One of the major challenges of plant biology is to understand and model plant molecular networks coordinating growth and development, in response to nutritional cues. Further, the predictions and/or the genetic modifications of these responses would have the potential of providing solutions to tweak nutrient inputs of the entire food chain. However, the functioning of a plant as a system is not the result of a simple network, but rather a combination of multiple, intertwined, dynamic, and linear or nonlinear interactions between its elements (DNA, RNA, proteins, metabolites, organelles, cell types, organs, etc.). In addition, this sessile system has to face a combination of fluctuating environmental conditions, including biotic and abiotic stresses. It becomes obvious that the comprehensive knowledge of how this plant system functions in its environment cannot be achieved by the sequential characterization of its elements one by one, or a single class of elements in isolation of the others. Thus, in plant science as well as in other fields of fundamental biology, a paradigm shift is needed. The complexity of the system is so large that understanding it as a whole is without any doubt a necessity to further understand biological systems. Indeed, the reductionist approach is mainly based on reverse genetics (use of mutants or overexpressors to decipher gene function). Several articles cite the fact that the reductionist approach, the one used by biologists on a daily basis, would fail, if it was applied to the study of complex physical system such as a radio (Lazebnik, 2002). Removing components of a radio would probably identify the major components of the studied systems (e.g. speakers or antenna), but it certainly will never give a global understanding of how the radio receives an electromagnetic signal and transform it into audible sounds. This change in our point of view needs to be applied to biological systems, and in particular to plant systems biology.
During these most recent years, the new field of plant systems biology has emerged that has been developed to resolve some key questions using a maximum level of integration: the plant as a whole. We can still think that the ultimate goal of these new approaches and associated tools is to build a model of the system precisely enough that we could build a virtual plant. Developing a virtual plant systems model can be used to (1) explain mechanistically how molecular network changes evoke responses and (2) predict molecular network states under untested conditions or in response to environmental or genetic modifications. In the long term, this systems approach should enable researchers to test biotechnological strategies for gene modification in silico, prior to implementation in transgenic crop plants.
The genesis of systems biology is linked to technological advances that were able to draw a broad picture (if not exhaustive) of each level of integration. The first step was clearly done when high-throughput technologies allowed the acquisition of omics data, i.e. genomic, transcriptomic, proteomic, and metabolomic. For (plant) biologists, this is the opportunity to collect large sets of comprehensive and quantitative data, in a large variety of conditions. The second step is the possibility to store these large data sets, compute them, and infer biological meaning by a click integrative analysis using web-based tools. This critical step in the post-genomic area has implied the opening of a dialog between mathematicians, statisticians, computer scientists, and biologists (Shasha, 2003). This dialog has been successful if we consider the number of programs and resources that have been developed these last years (for review, see Brady and Provart, 2009).
To date, plant systems biology is at a key step of its evolution, since presently most of the levels of omics data are being investigated and some studies have begun to integrate different dimensions of understanding. In this Update, we will first present the different levels/dimensions of understanding that can or have been integrated in modern plant biology. In the second part, we will present insights that systems approaches provided concerning plant nutrition.
DECIPHERING THE MULTIDIMENSIONAL PLANT SYSTEM
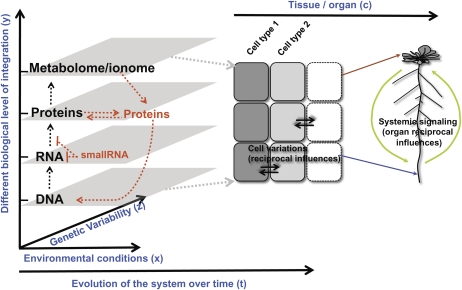
At present, the complexity of biological systems is largely misunderstood and underexploited. The limiting processes to this integrative knowledge are the development of tools able to give a broad enough picture of the system to integrate the different levels of information. In this section, we describe where the plant systems biology field stands concerning the different levels of integration that need to be studied and issues that need to be tackled to give a precise picture of the underlying molecular events occurring as a whole in plants. The examples will be taken as much as possible from data integrating nutritional signals in plants. We classified the elements of the plant system into five (nonexhaustive) dimensions (Fig. 1, in blue).
