Abstract
The Saccharomyces cerevisiae SEN1 gene codes for a nuclear-localized superfamily I helicase. SEN1 is an ortholog of human SETX (senataxin), which has been implicated in the neurological disorders ataxia-ocular apraxia type 2 and juvenile amyotrophic lateral sclerosis. Pleiotropic phenotypes conferred by sen1 mutations suggest that Sen1p affects multiple steps in gene expression. Sen1p is embedded in a protein–protein interaction network involving direct binding to multiple partners. To test whether the interactions occur independently or in a dependent sequence, we examined interactions with the RNA polymerase II subunit Rpb1p, which is required for transcription, and Rnt1p, which is required for 3′-end maturation of many noncoding RNAs. Mutations were identified that impair one of the two interactions without impairing the other interaction. The effects of the mutants on the synthesis of U5 small nuclear RNA were analyzed. Two defects were observed, one in transcription termination and one in 3′-end maturation. Impairment of the Sen1p–Rpb1p interaction resulted in a termination defect. Impairment of the Sen1p–Rnt1p interaction resulted in a processing defect. The results suggest that the Sen1p–Rpb1p and Sen1p–Rnt1p interactions occur independently of each other and serve genetically separable purposes in targeting Sen1p to function in two temporally overlapping steps in gene expression.
PROTEIN–protein interaction networks contribute tfhe underlying basis for phenotypic pleiotropy. In Saccharomyces cerevisiae, global studies suggest that each protein interacts on average with five other proteins (Grigoriev 2003), leading to a complex network of interactions involving at least 16,000 individual protein–protein interactions that influence the functions of wild-type proteins and the phenotypes of mutants. The essential S. cerevisiae SEN1 gene codes for a nuclear-localized nucleic acid helicase (DeMarini et al. 1992) that is embedded in a complex network of protein–protein interactions (Ursic et al. 2004). Furthermore, mutations in SEN1 confer pleiotropic phenotypes, including defects in transcription termination, RNA processing, and DNA repair (Steinmetz and Brow 1996, 1998; Rasmussen and Culbertson 1998; Steinmetz et al. 2001, 2006; Ursic et al. 2004). The study of SEN1 therefore provides a useful paradigm to examine the impact of protein–protein interactions on mutant phenotypes and function.
Mutations in human SETX (senataxin), the ortholog of yeast SEN1, cause two clinically distinct neurological diseases, ataxia-ocular apraxia 2 and juvenile amyotrophic lateral sclerosis (Chen et al. 2004, 2006; Moreira et al. 2004; Duquette et al. 2005; Suraweera et al. 2007; Suraweera et al. 2009). The yeast and human proteins are strikingly similar in their organization. Some of the human mutations cause changes in the ATP-helicase domain, whereas others cause changes in the N-terminal region where protein-binding domains reside. Some of the clinical differences might be caused by mutations that differentially affect the function of senataxin by disrupting different protein–protein interactions.
Sen1p interacts with the C-terminal domain of Rpb1p, the largest subunit of RNA polymerase II (RNAP II) (Myer and Young 1998); with Rad2p, a single-strand DNA endonuclease required for DNA repair (Habraken et al. 1993; Prakash and Prakash 2000); with Rnt1p, a double-strand RNA cleavage enzyme involved in 5′- or 3′-end processing (Elela et al. 1996; Chanfreau et al. 1997; Lamontagne et al. 2000); and with SmD3p (Fromont-Racine et al. 1997), a subunit of the heteroheptameric Sm complex that assembles small nuclear RNAs (snRNAs) into ribonucleoprotein particles required for pre-mRNA splicing (Roy et al. 1995; Kambach et al. 1999; Zhang et al. 2001). Recently, it was shown that Sen1p interacts with Glc7p, a protein phosphatase subunit of the cleavage/polyadenylation factor, and with Nab3p, a RNA-binding protein that interacts with other proteins involved in transcription termination of noncoding RNAs (Conrad et al. 2000; Nedea et al. 2008).
RNA processing, ribonucleoprotein assembly, and transcription-coupled DNA repair occur concomitantly with transcription (Komarnitsky et al. 2000; Maniatis and Reed 2002; Neugebauer 2002; Hanawalt and Spivak 2008), suggesting a complex interplay between protein–protein interactions that potentially orchestrate cotranscriptional pathways. The interactions of Sen1p with proteins involved in transcription, processing, and repair might occur independently of each other or they might occur in a dependent sequence of interactions.
To begin assessing the relationships between the different Sen1p protein–protein interactions, we analyzed the effects of sen1 mutations on the expression of SNR7, which codes for U5 snRNA. SNR7 serves as a diagnostic indicator of the relationship between Sen1p protein–protein interactions and Sen1p function because previous studies based on depletion assays suggested a role for SEN1 in U5 RNA 3′-end processing (Ursic et al. 2004). Other reports indicated that Sen1p plays a role in the transcription termination of noncoding RNAs (Steinmetz and Brow 1996, 1998; Rasmussen and Culbertson 1998; Steinmetz et al. 2001, 2006). Furthermore, the Sen1p-interacting partners Rpb1p and Rnt1p are required for U5 snRNA transcription and maturation, respectively (Myer and Young 1998).
The U5 snRNA transcript matures through a branched pathway leading to the production of two functional end products, U5L (214 nucleotides) and U5S (180 nucleotides) (Patterson and Guthrie 1987; Chanfreau et al. 1997) (see Figure 1B). During cotranscriptional maturation, Rnt1p cleaves at two locations in a stem–loop structure leading to accumulation of U5L-3′ RNA (240 nucleotides) and U5-3′a RNA (270 nucleotides). The exosome removes 3′ nucleotides from each of the cleavage products to form mature U5L and U5S RNA, respectively (Allmang et al. 1999). Despite this, a deletion of RNT1 affects only synthesis of U5L, indicating that a Rnt1-independent bypass pathway allows for U5S RNA synthesis in the absence of Rnt1p cleavage. Sen1p and Rnt1p are required for production of U5L RNA but not for U5S RNA (Ursic et al. 2004).
