/petals (in red) in the orchid flowers. During evolution, one of the short sepals/petals was converted into a well-expanded lip structure (in yellow) due to the loss of OMADS5 expression, resulting in the formation of one expanded lip (in yellow; OMADS3/8/9/X) and five short sepals/petals (in red; OMADS3/8/5/9/X), as seen in the modern O. Gower Ramsey. Unlike O. Gower Ramsey, there is only one paleoAP3 ortholog, LMADS1, and one PI ortholog, LMADS8, in lily. The complex containing LMADS1/8 and other A/E proteins (Y) may have the ability, similar to OMADS3/8/X, to convert the first and second whorl organs into well-expanded tepals (in white) in lily. The production of short tepals by suppressing cell proliferation has not been seen in lily, since no other paleoAP3 gene, such as OMADS5, has been identified in lily. [See online article for color version of this figure.]
This assumption was further examined using yeast two-hybrid analyses of the interactions among the orchid B class proteins OMADS3, OMADS5, OMADS8, and OMADS9. The result indicated that OMADS3 formed homodimers and heterodimers with OMADS5, OMADS8, and OMADS9. OMADS8 only formed heterodimers with OMADS3, whereas OMADS5 and OMADS9 formed homodimers individually and heterodimers with each other and with OMADS3 (Fig. 8). The ability to form homodimers supported the possibility that OMADS3, OMADS5, and OMADS9 also likely retained the characteristic of ancestral forms of the B genes for the paleoAP3 lineage in monocots, such as LMADS1 in lily (Tzeng and Yang, 2001). This result revealed a possible model for the interaction of these four B class proteins in regulation of the formation of sepals, petals, and lips in O. Gower Ramsey. As illustrated in Figure 10, a complex composed of at least OMADS3/8 and/or OMADS9 is needed for the formation of sepals, petals, and lips. Adding OMADS5 to the complex may convert organs into sepals/petals; any complex without OMADS5 controls lip formation. In addition to these B class proteins, other A/E class proteins are expected to participate in these complexes. For example, OMADS1, an AGL6-like gene of O. Gower Ramsey, is predominantly expressed in lips (Fig. 3) and could form heterodimers with OMADS3 (Hsu et al., 2003). High expression of OMADS1 is clearly needed for lip formation and was supported by the up-regulation of OMADS1 in both lip-like petals and lip-like sepals of peloric mutant flowers (Fig. 6F). This assumption was further supported by previous work demonstrating that the B class proteins DcAP3A/B and DcOPI of Dendrobium formed higher order complexes with the E class protein DcOSEP1 (Xu et al., 2006).
One interesting and critical question is what is the possible role for OMADS5 (clade 1) in negatively regulating lip formation. Since an increase in cell proliferation rather than cell expansion was the main reason for the large size of the lips in Oncidium (Fig. 7), it is reasonable to propose that a gene such as OMADS5 may play a role in suppressing lip formation by inhibiting cell proliferation in orchids like Oncidium (Fig. 10). Any protein complex that contains OMADS5 may inhibit cell proliferation and result in the formation of short organs, such as sepals and petals of Oncidium. When OMADS5 was absent in these complexes, the cells proliferated, resulting in the formation of large lips (Fig. 10). Based on this assumption, there were likely six lip-like organs in the ancestral O. Gower Ramsey before the gene duplication that generated OMADS5. It has also been proposed that the perianth of the most recent common ancestor of orchids and the rest of the Asparagales possibly is composed of six almost identical tepals in which an ancestral paleoAP3-like gene was uniformly expressed (Mondragón-Palomino and Theißen, 2008, 2009). In this case, the ancestral complex containing OMADS3/8 and other A/E proteins is sufficient to convert the first and second whorl organs into a well-expanded lip-like sepal/petal. After gene duplication, OMADS5 (clade 1) might be functionally altered (Fig. 10). This would have caused a possible reduction or loss of the ability to promote cell proliferation for any complex that includes OMADS5 and resulted in the formation of short sepals/petals in the flowers (Fig. 10). Later, OMADS5 expression in one of the short sepals/petals was altered and eliminated, resulting in the conversion of this organ into a well-expanded lip structure, as seen in the modern O. Gower Ramsey. One can expect that any one of the sepals or petals will be converted into a lip-like structure once expression of OMADS5 was reduced or eliminated, as seen in the peloric mutant flowers (Fig. 6B). Furthermore, the degree of the conversion was clearly correlated with the level of the reduction of OMADS5 expression, since OMADS5 expression was completely eliminated in the normal lips, which were relatively larger than both the lip-like petals and the lip-like lateral sepals in the peloric mutant flowers.