Figure 1.
Plant systems biology: a dimension-wise challenge. This scheme illustrates the different (nonexhaustive) dimensions (in blue) that can be integrated in systems approaches, to decipher emerging properties of plant regulatory networks. This work presents different studies that integrate those dimensions to enrich our knowledge of physiology of plant nutrition and to build predictive models with applications in agriculture and the environment.
First, the core of the actual systems biology is the integration of the current paradigm of the components of molecular biology. This consists of the sequential catalysis of DNA information into RNA and proteins (Fig. 1, axis y); the latter being involved in driving metabolism. To date, the best-documented dimension of the system is at the RNA level, since microarrays and new deep-sequencing methods can give an almost exhaustive view of the RNA pool of cells (Lister et al., 2008). The protein level is to some extent less accessible to high-throughput experimental procedures, compared to the RNA world. However, new advances such as protein chips are being used to identify protein interactions genome wide (Popescu et al., 2007) and platforms using enzymatic cycling assays to monitor enzymatic activities genome wide (Gibon et al., 2004) have begun to capture information about the protein world. At the next step, the exhaustive study of all metabolites profile, now provides a very good quantitative catalog of this layer of information (Schauer and Fernie, 2006). Many studies have begun to integrate metabolite studies together with mRNA accumulation analysis (e.g. Sulpice et al., 2009). Beside these biomolecules (RNA, proteins, and metabolites), the ionome (defined as the elemental composition of living organism) constitutes an additional level of the plant system state (Salt et al., 2008). For instance, ionomics can be used as a read out of the whole-plant physiological state (Baxter et al., 2008).
Second, a crucial level of understanding is to study the system behavior by varying the experimental conditions (Fig. 1, axis x). Most of the studies in plant systems biology now involve the study of one or more different nutrient parameters such as nitrogen, carbon, sulfate, and light treatments. For instance, transcriptome studies have been used to investigate the regulatory interaction between multiple inputs such as carbon and nitrogen (Price et al., 2004; Gutierrez et al., 2007), carbon and light (Thum et al., 2004, 2008), as well as carbon and circadian rhythms (Blasing et al., 2005), or more recently carbon-light-nitrogen in two different organs (Krouk et al., 2009). Interestingly, in the latter dataset this complete combinatorial experiment involving three inputs (carbon, light, and nitrogen), which enabled the measurement of mRNA accumulation with a potential high degree of freedom, highlighted the fact that multiple signals coordinate gene expression precisely and according to a constrained plan. Indeed, gene expression can be explained by a relatively limited number of behaviors/models of expression compared to the potential freedom given by the combinatorial experiment (Krouk et al., 2009). This kind of analysis has some potential to uncover the relationships between signal interactions in the control of gene expression. However, to our knowledge, this combinatorial approach has never been used to study other components of dimension number one (i.e. proteins or metabolites) in Figure 1. It might be of great interest to know if this constrained signaling structure is conserved at other levels of integration, such as metabolites for instance.
Third, the use of natural variation becomes an increasing new level of understanding in plants (e.g. genome-wide association mapping), in particular in conjunction with new deep-sequencing techniques (http://1001genomes.org/downloads/shore.html; Nordborg and Weigel, 2008; Fig. 1, axis z). Thus, natural or artificial genetic variability is an invaluable resource for systems biology. Indeed, the genetic perturbation of the biological system was, is, and will always be, a gold standard to experimentally validate hypothesis generated by systems approaches (Gutierrez et al., 2005). Moreover, to date, only very few studies reported the use of natural variation as support for the building of gene regulatory networks or as a source of variation needed to infer relationships between the different dimensions of the system as a whole (Sulpice et al., 2009).
Fourth, for multicellular organisms, the recent advances in the technique of cell sorting has opened the door to exciting quantitative understanding of how the different cell types coordinate gene expression responses (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008; Gifford et al., 2008; Fig. 1, axis c). For instance, the Petersson et al. (2009) study is a striking example of the possibility offered by the cell sorting to measure metabolite (here auxin accumulation and synthesis) and mRNA level (even if the data are not issued form the same set of experiments), to understand the possible connection between transcriptome and metabolome at a cell type level. Just a step further, is the integration of the signal in different organs. This organ specificity in signal transduction is of great interest, since it has been demonstrated that the same signals are differently transduced in different organs (Krouk et al., 2009).