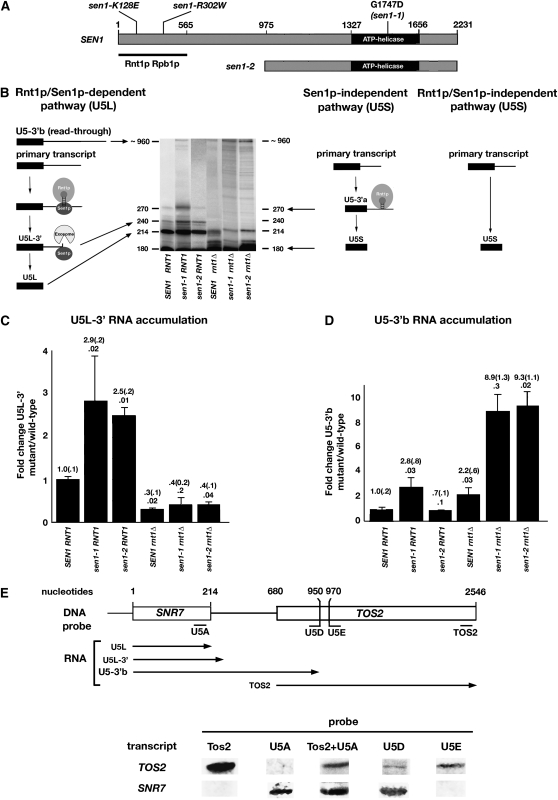
Figure 1.—
Sen1p mutations and interactions. (A) Schematic of Sen1p showing the locations of the interaction domains for Rbp1p and Rnt1p (Ursic et al. 2004; Nedea et al. 2008). Sen1 alleles that affect interactions (sen1-K128E, sen1-R302W, sen1-2) or function (sen1-1) are shown. Numbers refer to amino acids. (B) Alternative pathways for U5 snRNA synthesis. RNAs are named as previously described (Elela et al. 1996; Chanfreau et al. 1997; Lamontagne et al. 2000). The length of the primary transcript is unknown. Shown is a representative Northern blot in which RNAs were fractionated by PAGE and detected using a complementary probe spanning nucleotides 1–320 of the SNR7 gene, which codes for U5 RNA (materials and methods). (C) Effects of sen1-1 and sen1-2 on the relative accumulation of U5L-3′ RNA, an intermediate in 3′-end processing. (D) The effects of sen1-1 and sen1-2 on the relative accumulation of U5-3′b RNA, a readthrough transcript. Fold changes are indicated with standard error in parentheses. P-values are indicated below the fold changes. Methods of quantitation are described in materials and methods. (E) To delimit the location of the 3′-end of U5-3′b RNA, PAGE-fractionated RNAs from a sen1-1 rnt1Δ strain were analyzed by Northern blotting with probes complementary to segments as shown.
To distinguish whether the interactions with Rpb1p and Rnt1p have a dependent or independent relationship, sen1 mutations that impair one interaction without impairing the other interaction were identified. The phenotypes were assessed to determine their effects on SNR7 expression. Two defects were observed, one in transcription termination and one in 3′ processing. The genetic data support a model in which the interactions occur independently of each other.
MATERIALS AND METHODS
Strains, genetic methods, and plasmids:
Strains carrying sen1-1 were derived from FWY1 (MATa ura3-52 leu2-3, −112 pep4-3 trp1 sen1-1) (Ursic et al. 2004); sen1-2 from DDY86 (MATα ade2-101 his3-200 lys2-801 trp1-Δ1 ura3-52 leu2-Δ1∷sen1-2) (DeMarini et al. 1992); sen1-K128E from JFY41 (MATa leu2Δ ura3Δ his3Δ1 trp1Δ sen1-K128E) (this study); sen1-R302W from DUY1513 (MATa leu2Δ ura3Δ his3Δ1 met15Δ sen1-R302W) (this study); rrp6Δ from BY4742 (MATα his3Δ1 leu2Δ lys2Δ ura3Δ rrp6Δ∷KanMX4) (Open Biosystems); rnt1Δ from JFY5 (MATα ade2-101 his3-200 leu2-Δ1∷sen1-2 HIS3:pet56:rnt1) (Ursic et al. 2004); and mtr4-1 from YSL402 (MATa ura3-52 lys2-801 pep4∷HIS3 prb1-Δ1.6R mtr4-1) (Liang et al. 1996). Isogenic sets of strains were created by two-step gene replacement (Boeke et al. 1984). Standard yeast mating and dissection techniques were used to construct double mutants. Growth media were described previously (Ursic et al. 2004). Gene deletions were constructed using the PCR-based gene disruption method (Wach et al. 1994).
All strains were grown at 30°. Strains carrying sen1-K128E and sen1-R302W grow at normal rates. Strains carrying sen1-2 in single copy are viable but grow at a reduced rate. Strains carrying sen1-1 are temperature sensitive for growth. Thirty degrees is permissive for growth, but the changes in levels of accumulation of U5-related RNAs at 30° resemble the changes observed at a nonpermissive temperature of 37°.
The plasmids pU5mt and pU5wt contain DNA starting 500 nucleotides upstream of SNR7 and ending 500 nucleotides downstream of the TOS2 open reading frame (ORF). pU5mt contains GAAA in the stem-loop recognized by Rnt1p in place of AGUC in pU5wt (see Figure 4A). pJF89 contains the same DNA insert as in pU5wt except that the TOS2 ORF is replaced with the Escherichia coli lacZ ORF. Plasmids were introduced into strains by lithium acetate transformation (Gietz and Woods 2002).
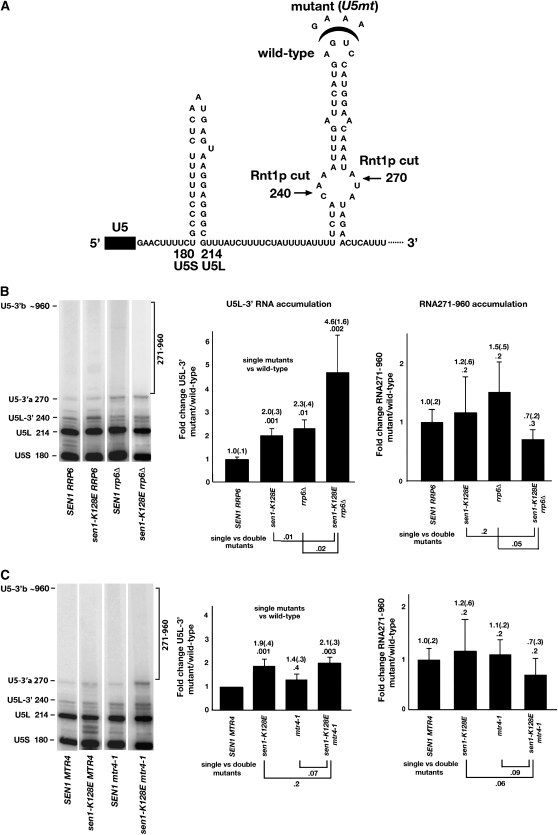
Figure 4.—
Interactions between Sen1p with the exosomal nuclease Rrp6p and the TRAMP helicase Mtr4p. (A) Structure of the 3′ extended region of U5 RNA. Nucleotide substitution of AGUC in wild-type SNR7 with GAAA at the top of the Rnt1p recognition stem-loop impairs the ability of Rnt1p to cleave the RNA (Elela et al. 1996; Chanfreau et al. 1997; Lamontagne et al. 2000). (B) The representative Northern blots and bar graphs show the effects of sen1-K128E and rrp6Δ alone and in combination on the relative accumulation of U5L-3′ RNA and the sum of RNAs in the 271–960 nucleotides size range (RNA271–960). (C) The Northern blots and bar graphs show the effects of sen1-K128E and mtr4-1 alone and in combination on RNA accumulation as described in B. Relative accumulation was quantitated as described in materials and methods. Fold changes are indicated with the standard error in parentheses. P-values are indicated below the fold changes. The brackets and numbers below the bar graphs indicate the P-values for single vs. double mutants.