This assumption can be further supported by the analysis of lily flowers that contained extremely similar sepals and petals, known as perianth segments or tepals, which are fully expanded organs (Fig. 10). There is only one paleoAP3 ortholog, LMADS1 (Tzeng and Yang, 2001; Tzeng et al., 2004), and two nearly identical PI orthologs, LMADS8 and LMADS9 (M.-K. Chen, W.-P. Hsieh, and C.-H. Yang, unpublished data), that have been identified and characterized in L. longiflorum so far. Interestingly, both lily LMADS1 (Tzeng and Yang, 2001) and LMADS8 and LMADS9 (M.-K. Chen, W.-P. Hsieh, and C.-H. Yang, unpublished data) were expressed in all four flower organs, similar to the orchid orthologs OMADS3 and OMADS8 (Fig. 3). LMADS1 was also able to form homodimers (Tzeng and Yang, 2001) and heterodimers with LMADS8 (M.-K. Chen, W.-P. Hsieh, and C.-H. Yang, unpublished data). This indicated that the lily paleoAP3 (LMADS1) and PI orthologs (LMADS8) retained similar functions and regulation as the ancestral B gene and the orchid orthologs OMADS3 and OMADS8 (Fig. 10). Therefore, the LMADS1/8 complex may have an ability similar to OMADS3/8 to convert the first and second whorl organs into well-expanded tepals in lily. Since there is no evidence for gene duplication to generate other paleoAP3 genes, such as OMADS5 in lily, the production of short tepals by possibly suppressing cell proliferation was not seen in lilies.
Not surprisingly, functional analysis produced useful results for interpretation of the possible roles of OMADS5, OMADS8, and OMADS9 in flower formation. The production of elongated petal-like sepals in 35S∷OMADS8 transgenic Arabidopsis plants was similar to that observed in transgenic Arabidopsis plants ectopically expressing PI (Lamb and Irish, 2003) or its orthologs, such as DcOPI of Dendrobium (Xu et al., 2006) and LMADS8 and LMADS9 of lily (M.-K. Chen, W.-P. Hsieh, and C.-H. Yang, unpublished data). This result provides further evidence to support the assumption that OMADS8 simply functions as a B gene in the PI lineage. By contrast, no homeotic conversion in the floral organs was observed in the flowers ectopically expressing either OMADS9 or OMADS5. One explanation is that the orchid OMADS9 and OMADS5 proteins are unable to interact with or insufficient for interaction with the Arabidopsis B/A/E class protein complex that regulates sepal and petal formation. This assumption might be supported by PI orthologs being more conserved in their protein sequences than AP3 orthologs in the orchid and Arabidopsis. When the sequence was compared, OMADS8 showed 59% identity to Arabidopsis PI, with 76% identity in the MADS box domain. OMADS5 and OMADS9 showed less than 47% identity to Arabidopsis AP3, with less than 71% identity in the MADS box domain. Furthermore, it has been reported that a mutant form of the lily paleoAP3 ortholog LMADS1 was able to form a heterodimer with Arabidopsis PI and generated an ap3-like dominant negative phenotype in transgenic Arabidopsis (Tzeng and Yang, 2001). When the sequences were compared, LMADS1 only showed 78%, 64%, and 61% identity to OMADS9, OMADS5, and OMADS3, respectively. This sequence diversity might result in the difference between the interactions of lily LMADS1 and orchid OMADS9 and OMADS5 proteins with the Arabidopsis B/A/E class protein complex that regulates sepal and petal formation.