To explore further the question of how a cellular territory can influence the response of another one (e.g. influence of epidermis on pericycle cells responses; influences of shoots on roots responses), it will be also a main challenge to integrate the reciprocal influences between molecular and macroscopic phenotypes (e.g. leaf development, architecture of root system). However, like systems molecular components, these morphological traits have a dynamic and quantitative nature, which needs to be integrated into the models. Then, the development of high-throughput phenotyping platforms allowing (1) the control of nutritional stress conditions, and (2) the simultaneous acquisition of morphological changes across the time for many plants has to be a main objective for plant systems biology (Granier et al., 2006; Montes et al., 2007).
Fifth, the plant system in its multidimensional space is not a fixed picture, but a dynamic one. Intuitively, if two components A and B (which can be any kind of molecular of physiological parameter described above) are correlated in a given data set, nothing precludes their potential cause/consequence relationships (it can be A→B or B→A). However, time wise, if A comes first and is still lag correlated to B, then the relationship inferred will be A→B. This simplistic example demonstrates that the time factor can greatly simplify inferred regulatory networks at all the integrative levels (Fig. 1; axis t). Surprisingly, only a very few reports use time-course analysis to infer regulatory/metabolic networks in plants in response to nutritional cues (Hirai and Saito, 2008; Usadel et al., 2008).
Together, these five dimensions of plant systems (Fig. 1) define a world of study far beyond from what our brain can intuitively process to create biological meaningful hypotheses. This is where computer science and mathematics can help to resolve this projection. All the currently developed informatic tools available for analyzing and integrating plant genomic data have recently been reviewed by Brady and Provart (2009). However, here we wish to emphasize some of them based of their capacity to bring together different layers/dimensions of information. First, since we consider that one purpose of systems approach is to operate a projection in understandable dimensions, one very popular technique to operate this delicate operation is the principal component analysis. Principal component analysis has been successfully used to evaluate the coordination of multidimensional dataset in studies involving, for example, carbon (Sulpice et al., 2009) or sulfur (Hirai et al., 2004). Second, another very popular tool is MapMan (Thimm et al., 2004), which helps to visualize metabolic pathways and other cellular processes. This tool is able to also incorporate metabolites accumulation data in the same pathways. This step is crucial to compare data from mRNA level and metabolites accumulation. Our lab has developed a tool embodied in the VirtualPlant platform, which enables the integration of all available information on Arabidopsis (Arabidopsis thaliana) genomic data into a multinetwork. This multinetwork embodies edges connecting gene nodes coming from multiple evidence, including: metabolic pathway connections, protein:protein and protein:DNA interactions, microarray data, microRNA:target datasets, and literature-based interactions (Gutierrez et al., 2007). This approach combined with correlation databased on mRNA accumulation levels lead to successful findings described below for their physiological consequences. Along parallel lines, a predicted protein interactome has been built for Arabidopsis (Geisler-Lee et al., 2007). This predicted interactome network offers the possibility to see if coexpressed genes are interologs (predicted interactions) or interactors (known interactions). Furthermore, biological function of unknown genes can be hypothesized according to the predicted interaction with a gene displaying a known function.