RNA methods:
Methods for RNA isolation and Northern blotting were described previously (Ursic et al. 2004). RNAs (10 μg) were fractionated on 2% agarose or 6% acrylamide:8 m urea (29:1) gels, transferred to GeneScreen Plus membranes (NEN Life Science Products), and crosslinked using a UV Stratalinker 2400 (Stratagene). Probes were labeled using T4 polynucleotide kinase (Pharmacia) in the presence of [γ-32P]ATP (Amersham). Riboprobes used in Figure 6B were prepared using an in vitro transcription kit (Promega) in the presence of [α-32P]CTP and [α-32P]UTP (3000 Ci/mmol) (Perkin-Elmer). Band intensities on the Northern blots were quantitated using a Typhoon 9200 Variable Mode Imager (Amersham Biosciences). Oligonucleotide probes (Thermo Scientific) used for Northern blotting to map the U5-3′b 3′-end were as follows: U5A (CGCCCTCCTTACTCATTG), U5D (TAATCCATCTTCGGTAAATAG), U5E (GCATTGCTGTCTGAGTTTG), and TOS2 (TTATACATGTACATTCTCG).
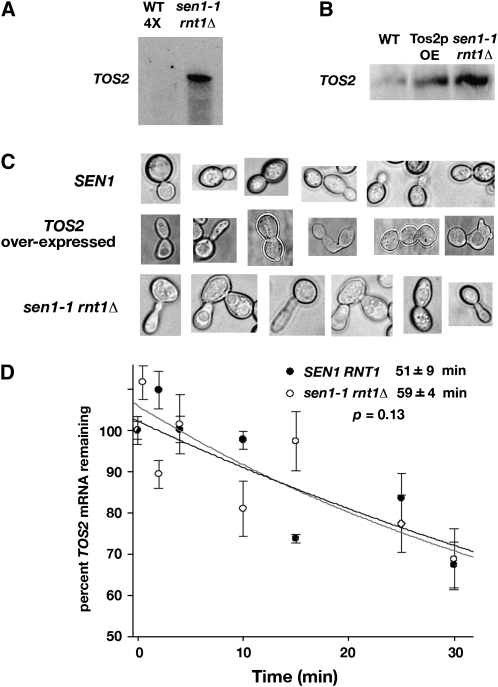
Figure 6.—
Effect of SNR7 transcriptional readthrough on expression of a downstream gene. (A) Increased expression of TOS2 mRNA in a sen1-1 rnt1Δ strain. RNA levels were examined by Northern blotting using an end-labeled probe that anneals to the TOS2 3′-end. (B) TOS2 RNA levels compared with overexpression of CUP1-TOS2 from a multi-copy plasmid (designated Tos2p OE). RNA levels were examined by Northern blotting using a riboprobe (materials and methods). (C) Cellular morphology of a strain that overexpresses CUP1-TOS2 from a multi-copy plasmid and a strain that carries sen1-1 rnt1Δ. (D) Half-life of TOS2 mRNA in SEN1 and sen1-1 rnt1Δ strains.
The rate of TOS2 mRNA decay was determined by measuring the temporal decline in mRNA by Northern blotting following transcription inhibition using 10 μg/ml thiolutin (Pfizer) (Guan et al. 2006). SCR1 RNA was monitored as a loading control. SigmaPlot was used to evaluate decay data using the following exponential decay formula: y = a × exp(−b × x). Estimations of b, designated as b, and corresponding standard errors, were used to calculate standard error [t1/2 = log(2)/b]. t1/2 ± SE(t1/2) was calculated as [log(2)/(b + SE(b)), log(2)/(b-SE(b))].
The relative levels of U5-related RNAs detected by Northern blotting were determined as follows. U5S RNA was used as an internal standard in each gel lane because U5S levels are not affected by mutations in SEN1 or RNT1 (Chanfreau et al. 1997, 1998; Ursic et al. 2004). U5-related RNAs were named as described previously (Chanfreau et al. 1997). The relative levels of U5L-3′ RNA, U5-3′b RNA, and the sum of the RNAs ranging in size from 271 to 960 nucleotides (designated RNA271–960) were measured by calculating the band intensities of the RNAs divided by the band intensity corresponding to U5S RNA in the same lane. Fold changes were calculated by dividing the ratios in mutant strains by the ratio in the corresponding wild-type strain.
Protein and immunological methods:
β-Galactosidase activity was measured as described previously (Stahl et al. 1995). Immunoprecipitation (IP) and Western blotting were described previously (Ursic et al. 2004). Primary antibodies that recognize epitope-tagged proteins were as follows: mouse monoclonal anti-HA antibody (clone HA-7, Sigma) recognizes Rpb1p-HA; mouse monoclonal anti-cMyc antibody (clone 9E10, Sigma) recognizes cMyc-Sen1p, cMyc-sen1-K128Ep, and cMyc-sen1-R302W; and rabbit anti-tandem affinity purification (TAP) antibody (Thermo Scientific) recognizes Rnt1p-TAP, Sen1p-TAP, sen1-K128Ep-TAP, and sen1-R302Wp-TAP. Membranes for Western blotting were probed with anti-mouse or anti-rabbit peroxidase-conjugated antibodies (Thermo Fisher Scientific). Protein bands were visualized by chemiluminescence (SuperSignal West Pico chemiluminescent substrate, Thermo Fisher Scientific) and quantified using a Typhoon 9200 Variable Mode Imager (Amersham Biosciences). For IP experiments, 10 μg/ml RNase A (Sigma) was added to pre-IP lysates. The relative amount of protein that copurified with an immunoprecipitated protein was determined by comparing the ratio of the band intensity of the immunoprecipitated protein to the band intensity of the copurifying protein detected by Western blotting of IP lysates. The effects of sen1 mutations on the extent of copurification were determined by calculating the ratio of the band intensities of the immunoprecipitated and co-immunoprecipitated proteins in the mutant divided by the ratio of the band intensities in wild type.