In summary, in addition to the clade 2 paleoAP3 gene OMADS3 characterized previously in our laboratory (Hsu and Yang, 2002), two more paleoAP3 genes, OMADS5 (clade 1) and OMADS9 (clade 3), and a PI gene, OMADS8, that specify flower development were characterized from the orchid O. Gower Ramsey in this study. Based on the expression patterns and the protein interactions among these four orchid B class genes, we propose that sepal and petal formation at least requires the presence of OMADS3/8/5 and/or OMADS9, whereas lip formation at least requires the presence of OMADS3/8/9 and the absence of OMADS5 in O. Gower Ramsey (Fig. 10). The characteristics of these four genes provide useful information for understanding the relationships among the orchid B class MADS box genes as well as their roles in regulating sepal/petal/lip formation. Since most of the A/E genes of O. Gower Ramsey have been isolated and characterized in our laboratory (Chang et al., 2009), further investigation of the role for these A/E genes in the interaction with the B genes characterized in this study should lead to deeper understanding of sepal/petal/lip formation in the orchid O. Gower Ramsey.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type and peloric mutant orchid plants used in this study were Oncidium Gower Ramsey, a hybrid (O. goldiana × O. guinea gold) introduced by Koh Keng Hoe in 1977 (Royal Horticultural Society, 1998; Fitch, 2004). Both O. goldiana (O. flexuosum × O. sphacelatum) and O. guinea gold (O. sphacelatum × O. varicosum) are also hybrids. Orchid plants were grown in the Tzu-Fu Chou orchid field in Houli, Taichung County, Taiwan. Seeds of Arabidopsis (Arabidopsis thaliana) were sterilized and placed on agar plates containing half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) at 4°C for 2 d. The seedlings were then grown in growth chambers under long-day conditions (16 h of light/8 h of dark) at 22°C for 10 d before being transplanted into soil. The light intensity of the growth chambers was 150 μE m−2 s−1.
Cloning of OMADS5, OMADS8, and OMADS9 cDNAs from O. Gower Ramsey
Total RNA was isolated from 5-mm-long floral buds as described by Hsu and Yang (2002). Flower bud RNA (1 μg) was used to synthesize cDNA using the ImProm-II RT System (Promega). B class gene-specific MADS box degenerate 5′ primer Kram1 (5′-GGGGTACCAAYMGICARGTIACITAYTCIAAGMGIMG-3′) and 3′ primers Kram2 (5′-TGIARRTTIFFITGIAWKGGITG-3′) and Kram3 (5′-CIAGICGIAGRTCRT-3′) were used for PCR amplification (Kramer et al., 1998). PCR products of 500 to 600 bp were cloned into the pGEM-T Easy vector (Promega) and sequenced. Partial sequences for OMADS5, OMADS8, and OMADS9 that showed high identity to B group MADS box genes were identified. The cDNAs containing the 5′ and 3′ ends of OMADS5, OMADS8, and OMADS9 were obtained by 5′ RACE and 3′ RACE using the SMART cDNA Amplification Kit (Clontech) and the following primers: internal 3′ gene-specific primers OM5-3 (5′-CTTGTAAGTGTCAGTTTGTGTG-3′) for OMADS5, OPI-3-6 (5′-CTCTGCGTTGATACCACTGCT-3′) for OMADS8, and OAP3-3-4 (5′-CCGATTCCTTACAAGCTTGAGGGCCTCA-3′) for OMADS9; internal 5′ gene-specific primers OM5-5 (5′-GATATATGAGAGGTACCAGATT-3′) for OMADS5, OPI-5-5 (5′-TGGTATCAACGCAGAGTACG-3′) for OMADS8, and OAP3-5-2 (5′-TCGAAGAGGAGAGCTGGGATCATGAAGAA-3′) for OMADS9. The longest cDNAs for OMADS5, OMADS8, and OMADS9 were obtained by PCR amplification using the following primers: OMADS5, p5-5 (5′-GGATCCGCAGAACAGAGCACGACAGAT-3′) and p5-3 (5′-GGATCCCCCATTAAATACAGCAACAA-3′); OMADS8, p8-5 (5′-TCTAGAATGGGGCGCGGAAAGATAGAG-3′) and p8-3 (5′-CTCGAGACCAACAAGGAGGAGTACAAT-3′); and OMADS9 p9-5 (5′-TCTAGAATGGGAAGAGGGAAGATTGAA-3′) and p9-3 (5′-CTCGAGTACTCAATGATGATGGTTCGC-3′). The specific 5′ and 3′ primers for OMADS5, OMADS8, and OMADS9 contained the BamHI (5′-GGATCC-3′; underlined), XbaI (5′-TCTAGA-3′; underlined), or XhoI (5′-CTCGAG-3′; underlined) recognition sites to facilitate cloning of the cDNAs.