BIOLOGICAL INSIGHTS FROM MULTIDIMENSIONAL APPROACHES
From DNA to Metabolome Integration (Fig. 1; Axis y)
Transcriptome and Metabolome Integration
To obtain a systems view of plant responses to nutrient stress, one widely used approach consists in integrating omics data in the context of known metabolic pathways. MapMan is one of these developed tools (Thimm et al., 2004). Using this software, a research group (M. Stitt, W.-R. Scheible, Y. Gibon, and collaborators) provided a large documentation of the transcriptome, metabolome, and even enzymatic activities coordination in response to different nutritional cues like carbon, nitrogen, potassium, and phosphate (Scheible et al., 2004; Morcuende et al., 2007; Osuna et al., 2007; Armengaud et al., 2009). For example, they have well-documented genome-wide reprogramming and metabolites changes in response to nitrate treatment of nitrogen-starved plants (Scheible et al., 2004). Nitrate addition led to a rapid (30-min) coordinated induction of genes for nitrate uptake and assimilation, genes required to provide reducing equivalents, and genes required for the synthesis of organic acids, whereas primary metabolites do not change significantly. However, after 3 h, the accumulation of central amino acids (Gln, Glu, Asp, Ala) is increased and while minor amino acids (e.g. Trp, Leu, Lys, etc.) decreased in levels. This primary metabolism modification is paralleled to the induction of genes assigned to amino acid and nucleotide biosynthesis, likely in coordination with the induction of genes required for RNA and protein synthesis. In addition to these responses of primary metabolism genes, this study also provided a broad view of how nitrate affects others cellular functions, including trehalose metabolism, hormone synthesis, signaling posttranslational modification, etc. Finally, in some cases, the level of change in gene expression was low, but their assignment as a group into known metabolic pathway allowed the analysis to highlight the regulation of relevant functional categories (e.g. RNA and protein synthesis; Scheible et al., 2004).
By integrating different levels of omics data onto known metabolic pathways, two research groups have provided comprehensive insights into the regulation and the coordination of sulfur metabolism at a system level (for review, see Hirai and Saito, 2008; Hoefgen and Nikiforova, 2008). This integration led to the reconstruction of a global scheme of coordination of plant response to short- and long-term hyposulfur stress (Hoefgen and Nikiforova, 2008). As an immediate response to sulfur starvation, modifications of metabolic pathways prioritization occurred. For example, the amounts of the first organic sulfur-containing compounds, like Cys or glutathione, decrease whereas precursors O-acetyl-l-Ser and Ser accumulate and then, Trp and Gly. Globally, these metabolic fluxes changes do not require the action of additional regulators. The major changes in transcript profiling occurred in the transition phase and late responses. For the long-term sulfur starvation, plant status is characterized by a significant rebalancing of metabolism. A network depicting mutual influence between sulfur assimilation, nitrogen imbalance, lipid breakdown, purine metabolism, and photorespiration has been assembled (Hoefgen and Nikiforova, 2008).
These two examples of responses to nitrate addition and sulfur starvation illustrate systems strategies for rapid adaptation to nutritional modifications. These results suggest that modifications of metabolic pathways are prioritized in response to nutrient limitation, whereas response to nutrient readdition is more characterized by a genome-wide transcriptional reprogramming.
Finally, beyond the purpose of modeling nutrient metabolism, the elucidation of gene-to-gene and metabolite-to-gene networks via these omics data integration also constitutes a strategy for the identification of novel gene functions (Saito et al., 2008). In this study, some integrated method, like batch-learning self-organizing mapping, allowed the researchers to test novel gene functions by clustering genes and metabolites based on their time-dependent expression/accumulation changes. For instance, a zoom into the glucosinolate biosynthesis during the sulfur starvation allowed the characterization of Myb transcription factors as positive regulatory factors for aliphatic glucosinolate biosynthesis (Hirai et al., 2007).
Integration of Transcriptomic Data into a Comprehensive Multinetwork Model
Integrative network biology is another interesting strategy to achieve omics data integration (Gutierrez et al., 2005). Recently, a multinetwork tool integrating information for cellular components interactions has been developed. This multinetwork integrates a variety of data like Arabidopsis metabolic pathways; known protein-protein, protein-DNA, and miRNA-RNA interactions; and predicted protein-protein and protein-DNA interactions (Gutierrez et al., 2007). Using the virtual plant software platform, quantitative transcriptome data are easily integrated to this qualitative model and visualized (http://www.virtualplant.org). In one study, this systems approach has been applied to understand how organic nitrogen (Glu or Glu-derived products) affects genome-wide expression and to infer the nutrient-dependent mechanisms controlling plant growth and development (Gutierrez et al., 2008). The transcriptome responses to nitrogen signals were integrated as regulatory edges into the multinetwork, and used to support predicted protein-DNA interactions as follows: Predictions for TF→target relationships required both overrepresentation of transcription factor-binding sites and high correlated expression of the transcription factors and the putative targets. This network analysis allowed the identification of a metabolic subnetwork of genes regulated by organic nitrogen and including the transcription factor CCA1, a key component of the Arabidopsis circadian clock. Genetic and chromatin immunoprecipitation experiments confirmed the predicted direct interaction of CCA1 with genes involved in nitrogen assimilation (Glu dehydrogenase, Gln synthetase, and Asn synthetase). This study illustrates how a systems approach identified emerging properties of a system. In this case, a feedback loop model of the interactions between the Arabidopsis circadian clock and nitrogen-assimilatory pathway has been proposed (Gutierrez et al., 2008).