RESULTS
Sen1p affects two steps in the expression of SNR7:
Two alleles of SEN1 that affect the expression of genes for noncoding RNAs were described previously (Figure 1A) (DeMarini et al. 1992). The sen1-1 mutation (G1747D) is located in a conserved motif in the ATP-helicase region of SEN1. On the basis of its location, sen1-1 most likely impairs helicase activity. The sen1-2 mutation is a partial deletion producing a stable, truncated protein lacking the first 975 amino acids of Sen1p. The deletion removes binding domains required for interaction with the RNase III cleavage enzyme Rnt1p and the largest RNA polymerase II subunit, Rpb1p (Ursic et al. 2004). Depletion of the sen1-2 protein causes a time-dependent accumulation of U5L-3′ RNA, the product of Rnt1p cleavage, at the expense of mature U5L RNA (Ursic et al. 2004). This establishes that elevated accumulation of U5L-3′ RNA is a diagnostic indicator of a Sen1-mediated processing defect.
Using a U5-specific probe, we examined the steady-state levels of U5-related RNAs by Northern blotting of RNA from strains carrying the sen1-1, sen1-2, and/or rnt1Δ mutations (Figure 1B). When the relative levels of U5L-3′ RNA were analyzed (Figure 1C), the sen1-1 mutation caused a 2.9 ± 0.2-fold increase in accumulation, whereas the sen1-2 mutation caused a 2.5 ± 0.2-fold increase. The strains carrying rnt1Δ caused a significant reduction in the level of U5L-3′ RNA since this RNA is the product of Rnt1p cleavage. A small amount of U5L-3′ RNA can still be detected in rnt1Δ strains, presumably due to inefficient degradation of a longer precursor by the exosome, which may stall at the stem-loop recognized by Rnt1p. These results are consistent with data showing that sen1-2 causes a time-dependent increase in the accumulation of U5L-3′ at the expense of U5L RNA but with no effect on U5S accumulation (Ursic et al. 2004).
The longest detectable U5-related RNA, U5-3′b, is ∼960 nucleotides in length (Figure 1B; see below). This RNA accumulated to a 2.8 ± 0.8-fold higher level in a sen1-1 strain (Figure 1D). However, excess accumulation of U5-3′b RNA was not observed in a sen1-2 strain. When double mutants were analyzed, U5-3′b RNA accumulated in excess in both sen1-1 rnt1Δ and sen1-2 rnt1Δ strains (8.9 ± 1.3-fold and 9.3 ± 1.1-fold, respectively). These results suggest that Rnt1p cleavage limits the accumulation of U5-3′b RNA and partially or completely masks the effects of sen1-1 and sen1-2 on accumulation. Overall, the results show that sen1-1 and sen1-2 affect the accumulation of two RNAs, U5L-3′ and U5-3′b. Furthermore, both the N-terminal and helicase regions of Sen1p are required for efficient expression of SNR7.
The distance between the 3′-end of U5L-3′ RNA and the beginning of the downstream TOS2 ORF is 466 nucleotides. The estimated length of U5-3′b RNA suggested that it might extend into the downstream TOS2 gene and could be a readthrough transcript. To test this possibility, the 3′-end of U5-3′b RNA was approximated by Northern blotting of RNA from a sen1-1 rnt1Δ strain using probes complementary to sequences spanning the region from the SNR7 gene to the end of the TOS2 ORF (Figure 1E). Probe U5A, which is complementary to sequences near the 3′-end of mature U5L RNA, hybridized to U5-3′b RNA, whereas the TOS2 probe detected TOS2 mRNA but not U5-3′b RNA. When the two probes were mixed, both RNAs were detected, verifying that RNAs of different sizes were detected with U5-3′b RNA being the smaller of the two. The approximate 3′-end of U5-3′b RNA was located by observing that probe U5D (3′-end at nucleotide 950) hybridized to U5-3′b and TOS2 RNA, whereas probe U5E (5′-end at nucleotide 970) hybridized only to TOS2 mRNA.
The 3′-end of U5-3′b RNA is therefore likely to be located at a position between 940 and 980 nucleotides from the beginning of transcription of SNR7. Since the 3′-end is located between 260 and 300 nucleotides downstream of the beginning of the TOS2 ORF, U5-3′b RNA is most likely a readthrough transcript that accumulates in sen1-1 rnt1Δ strains. Overall, these results suggest that sen1-1 and sen1-2 affect accumulation of two RNAs: an intermediate in RNA processing and a readthrough transcript presumed to result from impaired termination of transcription.
Impact of Sen1p–Rnt1p and Sen1p–Rpb1p interactions on Sen1p function:
Mutations in SEN1 that reduce the efficacy of specific protein–protein interactions might provide insights into the role of the interactions in Sen1p function. To pursue the genetic approach, the boundaries of the binding domains for interaction with Rnt1p and Rpb1p were approximated using polypeptide fragments of Sen1p in two-hybrid studies (Figure 1A). Direct physical interactions of Sen1p with Rnt1p and Rpb1p were previously demonstrated using two-hybrid and co-IP analyses (Ursic et al. 2004). Phylogenetic comparisons of sequences within the domains were used to identify conserved sites potentially important for each interaction. By screening a collection of candidate mutations for their effects in two-hybrid tests, it was found that sen1-K128E and sen1-R302W impaired the Sen1p–Rnt1p and Sen1p–Rpb1p interactions, respectively.
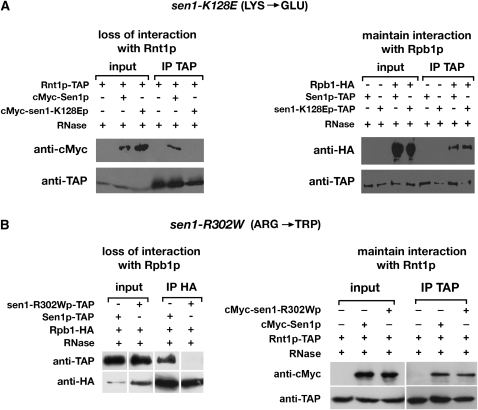
The alleles were examined by co-IP of proteins in native complexes (Figure 2). To examine sen1-K128E, protein extracts from strains expressing epitope-tagged cMyc-Sen1p or cMyc-sen1-K128Ep with Rnt1p-TAP were analyzed (Figure 2A, left). Proteins were bound to anti-TAP antibodies, eluted from beads, and analyzed by Western blotting using anti-TAP and anti-cMyc antibodies. The results indicate that wild-type Sen1p copurified with wild-type Rnt1p. However, the K128E mutation abolished the ability of the proteins to copurify. To test whether the loss of copurification was specific for the interaction with Rnt1p, protein extracts from strains expressing Sen1-TAP or sen1-K128E-TAP and Rpb1p-HA were analyzed (Figure 2A, right). Western blotting of the IP lysates shows that the K128E mutation had no discernible effect on the extent of copurification of Sen1p and Rpb1p.