RT-PCR
Total RNA isolated from various organs of the orchid or from leaves of Arabidopsis plants were used for cDNA synthesis. Five microliters of cDNA sample from the RT reaction was used for 25 cycles of PCR consisting of denaturation at 94°C (30 s), annealing at 55°C (30 s), and extension at 72°C (60 s). The PCR product (25 μL) in each reaction was analyzed by electrophoresis on 1.5% agarose gels. Primers specific for OMADS1, OMADS3, OMADS5, OMADS8, and OMADS9 used in RT-PCR are as follows: OMADS1, OAGL6-8-1 (5′-GAGCAAATGGTGGGTCATCG-3′) and O1RT-2 (5′-ATGGTTGCTTCAGAAGAGACATATTG-3′); OMADS3, OAP3-5-1 (5′-AAGGAGCTGCGCGGTCTTGA-3′) and OAP3-3-1 (5′-ATAGGGCTGGGTACATTTCTCTAG-3′); OMADS5, OM5-3 and OM5-5; OMADS8, OPI-3-6 and OPI-5-5; and OMADS9, OAP3-3-4 and OAP3-5-2. The O. Gower Ramsey α-tubulin gene and the Arabidopsis ACTIN gene were used as internal controls for the orchid tissue and Arabidopsis tissue, respectively, with the corresponding primers as follows: α-tubulin forward primer (5′-AGGGCTTTCTGGTTTTCAATGCTGT-3′) and reverse primer (5′-CGGCGACGGCAGTGTTGTT-3′); ACTIN forward primer (5′-ATGAAGATTAAGGTCGTGGCA-3′) and reverse primer (5′-TCCGAGTTTGAAGAGGCTAC-3′).
Quantitative Real-Time PCR
Quantitative real-time PCR was conducted using a Mini Opticon Real-Time PCR Detection System and the Optical System Software version 3.0a (Bio-Rad Laboratories) and the SYBR Green Master Mix (Toyobo) for transcript measurements. The reaction mixture was cycled as follows: one cycle at 95°C for 1 min, then 40 cycles of 95°C (15 s), 58°C (15 s), and 72°C (30 s), and plate reading after each cycle. The following gene-specific primers were used: OMADS1, O1-RT-1 (5′-TGGAGACTTTTCGTAATCATTCAAATAAC-3′) and O1-RT-2 (5′-ATGGTTGCTTCAGAAGAGACATATTG-3′); OMADS3, O3-RT-1 (5′-GCGAATCAGAGGTTAGCACATTG-3′) and O3-RT-2 (5′-AAATTACTGGTCTATACATGAGGAAAGG-3′); OMADS5, O5-RT-1 (5′-ACGAACTGGAGATGAAAGACGAG-3′) and O5-RT-2 (5′-GTAGATTGGGTTGAATTGGCTGAG-3′); OMADS8, O8-RT-1 (5′-ATGGAAGGCAGCATGAGAGAAC-3′) and O8-RT-2 (5′-AAAGCGTTAGCATTGTTACTTGTTT-3′); and OMADS9, O9-RT-1 (5′-TGATGATCCGAACAATTATGATGGTG-3′) and O9-RT-2 (5′-TTTGGCTGGCTTGGTTGGG-3′). The O. Gower Ramsey α-tubulin gene was used as a normalization control with the primers OnAT-RT-3 (5′-GGATTAGGCTCTCTGCTGTTGG-3′) and OnAT-RT-4 (5′-GTGTGGATAAGACGCTGTTGTATG-3′). Data were analyzed using the Gene Expression Macro software (version 1.1; Bio-Rad).