How Do Plants Integrate Their Multisignal Environment? (Fig. 1; Axis x)
A second challenge of systems biology is to understand how plants integrate multiple and combined variations of interacting signals to effect changes in gene expression, protein activities, or metabolite accumulation, and to direct new programs for development. To date, few reports have addressed the interaction between nutrient signaling on a genome-wide scale. Several reports on such integration from our own group have focused on the integration of three important signals: nitrogen, carbon, and light (Gutierrez et al., 2007; Thum et al., 2008; Krouk et al., 2009). In one of these studies, the authors explored global gene responses of roots exposed to a matrix of carbon and nitrogen treatments and integrated those data using the qualitative multinetwork approach described above (Gutierrez et al., 2007). Using ANOVA analysis, they defined quantitative models of carbon-nitrogen regulation for all detected genes to identify the main effects of carbon and/or nitrogen as well as the interaction between these two signals. First, in this carbon-nitrogen matrix, this modeling showed that among all the regulated genes, carbon alone and carbon-nitrogen (independently of the ratio) are the main factors driving gene regulation. Second, many genes previously identified as nitrogen or carbon responsive were found to be regulated by carbon-nitrogen interactions in this analysis. Finally, an Arabidopsis subnetwork controlled by carbon, nitrogen, and carbon-nitrogen has been built. For example, in this network, biological modules responding to carbon and/or nitrogen have been identified; e.g. the proteasome machinery is mainly regulated by carbon only (Gutierrez et al., 2007).
To go further into genome-wide signal integration, our group investigated the interactions between carbon, nitrogen, and light in two organs, roots, and shoots (four factors: carbon, nitrogen, light, organs; Krouk et al., 2009). To mine these complex data, the response of each gene has been first quantitatively modeled (ANOVA) as a function of each factor and all possible interactions. By analyzing the number of genes controlled by all signals (carbon, nitrogen, light, organs) or composite signals (e.g. carbon-nitrogen, carbon-light, carbon-organs, etc.), it has been proposed that a code governs signal integration at the organism level and some rules for this integration have been drawn. For example, in leaves light is the predominant signal controlling gene expression, coherently with the role of leaves as a light energy converter. By contrast, in roots, the signal integration is stronger since 90% of the genes are controlled by more than one signal or composite signal. For example in roots, light responses are related to carbon responses, suggesting that light is mainly sensed as an internal carbon signal in roots. Similarly, the effect of nitrogen is mainly observed in interaction with carbon and light, and very poorly controls gene expression on its own. Finally, a general model of signal integration in roots has been proposed. This model includes a gene module (containing 136 genes) that is regulated when carbon is provided alone, but not regulated when carbon is provided in combination with nitrogen. This module is thus likely to be under the control of a yet to be defined carbon-nitrogen balance sensing system (Krouk et al., 2009).
These two examples above illustrate how mathematical mining of complex transcriptome data sets provided insights into the integration of nutrient signal transduction pathways by reducing the complexity of the regulation of approximately 22,000 individual genes in a reasonable number of models of regulation.