Figure 2.—
Effects of amino acid substitutions on the interactions of Sen1p with Rnt1p and Rpb1p. (A) sen1-K128E impairs the interaction of Sen1p with Rnt1p (left) but does not affect the interaction with Rpb1p (right). Proteins were immunoprecipitated from cell lysates in the presence of 10 μg/ml RNase A using anti-TAP antibodies. The pre- and post-IP lysates were assayed by Western blotting using anti-cMyc and anti-TAP antibodies (materials and methods). (B) sen1-R302W impairs the interaction of Sen1p with Rpb1p (left) but does not affect the interaction with Rnt1p (right). Experiments were performed as described in A except that the pre- and post-IP lysates were assayed by Western blotting using anti-HA and anti-TAP antibodies.
Similar experiments were performed to analyze the effects of sen1-R302W. Protein extracts from strains expressing Sen1p-TAP or sen1-R302W-TAP with Rpb1p-HA were examined by co-IP of proteins bound to anti-HA antibodies (Figure 2B, left). Western blotting of the IP lysates indicated that wild-type Sen1p copurified with Rpb1p, but sen1-R302Wp failed to copurify. However, when protein extracts from strains expressing cMyc-Sen1p or cMyc-sen1-R302Wp with Rnt1p-TAP were analyzed (Figure 2B, right), the R302W mutation had no discernible effect on copurification.
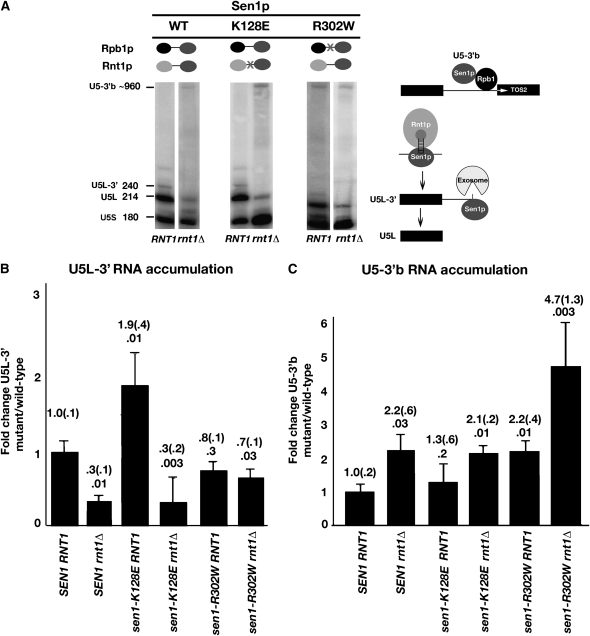
Although the results described above do not necessarily indicate that protein–protein binding is completely abolished, it was reasoned that the interactions might be sufficiently impaired that the mutations would have unique phenotypic effects on Sen1p function. To assess the functional consequences, strains expressing chromosomally integrated sen1-K128E and sen1-R302W alleles were analyzed by Northern blotting with a U5-specific probe (Figure 3A). The K128E mutation caused a 1.9 ± 0.4-fold increase in the relative accumulation of U5L-3′ RNA, the product of Rnt1p cleavage (Figure 3B). The increase was abolished in a sen1-K128E rnt1Δ double mutant. In a sen1-K128E RNT1 strain, there was no significant effect on the relative accumulation of U5-3′b RNA, the readthrough product (Figure 3C). In the sen1-K128E rnt1Δ double mutant, a 2.1 ± 0.2-fold increase was observed. This was similar to that observed for a SEN1 rnt1Δ strain, indicating that the increase is attributable to rnt1Δ and not to sen1-K128E.
Figure 3.—
Effects of sen1-K128E and sen1-R302W on U5 snRNA synthesis. (A) A representative Northern blot in which RNAs from RNT1 and rnt1Δ strains carrying alleles of SEN1 were analyzed as described in the Figure 1 legend. The effects of sen1-K128E and sen1-R302W on the relative accumulation of the processing intermediate U5L-3′ RNA (B) and the readthrough RNA U5-3′b (C) were quantitated as described in materials and methods. Fold changes are indicated with the standard error in parentheses. P-values are indicated below the fold changes.
The R302W mutation had no effect on the relative accumulation of the U5L-3′ RNA-processing intermediate in RNT1 or rnt1Δ strains (Figure 3B). When the accumulation of the U5-3′b readthrough RNA was examined, sen1-R302W caused a 2.2 ± 0.4-fold increase in a RNT1 strain (Figure 3C). A similar increase of 2.2 ± 0.6-fold was observed in a SEN1 rnt1Δ double mutant, and a 4.7 ± 1.3-fold increase was observed in a sen1-R302W rnt1Δ double mutant (Figure 3C). The synthetic phenotype of the double mutant suggests that the R302W amino acid substitution specifically affects accumulation of the readthrough RNA. Overall, the results suggest that sen1-R302W causes a defect in transcription termination without affecting 3′-end processing.
Functional relationship between Sen1p and the exosome:
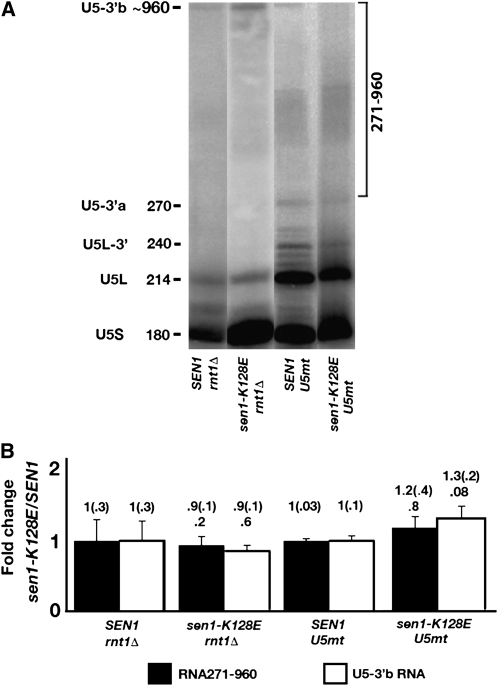
Since the product rather than the substrate of Rnt1p cleavage accumulates in strains carrying sen1-2 and sen1-K128E, the Sen1p–Rnt1p interaction is not functionally related to cleavage itself. We reasoned that the interaction could leave Sen1p, a 5′–3′ helicase, bound to the 3′-end of the RNA cleavage product. From that location, Sen1p could unwind the stem-loop (Figure 4A) and separate the cleaved RNAs. This might allow more efficient access for the exosome, which processes the 3′-end of the Rnt1p cleavage product in conjunction with the Trf4p/Air2p/Mtr4p polyadenylation complex (TRAMP) (Liang et al. 1996; Lacava et al. 2005; Milligan et al. 2005). To test potential relationships of Sen1p with the exosome and TRAMP, we analyzed the phenotypes of sen1-K128E combined with rrp6Δ, a viable deletion of RRP6, which codes for an exosomal subunit required in U5 snRNA processing (Allmang et al. 1999), and mtr4-1, an allele of MTR4, which codes for a 3′–5′ helicase subunit of TRAMP (Liang et al. 1996).