Plant Transformation and Transgenic Plant Analysis
The full-length cDNAs of OMADS5, OMADS8, and OMADS9 were cloned into binary vector PBI121 (Clontech) under the control of the cauliflower mosaic virus 35S promoter. These constructs were transformed into Arabidopsis plants using the floral dip method as described elsewhere (Clough and Bent, 1998). Transformants were selected in medium containing 50 μg mL−1 kanamycin and were further verified by PCR and RT-PCR analyses.
Yeast Two-Hybrid Analysis
The cDNA truncations without the MADS box regions for OMADS3, OMADS5, OMADS8, and OMADS9 were amplified by PCR. The primers for OMADS3 were O3-Y-5 (5′-CATATGCCTTCTACAGAAATCAAAGATGCG-3′) and O3-Y-3 (5′-GGATCCTATCCACAGGCAATGTAAAAT-3′); for OMADS5 were O5-Y-5 (5′-CATATGTACCAGATTGTAACTGGTAT-3′) and O5-Y-3 (5′-GGATCCCCCATTAAATACAGCAACAA-3′); for OMADS8 were O8-Y-5 (5′-CATATGCCATCGACAACGTTGTCGAAGATGT-3′) and O8-Y-3 (5′-GGATCCACCAACAAGGAGGAGTACAAT-3′); and for OMADS9 were O9-Y-5 (5′-CATATGACTACTGACACGAAGAGCATA-3′) and O9-Y-3 (5′-GGATCCTACTCAATGATGATGGTTCGC-3′). Specific 5′ and 3′ primers contained the NdeI (5′-CATATG-3′; underlined) or BamHI (5′-GGATCC-3′; underlined) recognition site to facilitate cloning of the cDNAs. PCR fragments were ligated into the plasmid pGBKT7 (binding domain vector) or pGADT7 (activation domain vector) provided by the Matchmaker Two-Hybrid System 3 (Clontech). Recombinant plasmids were transformed into yeast using the lithium acetate method (Gietz et al., 1992). The transformants were selected on selection medium according to the manufacturer's instructions. β-Galactosidase activity in the transformants was analyzed as described by Tzeng and Yang (2001) and calculated according to Miller (1992).
Confocal Laser Scanning Microscopy
Flower tissues were imaged by an Olympus FV1000 confocal microscope. The plant cell wall was stained with 40 μg mL−1 propidium iodide (Molecular Probes). Propidium iodide was excited by a 543-nm helium/neon laser line, and emission was collected at 555 to 655 nm. Three-dimensional reconstruction was performed using the Fluoview 1000 software.
Scanning Electron Microscopy
Scanning electron microscopy was performed according to the methods of Hsu and Yang (2002) and Tzeng and Yang (2001). Various floral organs were fixed in 2% glutaraldehyde in 25 mm sodium phosphate buffer (pH 6.8) at 4°C overnight. After dehydration in a graded ethanol series, specimens were critical point dried in liquid CO2. The dried materials were mounted and coated with gold-palladium in a JBS sputter coater (model 5150). Specimens were examined with a Topcon scanning electron microscope (model ABT-150S) with an accelerating voltage of 15 kV.