Integration of Genetic Variability in Systems Approach (Fig. 1; Axis z)
The integration of genetic variability provides an additional dimension in our understanding of the system. Below, we illustrate this point using two examples that involve experimentally induced and naturally occurring genetic variability, respectively. In the first study, an Arabidopsis mutant has been used, named cli186 for carbon and light insensitive, which was identified on the basis of the misregulation of the Asn synthase gene (ASN1) by carbon and light (Thum et al., 2008). This genetic approach has been integrated with a systems approach, including the generation of transcriptomic data, a combination of carbon and light treatments, and the use of the Arabidopsis qualitative multinetwork described above (Thum et al., 2008). The modeling of the gene expression data revealed that 20% of the genes regulated by carbon and/or light in the wild type are misregulated in the cli186 mutant. All regulated genes have been interpreted in the context of metabolic and regulatory network using the multinetwork. A subnetwork of all genes misregulated in cli186 revealed that carbon-compound metabolism, amino acid metabolism (e.g. ASN1), and glycolysis/gluconeogenesis are the main perturbed processes and likely coordinated by a homeobox-Leu zipper transcription factor, HAT22. This work illustrates how the combination of genetic, genomic, and systems approaches allows the identification of an upstream main integrator of nutritional signal and propose the network allowing the signal transduction until the regulation of the metabolic targets.
In the second study, natural genetic variability in Arabidopsis has been used to identify metabolic networks (metabolites and/or genes) that coordinate metabolism and growth, as potential targets for improvement or prediction of plant biomass (Sulpice et al., 2009). First, in a core collection of 94 Arabidopsis accessions, sophisticated correlation analysis between biomass and metabolite content (carbohydrates, organic acids, amino acids, etc.) showed that starch is the major integrator of the metabolic status in a wide range of environmental conditions. The fact that starch is negatively regulated with plant biomass could be explained by regulatory network programs that maximize growth at the expense of carbon reserves. Then, in a subset of 21 biomass-contrasted accessions, potential changes in a robust core sugar-responsive gene network have been tested and correlations with metabolites and biomass have been defined. Two genes, coding for a Kelch repeat F-box protein and a myoinositol-1-P synthase1, have been identified as contributors of the regulation of carbon partitioning and growth because their transcript levels correlate with rosette biomass and association mapping linked polymorphisms in these genes with rosette biomass and starch. This study is an elegant example of the valuable dimension that natural genetic variability offers to dissect networks controlling complex traits and to propose robust biomarker for prediction of agronomical traits. Overall, this study is a landmark in data integration, since it gathers almost all the dimensions depicted in Figure 1, including: RNA and metabolites accumulation, environmental conditions, natural genetic variability, and evolution of the system over time.
Cell Specificity in Systems Approach (Fig. 1; Axis c)
Developmental programs are processes driven for a large part by the regulation of different gene networks whereby cells acquire their functional specificity (for review, see Iyer-Pascuzzi and Benfey, 2009). To achieve a system-level understanding of developmental responses to nutritional cues requires that one integrate the cellular complexity of organs. However, so far few studies have dissected the root responses of gene networks in different cell types, likely due to the requirement of sophisticated technologies. A comprehensive view of how specific root tissues respond to nitrate signal on a global scale has been obtained (Gifford et al., 2008). This study reveals that a high level (86%) of the nitrate-responsive genes are specific for each of the cell types examined (i.e. lateral root cap, epidermis/cortex, endodermis/pericycle, stele, and pericycle founder cells). This result highlighted that responses to nitrogen are remarkably fine tuned within the root. Moreover, this approach uncovered an unknown aspect of nitrogen responses since 76% of these genes were not identified in previous studies of whole roots. Focusing on hormone-responsive transcription factors, the Gifford et al. (2008) study identified the pericycle and the lateral root cap tissues as the most likely candidate places for the integration of the nitrogen signal leading to an adaptive developmental response. Following this analysis, the authors showed that the balance between lateral root initiation and emergence in response to organic nitrogen signaling is controlled by a network involving the transcription factor ARF8 (for auxin response factor) that is posttranscriptionally controlled by the microRNA 167 (Gifford et al., 2008).