Single-mutant strains carrying sen1-K128E or rrp6Δ were compared with a sen1-K128E rrp6Δ double mutant by assaying effects on the accumulation of two sets of RNAs: the processing intermediate U5L-3′ RNA and the sum of RNAs in the 271–960 nucleotide size range, which includes the U5-3′b readthrough RNA. For the latter, we settled on the broader range of larger RNAs to include intermediates in exosomal degradation of the readthrough transcript.
A synthetic phenotype was observed for U5L-3′ RNA accumulation in the K128E rrp6Δ double mutant (Figure 4B). In the single mutants, the sen1-K128E mutation caused a 2.0 ± 0.3-fold increase in the relative accumulation of U5L-3′ RNA, whereas rrp6Δ caused a 2.3 ± 0.4-fold increase. However, a 4.6 ± 1.6-fold increase was observed in the double mutant, which is significantly higher than in either single mutant. When RNAs in the 271–960 nucleotide size range were examined, the sen1-K128E and rrp6Δ single mutants had no significant effect on relative accumulation. In the double mutant, no synthetic increase was observed. If anything, accumulation was marginally reduced, indicating that the synthetic interaction is restricted to effects on the U5L-3′ processing intermediate.
Similar experiments were performed to compare sen1-K128E and mtr4-1 single mutants with a sen1-K128E mtr4-1 double mutant (Figure 4C). Consistent with previous results, U5L-3′ RNA increased 1.9 ± 0.4-fold in the sen1-K128E single mutant. No significant increase was observed in the mtr4-1 single mutant. In the double mutant, U5L-3′ RNA accumulation increased 2.1 ± 0.3-fold, which was statistically the same as that observed in the sen1-K128E single mutant. No significant effects on the accumulation of RNAs in the 271–960 nucleotide size range were observed in either the single or the double mutants.
Overall, the results reveal a synthetic interaction between sen1-K128E and rrp6Δ that is related to RNA processing and unrelated to transcription termination, suggesting that the Sen1p–Rnt1p protein–protein interaction may serve a role in promoting TRAMP-independent exosomal processing of the Rnt1p cleavage product. If this model is correct, then sen1-K128E should have no effect on the expression of U5 RNA when RNT1 is deleted or when U5 RNA is altered by nucleotide substitutions that prevent Rnt1p recognition of the stem-loop in which Rnt1p cleaves. To test this, an allele of SNR7 was created in which four nucleotides, AGUC, at the top of the Rnt1p recognition loop, were changed to GAAA (U5mt, Figure 4A). It was shown previously that this alteration prevents Rnt1p cleavage (Chanfreau et al. 2000). The wild-type and mutant versions of SNR7 were expressed from plasmids in a strain carrying the wild-type SNR7 gene. Due to overexpression, the mutant snr7 RNAs outnumber the wild-type RNAs in strains producing U5mt from the plasmid.
The effects of sen1-K128E on the accumulation of the readthrough transcript and on RNAs in the 271–960 nucleotide size range were examined in strains carrying rnt1Δ or expressing cleavage-defective U5 RNA (U5mt) (Figure 5). The results indicate that sen1-K128E has no significant effect on either set of RNAs in rnt1Δ- or U5mt-expressing strains. The results suggest that the effects of sen1-K128E on SNR7 expression are limited to exosomal processing of the immediate product of Rnt1p cleavage.
Figure 5.—
Effects of sen1-K128E on SNR7 expression in Rnt1p cleavage-defective strains. (A) RNA from strains carrying SEN1 or sen1-K128E and either rnt1Δ or a plasmid expressing U5mt were analyzed by Northern blotting using the probe described in Figure 1B. A representative Northern blot is shown (see Figure 1 legend and materials and methods). (B) The bar graph compares the effects on the accumulation of U5-3′b RNA and the sum of RNAs in the 271–960 size range (RNA271–960). Relative accumulation was quantitated as described in materials and methods. Fold changes are indicated with the standard error in parentheses. P-values are indicated below the fold changes.
SNR7 transcriptional readthrough increases expression of downstream TOS2:
Mutations in SEN1 typically decrease the expression of downstream genes due to readthrough. For example, sen1-mediated readthrough of the small nucleolar RNA (snoRNA) gene SNR13 reduces expression of the downstream gene, TRS31 (Steinmetz and Brow 1996, 1998; Steinmetz et al. 2001, 2006). Since the SNR7 readthrough transcripts end in the ORF of downstream TOS2 (Figure 1E), we examined the effect of readthrough on TOS2 expression.
In a SEN1 RNT1 strain, TOS2 mRNA was below the level of detection even when four times the standard 10 μg of RNA was loaded on the gel (Figure 6A). However, in a sen1-1 rnt1Δ strain, TOS2 mRNA was readily detected, suggesting that the accumulation of U5-3′b RNA that occurs in sen1-1 rnt1Δ strains might cause increased expression of TOS2. To test this hypothesis, TOS2 expression levels were compared by Northern blotting with a high specific activity riboprobe in a wild-type strain, a strain carrying a multi-copy CUP1-TOS2 plasmid, and a strain carrying sen1-1 rnt1Δ (materials and methods; Figure 6B). TOS2 was detected at a low level in the wild-type strain. TOS2 mRNA expressed from the CUP1-TOS2 plasmid was detected at levels comparable to the level observed in a sen1-1 rnt1Δ strain lacking the plasmid.
Overexpression of TOS2 from a multi-copy plasmid disrupts cytokinesis, leading to aberrant cell morphology, including multiple elongated buds (Gandhi et al. 2006). When the cell morphology of a wild-type strain was compared to that of a strain expressing CUP1-TOS2 from a multi-copy plasmid and a strain carrying sen1-1 rnt1Δ, the elongated, multi-bud morphology was observed for the latter two strains compared to wild type (Figure 6C). Collectively, these results suggest that readthrough causes increased expression of TOS2.
TOS2 mRNA levels might be elevated in sen1-1 rnt1Δ strains as the result of an increased rate of TOS2 transcription or a decreased rate of mRNA decay. To distinguish between these models, the half-life of TOS2 mRNA was determined. Transcription was inhibited with 10 μg/ml thiolutin. Northern blotting with a TOS2-specific end-labeled probe was used to monitor the disappearance of the preexisting mRNA at time intervals following inhibition (Figure 6D). The half-life of TOS2 mRNA in a wild-type strain is 51 ± 9 min, which is statistically indistinguishable from the 59 ± 4 min (P = 0.13) observed in the sen1-1 rnt1Δ strain. This indicates that readthrough has no effect on the stability of TOS2 mRNA.