Phylogenetic Analyses
Predicted amino acid sequences of B class genes were obtained from the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/) database. These were aligned using ClustalW, and a phylogenetic tree was constructed using Bayesian analyses. To select an appropriate protein model for subsequent phylogenetic analyses, the program ProtTest (Abascal et al., 2005) was used. The JTT + G + I model with among-site rate heterogeneity (α = 1.58) and invariable sites (pinvar = 0.05) was selected as the most fit model using a test of the Akaike Information Criterion framework (Abascal et al., 2005). Phylogenetic trees were reconstructed with Bayesian, and the numbers on every node indicate the Bayesian posterior probabilities.
The Bayesian method was conducted with the program MrBayes (Huelsenbeck and Ronquist, 2001). In the Bayesian analysis, one million generations of Markov chains were run. Trees were saved every 100 generations for a total size of 20,000 in the initial samples. Stationary phase of log likelihood was reached within 500,000 generations. Thus, burin-in (numbers of the initial trees were discarded) was set to 5,000. A majority rule consensus tree was constructed from the 15,000 remaining trees.
Acknowledgments
We thank Mr. T.-F. Chou for helping us grow the orchid plants used in this research in the field. We also thank Dr. H.-W. Kao (Department of Life Science, National Chung Hsing University) for helping to generate the phylogenetic tree in this study.
This work was supported by the National Science Council of Taiwan (grant nos. NSC95–2317–B–005–006 and NSC96–2317–B–005–019 to C.-H.Y.) and by the Ministry of Education (5Y/50B grant).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Chang-Hsien Yang (chyang@dragon.nchu.edu.tw).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21 2104–2105 [DOI] [PubMed] [Google Scholar]
- Adam H, Jouannic S, Orieux Y, Morcillo F, Richaud F, Duval Y, Tregear JW (2007) Functional characterization of MADS box genes involved in the determination of oil palm flower structure. J Exp Bot 58 1245–1259 [DOI] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5 569–579 [DOI] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Bussher M, Colombo L, van Tunen AJ (1993) Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J 4 101–112 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Romera-Branchat M, Pelaz S (2005) A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J 43 586–596 [DOI] [PubMed] [Google Scholar]
- Chang YY, Chiu YF, Wu JW, Yang CH (2009) Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol 50 1425–1438 [DOI] [PubMed] [Google Scholar]
- Clark JI, Coen ES (2002) The cycloidea gene can respond to a common dorsoventral prepattern in Antirrhinum. Plant J 30 639–648 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37 [DOI] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF (2006) Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stilio VS, Kramer EM, Baum DA (2005) Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae): a new model for the study of dioecy. Plant J 41 755–766 [DOI] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14 1935–1940 [DOI] [PubMed] [Google Scholar]
- Doyle JJ (1994) Evolution of a plant homeotic multigene family: toward connecting molecular systematic and molecular developmental genetics. Syst Biol 43 307–328 [Google Scholar]
- Drea S, Hileman LC, de Martino G, Irish VF (2007) Functional analyses of genetic pathways controlling petal specification in poppy. Development 134 4157–4166 [DOI] [PubMed] [Google Scholar]
- Fitch CM (2004) Encyclopedia of orchids for indoors: Oncidium alliance. In CM Fitch, ed, The Best Orchids for Indoors. Brooklyn Botanic Garden, New York, pp 62–69
- Gietz D, St Jean A, Woods RA, Schiesti RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8 1548–1560 [DOI] [PubMed] [Google Scholar]
- Guo B, Hexige S, Zhang T, Pittman JK, Chen D, Ming F (2007) Cloning and characterization of a PI-like MADS-box gene in Phalaenopsis orchid. J Biochem Mol Biol 40 845–852 [DOI] [PubMed] [Google Scholar]
- Hernandez-Hernandez T, Martinez-Castilla LP, Alvarez-Buylla ER (2007) Functional diversification of B MADS-box homeotic regulators of flower development: adaptive evolution in protein-protein interaction domains after major gene duplication events. Mol Biol Evol 24 465–481 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529 [DOI] [PubMed] [Google Scholar]
- Hsu HF, Huang CH, Chou LT, Yang CH (2003) Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 44 783–794 [DOI] [PubMed] [Google Scholar]