Iron deprivation and high-salinity treatments have been used to address the question of the influence of cell identity on abiotic stress (Dinneny et al., 2008). In this study, the dissection of the roots into a longitudinal and radial axis combined to a time-course analysis has increased the resolution of the gene networks. In brief, (1) transcriptional responses to iron deprivation occurred mainly in the maturation zone and the stele of the roots where iron is predominantly circulated in mature plants, (2) the response in stele is characterized by the regulation of generalized stress-responsive genes, whereas (3) the epidermis is characterized by the activation of genes coding for metal-ion transporter and nicotianamine synthesis and the repression of genes coding for cell wall biogenesis and organization, likely to explain the shortness and misshapenness of root hairs. Interestingly but counterintuitively, the comparison between iron and salt stresses tends to show that cell-specific responses correspond to a general stress response whereas generalized responses to all cells layers are more stimuli specific (Dinneny et al., 2008).
Undeniably, the cell-specific approach offers a new perspective in the identification of relevant transcriptional network for our understanding of the developmental response of multicellular organism-like plants without substituting whole organ approach.
A Dynamic View of the System (Fig. 1; Axis t)
Finally, systems approach cannot do without the temporal dimension because (1) the reconstitution of gene to metabolite networks across a time course allows the implementation of directionality; means that A-B becomes A→B and, (2) metabolic and physiological processes cannot be fully understood if, for example, the influence of diurnal cycles on the regulation of gene to metabolite networks is missing.
For instance, responses to sulfur deprivation have been studied in a dynamic point of view by applying modeling methods to time-series data (see “From DNA to Metabolome Integration”). In one of these studies, the authors reconstructed a gene-metabolite network of correlation driven by a cause-to-effect directionality. For example, paths including proteins involved in auxin biosynthesis and signaling have related the primary cause (sulfur deficiency) to a physiological end point (enhanced root formation; Nikiforova et al., 2005).
Typically, our understanding of carbon metabolism in plants is not separable from the diurnal cycles. Indeed, plants have constantly to deal with carbon surplus in the light and with carbon depletion at night. To overcome these imbalances, plants store carbon surplus as starch and trigger strategic remobilization of this starch for Suc synthesis and export (Smith and Stitt, 2007). A comprehensive analysis of the global response of transcripts and metabolites to progressive carbon depletion has been done to elucidate the mechanisms underlying the strategic carbon remobilization during the night (Usadel et al., 2008). This analysis showed that a major transcriptional reprogramming, including genes involved in biosynthesis and cellular growth, is triggered by small changes of the carbon status. However, this reprogramming is sequential with the regulation of different group of genes across the time. Then, these temporal responses allowed the identification of genes that are potential upstream components of a larger carbon-responsive network and the modeling of these responses by a simple linear model, which captures interactions between the carbon, the light, and the clock (Usadel et al., 2008).
These examples above illustrate that the regulation of genes and/or metabolites in a temporal dimension is an essential measurement to include to our systems approach given that it is a critical input in data modeling.
CONCLUSION AND VISIONS: PREDICTIVE MODELS
Systems biology is not a newborn concept, but the technology needed to practice it on a large scale is just emerging. In plants, as well in other organisms, we now face the challenge to elaborate a projection of the multidimensional systems that we have described herein to understand the system as a whole. This projection needs intense intervention of computer science and math with biology to create new hypothesis and even new findings, as it has been illustrated in this systems analysis from the perspective of plant nutrition. These exciting approaches will likely speed up the findings of emerging properties of molecular networks and plant molecular physiology.
In this review, we essentially illustrated the power of systems approach citing studies that use a top-down approach, which is the most routinely chosen in systems biology. As the name suggests, this approach is looking at the system from the top and consists of an exhaustive description of the interactions among the biological components that will eventually lead to decompose the system to smaller parts. However, a bottom-up approach is also essential to achieve systems modeling, because it aims at reconstituting larger parts from elemental steps (Katagiri, 2003). For instance, the Asp-derived amino acid pathway in Arabidopsis has been chosen as a model system for understanding regulatory mechanisms in branched metabolic pathways. Then, the first detailed kinetic model of a branched metabolic pathway, taking into account all the complexity of the system (the allosteric controls and the different isoforms of the enzymes) has been determined. For example, this model can be used to interpret the effect of genome changes on metabolism (Curien et al., 2009).