One way that TOS2 expression might be elevated is through positive auto-regulation mediated by Tos2p itself. To test this model, pJF89, which expresses DNA including the SNR7-TOS2 region but with lacZ replacing the TOS2 ORF (materials and methods), was transformed into strains carrying SEN1 and sen1-1. β-Galactosidase assays revealed that the sen1-1 strain had a 4.3 ± 2.3-fold increase in activity compared to wild type. Northern blotting revealed a 7.4 ± 1.8-fold increase in accumulation of the lacZ transcript in a sen1-1 strain compared to wild type. These results indicate that increased expression is not dependent on the TOS2 ORF and must therefore depend on upstream sequences.
DISCUSSION
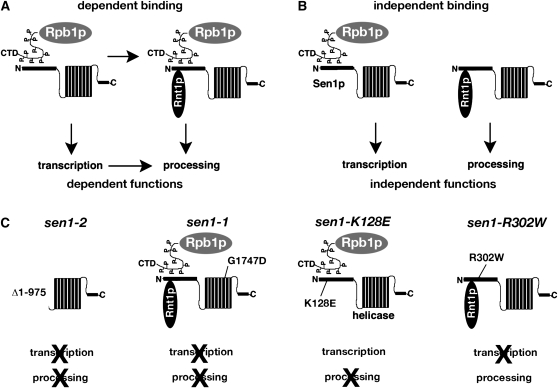
The goal of this study was to assess whether a genetic approach could be used to distinguish between alternative models for the functional relationships between protein–protein interactions. According to one model, interactions could occur in a dependent sequence of protein–protein binding reactions (Figure 7A). One way in which this could be achieved is if two binding domains overlap such that occupation of the domain for one protein precludes occupation by a second protein. Alternatively, protein–protein interactions might occur independently of each other, and there might be no obligate order of binding (Figure 7B). For a protein that interacts with multiple partners, coexistence of dependent and independent sets of binding reactions is possible.
Figure 7.—
Protein–protein interactions and their relationships to function. (A and B) Summaries of models for the relationship between the binding of Rpb1p and Rnt1p to Sen1p and the corresponding functional roles for Sen1p in transcription and processing. Sen1p, which binds to the C-terminal domain of Rpb1p (Ursic et al. 2004), is depicted as a helicase domain composed of stacked α-helices and an N-terminal segment containing multiple protein-binding domains. (C) Relationship between protein–protein interactions and function based on the phenotypes of sen1 mutants.
To determine whether the Sen1p–Rnt1p and Sen1p–Rpb1p interactions depend on each other, we asked how they affect the function of Sen1p in the expression of SNR7 coding for U5 snRNA. Two previously described alleles, sen1-1, which contains a point mutation in a conserved helicase motif, and sen1-2, which is missing DNA coding for the first 975 amino acids (Ursic et al. 2004), each cause two defects in SNR7 expression: one in transcription termination and the other in 3′-end processing. Two new alleles described in this study, sen1-K128E and sen1-R302W, specifically impair the interactions with Rnt1p and Rpb1p, respectively. The mutants served as primary tools for asking how the termination and processing defects might be related. As summarized in Figure 7C, sen1-K128E caused elevated accumulation of the U5L-3′ RNA without detectable accumulation of the readthrough RNA, whereas sen1-R302W caused accumulation of the readthrough RNA without excess accumulation of U5L-3′ RNA. The evidence supports a model for independent binding and independent function in transcription and processing since the phenotypic effects of mutations are genetically separable (Figure 7B).
It is likely that the defect in SNR7 expression caused by sen1-K128E is entirely related to impaired Sen1p–Rnt1p binding because sen1-K128E had no phenotypic effects on RNA accumulation when the RNT1 gene was deleted or when SNR7 RNA was altered to a form that is immune to Rnt1p cleavage. On the basis of this and the synthetic effect of a sen1-K128E rrp6Δ exosome-defective double mutant on U5L-3′ RNA accumulation, we suggest that the primary purpose of the Sen1p–Rnt1p interaction is to assist the exosome in the processive shortening of U5L-3′ RNA to mature U5L by the exosome. The binding of Sen1p to Rnt1p might place Sen1p in physical proximity to the 3′-end of the exosomal substrate. The interaction could play a similar role in the processing of other noncoding RNAs that depend on Sen1p, Rnt1p, and the exosome.
Sen1p is a 5′–3′ helicase (Kim et al. 1999), whereas Mtr4p, a component of TRAMP that assists in exosomal degradation, is a 3′–5′ helicase (Liang et al. 1996; Lacava et al. 2005; Milligan et al. 2005). No synthetic interaction was observed in sen1-K128E mtr4-1 double mutants. We propose that Mtr4p helicase activity is sufficient to aid the exosome in degrading most RNA structures. However, following Rnt1p cleavage, the RNA products may remain based-paired in the stem-loop region of the Rnt1p-binding/cleavage domain. Because of this, the 5′–3′ helicase activity of Sen1p may promote unwinding of the stem to force separation of the cleaved RNAs, allowing for efficient 3′ access to the exosome. Sen1p could assist the exosome in other regions of the RNA by providing a 5′–3′ unwinding activity, but it is not likely that this would depend on the Sen1p–Rnt1p protein–protein interaction.
The Sen1p–Rnt1p interaction may play a role in other RNA biosynthetic pathways in addition to U5 snRNA. We observed that both sen1 and rnt1 mutations cause changes in the accumulation of RNAs detected with probes complementary to the snoRNAs SNR40 and SNR47. Furthermore, sen1 rnt1 double mutants exhibited novel patterns of accumulation for SNR40- and SNR47-related RNAs (J. S. Finkel, unpublished observations), suggesting a potential role for the Sen1p–Rnt1p interaction in these pathways. However, other snoRNAs such as SNR13 and SNR10 were affected only by sen1 mutations, and the effects are most likely restricted to defects in transcription termination (Steinmetz and Brow 1996, 1998; Rasmussen and Culbertson 1998; Steinmetz et al. 2001, 2006; Ursic et al. 2004).
It seemed possible that all functional roles for Sen1p during the transcription cycle might depend on the binding of Sen1p to the RNAP II subunit Rpb1p. Our genetic evidence argues against this model. Strains carrying sen1-R302W produce a protein that fails to bind to Rpb1p, but there is no observable defect in the processing of U5L-3′ RNA. Thus, despite the fact that transcription and processing are temporally coupled, the genetic data indicate that coupling is not enforced by a dependent sequence of protein–protein interactions. This does not necessarily mean that dependent sequences do not exist. Although it has not yet been tested, the interactions of Sen1p with Rpb1p and Rad2p (Ursic et al. 2004) could form a dependent sequence in which transcription-coupled DNA repair might require binding to Rpb1p as a prerequisite to Sen1p–Rad2p binding. The protein–protein interaction network in which Sen1p is embedded could be composed of sets of proteins that form both dependent and independent interactional and functional relationships. The broader possibilities remain to be tested.