- Hsu HF, Yang CH (2002) An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol 43 1198–1209 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755 [DOI] [PubMed] [Google Scholar]
- Irish VF (2009) Evolution of petal identity. J Exp Bot 60 2517–2527 [DOI] [PubMed] [Google Scholar]
- Jack T (2004) Molecular and genetic mechanisms of floral control. Plant Cell (Suppl) 16 S1–S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A, Nakada M, Akita Y, Hirai M (2007) Class B gene expression and the modified ABC model in nongrass monocots. ScientificWorldJournal 19 268–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A, Saeki H, Kameya T, Saedler H, Theißen G (2003) Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52 831–841 [DOI] [PubMed] [Google Scholar]
- Kim S, Yoo MJ, Albert VA, Farris JS, Soltis PS, Soltis DE (2004) Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am J Bot 91 2102–2118 [DOI] [PubMed] [Google Scholar]
- Kim SY, Yun PY, Fukuda T, Ochiai T, Yokoyama J, Kameya T, Kanno A (2007) Expression of a DEFICIENS-like gene correlates with the differentiation between sepal and petal in the orchid, Habenaria radiata (Orchidaceae). Plant Sci 172 319–326 [Google Scholar]
- Kramer EM (2009) Aquilegia: a new model for plant development, ecology, and evolution. Annu Rev Plant Biol 60 261–277 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Di Stilio VS, Schlüter PM (2003) Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int J Plant Sci 164 1–11 [Google Scholar]
- Kramer EM, Dorit RL, Irish VF (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149 765–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM (2007) Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19 750–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Irish VF (1999) Evolution of genetic mechanisms controlling petal development. Nature 399 144–148 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM (1996) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122 11–22 [DOI] [PubMed] [Google Scholar]
- Lamb RS, Irish VF (2003) Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc Natl Acad Sci USA 100 6558–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54 701–711 [DOI] [PubMed] [Google Scholar]
- Lu HC, Chen HH, Tsai WC, Chen WH, Su HJ, Chang DC, Yeh HH (2007) Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol 143 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZX, Wu M, Loh CS, Yeong CY, Goh CJ (1993) Nucleotide sequence of a flower-specific MADS box cDNA clone from orchid. Plant Mol Biol 23 901–904 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360 273–277 [DOI] [PubMed] [Google Scholar]
- Miller JH (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, New York
- Mondragón-Palomino M, Hiese L, Härter A, Koch MA, Theißen G (2009) Positive selection and ancient duplications in the evolution of class B floral homeotic genes of orchids and grasses. BMC Evol Biol 9 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Theißen G (2008) MADS about the evolution of orchid flowers. Trends Plant Sci 13 51–59 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Theißen G (2009) Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann Bot (Lond) 104 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YH, Jung JY, Kang HG, An G (1999) Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol Biol 40 167–177 [DOI] [PubMed] [Google Scholar]
- Münster T, Wingen LU, Faigl W, Werth S, Saedler H, Theissen G (2001) Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 262 1–13 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–479 [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-Lopez R, Alvarez-Buylla ER, Yanofsky MF (2001) Conversion of leaves into petals in Arabidopsis. Curr Biol 11 182–184 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta DS, Savidge B, Liljergren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K, Vijayraghavan U (2003) Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorlspecific function in floral organ patterning. Genetics 165 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Hileman LC (2009) Developmental genetics of floral symmetry evolution. Trends Plant Sci 14 147–154 [DOI] [PubMed] [Google Scholar]
- Purugganan MD (1997) The MADS-box floral homeotic gene lineages predate the origin of seed plants: phylogenetic and molecular clock estimates. J Mol Evol 45 392–396 [DOI] [PubMed] [Google Scholar]
- Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF (1995) Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal Horticultural Society (1998) RHS Orchids 98 (CD-ROM). Royal Horticultural Society, Singapore
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig WE, Saedler H, Sommer H (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Su K, Lu W, Kong H, Chen Z, Meng Z (2006) Conservation and divergence of candidate class B genes in Akebia trifoliata (Lardizabalaceae). Dev Genes Evol 216 785–795 [DOI] [PubMed] [Google Scholar]
- Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, Schwarz-Sommer Z (1990) Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J 9 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellari GM, Jaramillo MA, Kramer EM (2004) Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Mol Biol Evol 21 506–519 [DOI] [PubMed] [Google Scholar]
- Theißen G (2001) Development of floral organ identity: stories from MADS house. Curr Opin Plant Biol 4 75–85 [DOI] [PubMed] [Google Scholar]
- Theißen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42 115–149 [PubMed] [Google Scholar]
- Theißen G, Kim JT, Saedler H (1996) Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol 43 484–516 [DOI] [PubMed] [Google Scholar]
- Theißen G, Saedler H (1995) MADS-box genes in plant ontogeny and phylogeny: Haeckel's ‘biogenetic law’ revisited. Curr Opin Genet Dev 5 628–639 [DOI] [PubMed] [Google Scholar]
- Theißen G, Saedler H (2001) Floral quartets. Nature 409 469–471 [DOI] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z (1992) GLOBOSA: a homeotic gene which interacts with DEFICIENS in control of Antirrhinum floral organogenesis. EMBO J 11 4693–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Kuoh CS, Chuang MH, Chen WH, Chen HH (2004) Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol 45 831–844 [DOI] [PubMed] [Google Scholar]
- Tsai WC, Lee PF, Chen HI, Hsiao YY, Wei WJ, Pan ZJ, Chuang MH, Kuoh CS, Chen WH, Chen HH (2005) PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol 46 1125–1139 [DOI] [PubMed] [Google Scholar]
- Tsai WC, Pan ZJ, Hsiao YY, Jeng MF, Wu TF, Chen WH, Chen HH (2008) Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol 49 814–824 [DOI] [PubMed] [Google Scholar]
- Tzeng TY, Liu HC, Yang CH (2004) The C-terminal sequence of LMADS1 is essential for the formation of homodimers for B function proteins. J Biol Chem 279 10747–10755 [DOI] [PubMed] [Google Scholar]
- Tzeng TY, Yang CH (2001) A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol 42 1156–1168 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T (2004) The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol AR, Brunelle A, Tsuchimoto S, Chua NH (1993) Functional analysis of petunia floral homeotic MADS box gene Pmads1. Genes Dev 7 1214–1228 [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78 203–209 [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Zanis MJ, Kellogg EA, Schmidt RJ (2007) Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc Natl Acad Sci USA 104 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theißen G (1999) MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc Natl Acad Sci USA 96 7342–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter KU, Weiser C, Kaufmann K, Bohne A, Kirchner C, Kanno A, Saedler H, Theißen G (2002) Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol Biol Evol 19 587–596 [DOI] [PubMed] [Google Scholar]
- Xu Y, Teo LL, Zhou J, Kumar PP, Yu H (2006) Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J 46 54–68 [DOI] [PubMed] [Google Scholar]
- Yadav SR, Prasad K, Vijayraghavan U (2007) Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39 [DOI] [PubMed] [Google Scholar]
- Yao SG, Ohmori S, Kimizu M, Yoshida H (2008) Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol 49 853–857 [DOI] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pollanen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH (1999) Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J 17 51–62 [DOI] [PubMed] [Google Scholar]
- Yu H, Goh CJ (2000) Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol 123 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Yang SH, Goh CJ (2002) Spatial and temporal expression of the orchid floral homeotic gene DOMADS1 is mediated by its upstream regulatory regions. Plant Mol Biol 49 225–237 [DOI] [PubMed] [Google Scholar]
- Zahn LM, Leebens-Mack J, Depamphilis CW, Ma H, Theißen G (2005) To B or not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J Hered 96 225–240 [DOI] [PubMed] [Google Scholar]