Overall, the analysis and the modeling of the whole system or any system in the system open some exciting experimental perspectives. Indeed, the goal of systems biology is to model the systems precisely enough that it enables to predict its reaction in untested conditions. This level of understanding has already been successfully reached in bacteria (Bonneau et al., 2007) exposed to distinct (including nutritional) environments and provides substantial vision for predictive modeling approaches in plant systems biology.
Such predictive modeling in plants can be used to predict how a plant will react to a changing environment, and/or to perturbations in its genome. Thus, in silico tests can be used to identify which genes in the system to change in transgenic plants, to effect changes in response and use of nutrients in the soil, among many other applications.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences (grant no. GM032877), the National Science Foundation Arabidopsis 2010 (grant no. MCB–0929338), the National Science Foundation database activities (grant no. DBI–0445666), and the Department of Energy (grant no. DEFG02–92ER20071). G.K. is supported by a European-FP7-International Outgoing Fellowship (Marie Curie; AtSYSTM-BIOL; grant no. PIOF–GA–2008–220157).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gloria M. Coruzzi (gloria.coruzzi@nyu.edu).
References
- Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y (2009) Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol 150 772–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, Guerinot ML, Salt DE (2008) The leaf ionome as a multivariable system to detect a plant's physiological status. Proc Natl Acad Sci USA 105 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, et al (2007) A predictive model for transcriptional control of physiology in a free living cell. Cell 131 1354–1365 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Brady SM, Provart NJ (2009) Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G, Bastien O, Robert-Genthon M, Cornish-Bowden A, Cardenas ML, Dumas R (2009) Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol Syst Biol 5 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320 942–945 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O'Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, et al (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169 623–635 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Lejay L, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8 R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Shasha DE, Coruzzi GM (2005) Systems biology for the virtual plant. Plant Physiol 138 550–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Saito K (2008) Analysis of systemic sulfur metabolism in plants using integrated ‘-omics’ strategies. Mol Biosyst 4 967–973 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefgen R, Nikiforova VJ (2008) Metabolomics integrated with transcriptomics: assessing systems response to sulfur-deficiency stress. Physiol Plant 132 190–198 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Benfey PN (2009) Transcriptional networks in root cell fate specification. Biochim Biophys Acta 1789 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F (2003) Attacking complex problems with the power of systems biology. Plant Physiol 132 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Tranchina D, Lejay L, Cruikshank AA, Shasha D, Coruzzi GM, Gutierrez RA (2009) A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLoS Comput Biol 5 e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik Y (2002) Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer Cell 2 179–182 [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes JM, Melchinger AE, Reif JC (2007) Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci 12 433–436 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30 85–112 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R (2005) Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J Exp Bot 56 1887–1896 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Weigel D (2008) Next-generation genetics in plants. Nature 456 720–723 [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Blasing OE, Hohne M, Gunter M, Kamlage B, Trethewey R, Scheible WR, et al (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49 463–491 [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 104 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin KS, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K (2008) Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends Plant Sci 13 36–43 [DOI] [PubMed] [Google Scholar]
- Salt DE, Baxter I, Lahner B (2008) Ionomics and the study of the plant ionome. Annu Rev Plant Biol 59 709–733 [DOI] [PubMed] [Google Scholar]
- Schauer N, Fernie AR (2006) Plant metabolomics: towards biological function and mechanism. Trends Plant Sci 11 508–516 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shasha DE (2003) Plant systems biology: lessons from a fruitful collaboration. Plant Physiol 132 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30 1126–1149 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Thum K, Shin M, Palenchar P, Kouranov A, Coruzzi G (2004) Genome-wide investigation of light and carbon signaling interactions in Arabidopsis. Genome Biol 5 R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum KE, Shin MJ, Gutierrez RA, Mukherjee I, Katari MS, Nero D, Shasha D, Coruzzi GM (2008) An integrated genetic, genomic and systems approach defines gene networks regulated by the interaction of light and carbon signaling pathways in Arabidopsis. BMC Syst Biol 2 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Blasing OE, Gibon Y, Retzlaff K, Hohne M, Gunther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]