The binding of Sen1p to Rpb1p is required for termination of transcription of the SNR7 gene, since sen1-R302W causes accumulation of a readthrough transcript that extends into the downstream TOS2 gene. Similar effects of sen1 mutants on transcriptional termination have been reported for many other noncoding RNA genes (Steinmetz and Brow 1996, 1998; Rasmussen and Culbertson 1998; Steinmetz et al. 2001). What distinguishes the readthrough effect for SNR7 is that the extended transcript causes increased rather than decreased expression of TOS2.
Typically, the expectation is that promoter occlusion would decrease expression of the downstream gene. We ruled out the possibility that increased expression is mediated at the level of mRNA stability or by Tos2p-mediated auto-regulation because there was no effect on the TOS2 mRNA half-life and because increased expression was observed when the TOS2 ORF was replaced with the lacZ ORF. Since increased expression must involve sequences upstream of the TOS2 ORF, an elevated rate of TOS2 transcription is likely. One way this could occur is if the readthrough transcript displaces a transcriptional repressor. Such displacement could override the potential effects of promoter occlusion, leading to a net increase in transcription.
Acknowledgments
This research was supported by the University of Wisconsin College of Agricultural and Life Sciences, the School of Medicine and Public Health, National Institutes of Health grant GM65172 (M.R.C.), National Science Foundation grant MCB 0744017 (M.R.C.), and Kirschstein National Research Service Award Individual predoctoral fellowship F31 GM077078 (J.S.F.). This is Laboratory of Genetics paper no. 3645.
References
- Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski et al., 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., S. A. Elela, M. Ares, Jr. and C. Guthrie, 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 11 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., P. Legrain and A. Jacquier, 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., M. Buckle and A. Jacquier, 2000. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. USA 97 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. Z., C. L. Bennett, H. M. Huynh, I. P. Blair, I. Puls et al., 2004. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74 1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. Z., S. H. Hashemi, S. K. Anderson, Y. Huang, M. C. Moreira et al., 2006. Senataxin, the yeast Sen1p orthologue: characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol. Dis. 23 97–108. [DOI] [PubMed] [Google Scholar]
- Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow et al., 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini, D. J., M. Winey, D. Ursic, F. Webb and M. R. Culbertson, 1992. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 12 2154–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette, A., K. Roddier, J. McNabb-Baltar, I. Gosselin, A. St-Denis et al., 2005. Mutations in senataxin responsible for Quebec cluster of ataxia with neuropathy. Ann. Neurol. 57 408–414. [DOI] [PubMed] [Google Scholar]
- Elela, S. A., H. Igel and M. Ares, Jr., 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85 115–124. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., J. C. Rain and P. Legrain, 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16 277–282. [DOI] [PubMed] [Google Scholar]
- Gandhi, M., B. L. Goode and C. S. Chan, 2006. Four novel suppressors of gic1 gic2 and their roles in cytokinesis and polarized cell growth in Saccharomyces cerevisiae. Genetics 174 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods Enzymol. 350 87–96. [DOI] [PubMed] [Google Scholar]
- Grigoriev, A., 2003. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 31 4157–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Q., W. Zheng, S. Tang, X. Liu, R. A. Zinkel et al., 2006. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2 1924–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken, Y., P. Sung, L. Prakash and S. Prakash, 1993. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature 366 365–368. [DOI] [PubMed] [Google Scholar]
- Hanawalt, P. C., and G. Spivak, 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9 958–970. [DOI] [PubMed] [Google Scholar]
- Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle et al., 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96 375–387. [DOI] [PubMed] [Google Scholar]
- Kim, H. D., J. Choe and Y. S. Seo, 1999. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38 14697–14710. [DOI] [PubMed] [Google Scholar]
- Komarnitsky, P., E. J. Cho and S. Buratowski, 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson et al., 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121 713–724. [DOI] [PubMed] [Google Scholar]
- Lamontagne, B., A. Tremblay and S. Abou Elela, 2000. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 20 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, S., M. Hitomi, Y. H. Hu, Y. Liu and A. M. Tartakoff, 1996. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol. Cell. Biol. 16 5139–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., and R. Reed, 2002. An extensive network of coupling among gene expression machines. Nature 416 499–506. [DOI] [PubMed] [Google Scholar]
- Milligan, L., C. Torchet, C. Allmang, T. Shipman and D. Tollervey, 2005. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol. 25 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, M. C., S. Klur, M. Watanabe, A. H. Nemeth, I. Le Ber et al., 2004. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 36 225–227. [DOI] [PubMed] [Google Scholar]
- Myer, V. E., and R. A. Young, 1998. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 273 27757–27760. [DOI] [PubMed] [Google Scholar]
- Nedea, E., D. Nalbant, D. Xia, N. T. Theoharis, B. Suter et al., 2008. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell 29 577–587. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K. M., 2002. On the importance of being co-transcriptional. J. Cell Sci. 115 3865–3871. [DOI] [PubMed] [Google Scholar]
- Patterson, B., and C. Guthrie, 1987. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell 5 613–624. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2000. Nucleotide excision repair in yeast. Mutat. Res. 451 13–24. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T. P., and M. R. Culbertson, 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 6885–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, J., B. Zheng, B. C. Rymond and J. L. Woolford, Jr., 1995. Structurally related but functionally distinct yeast SmD core small nuclear ribonucleoprotein particle proteins. Mol. Cell. Biol. 15 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, G., L. Bidou, J. P. Rousset and M. Cassan, 1995. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, E. J., and D. A. Brow, 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16 6993–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, E. J., and D. A. Brow, 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95 6699–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, E. J., N. K. Conrad, D. A. Brow and J. L. Corden, 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413 327–331. [DOI] [PubMed] [Google Scholar]
- Steinmetz, E. J., S. B. Ng, J. P. Cloute and D. A. Brow, 2006. Cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol. 26 2688–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera, A., O. J. Becherel, P. Chen, N. Rundle, R. Woods et al., 2007. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J. Cell Biol. 177 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera, A., Y. Lim, R. Woods, G. W. Birrell, T. Nasim et al., 2009. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet. 18 3384–3396. [DOI] [PubMed] [Google Scholar]
- Ursic, D., K. Chinchilla, J. S. Finkel and M. R. Culbertson, 2004. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 32 2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pöhlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Zhang, D., N. Abovich and M. Rosbash, 2001. A biochemical function for the Sm complex. Mol. Cell 7 319–329. [DOI] [PubMed] [Google Scholar]