Abstract
Advances in next-generation sequencing technology have facilitated the discovery of single nucleotide polymorphisms (SNPs). Sequenom-based SNP-typing assays were developed for 1359 maize SNPs identified via comparative next-generation transcriptomic sequencing. Approximately 75% of these SNPs were successfully converted into genetic markers that can be scored reliably and used to generate a SNP-based genetic map by genotyping recombinant inbred lines from the intermated B73 × Mo17 population. The quantitative nature of Sequenom-based SNP assays led to the development of a time- and cost-efficient strategy to genetically map mutants via quantitative bulked segregant analysis. This strategy was used to rapidly map the loci associated with several dozen recessive mutants. Because a mutant can be mapped using as few as eight multiplexed sets of SNP assays on a bulk of as few as 20 mutant F2 individuals, this strategy is expected to be widely adopted for mapping in many species.
WITH the availability of a sequenced genome it is feasible to undertake chromosome walking projects to clone genes responsible for mutant phenotypes (Alleman et al. 2006; Briggs et al. 2007; Menzel et al. 2007; Song et al. 2007) and quantitative trait loci (QTL) (Glazier et al. 2002; Korstanje and Paigen 2002; Salvi et al. 2007). However, it can be logistically difficult and time-consuming to map mutants with current technologies. A high-throughput system to map phenotypic mutants would be very useful in converting the wealth of phenotypic mutants into an understanding of the molecular basis.
Single nucleotide polymorphisms (SNPs) can be converted into genetic markers that are scored in mapping populations using various high-throughput SNP-typing technologies (Gabriel and Ziaugra 2004; Gunderson et al. 2005; Hui et al. 2008). High-throughput SNP discovery (Marth et al. 1999; Weckx et al. 2005; Zhang et al. 2005; Barbazuk et al. 2007; H. Li et al. 2008; R. Li et al. 2009) and genotyping technologies have simplified the generation of genetic maps and the analysis of recombinants (Shifman et al. 2006). Dense maps in economically important crops will be invaluable for marker-assisted selection programs (Prigge et al. 2009), analyzing linkage disequilibrium (Kruglyak 2008; Wang et al. 2008), detection of intraspecies cis-regulatory variation (Stupar and Springer 2006), and other quantitative genetic studies (Cookson et al. 2009).
Maize (Zea mays L.) is an important model organism with substantial economic value. In this species, SNPs occur at a rate of one per 28–214 bp (Tenaillon et al. 2001; Barbazuk et al. 2007). Using our 454-based SNP discovery pipeline, we identified >7000 putative SNPs, >85% (94/110) of which could be validated via Sanger sequencing (Barbazuk et al. 2007). Here, we report the analysis of 1359 of these putative SNPs. Approximately 75% of the tested SNPs could be converted into genetic markers, and only ∼3% were deemed to be false positives. These SNP-based markers were used to construct a genetic map that can be used to address diverse biological questions. Finally, we apply the combination of quantitative SNP typing and bulked segregant analysis (BSA) (Michelmore et al. 1991) to efficiently map phenotypic mutants.
MATERIALS AND METHODS
Genetic materials:
Using a high-throughput protocol (Dietrich et al. 2002), leaf DNA was extracted from the inbred lines B73 and Mo17 [the parents of the intermated B73 × Mo17 (IBM) recombinant inbred lines (RILs)], the 297 IBM RILs (Fu et al. 2006) (Table S1, supporting information), the 25 non-B73 parents of the nested association mapping (NAM) population (Yu et al. 2008), and mutant and non-mutant pools of DNAs for BSA. For BSA, tissues from all mutant (or non-mutant) individuals from within the same F2 family were pooled, and a single DNA isolation was performed or DNA was isolated from each individual and then equal amounts from each individual were bulked. The two methods gave similar results.
SNP typing:
A total of 1393 putative SNPs flanked by ∼60 bp on each side (121 bp total) were submitted to Sequenom's primer design software (MassARRAY Assay Design 3.0). Of these, it was possible to design primers for 1359 (98%) SNPs that were grouped into 48 multiplex assays. The majority of these multiplex assays (41/48) contained 29 SNPs; the remaining seven multiplexes each contained between 18 and 28 SNPs (Table S2). These 1359 SNP assays were used to genotype B73, Mo17, 297 IBM RILs, and the 25 NAM parents. Experiments were conducted following the Sequenom iPLEX Assay application note (Oeth et al. 2005). Genotyping data were acquired using the Sequenom MassARRAY and processed using Sequenom Typer3.4 software.
Map construction:
A genetic map was constructed using the 1049 dominant and codominant markers (Table 1) that yielded genotyping scores for at least 268 of the 297 IBM RILs (Fu et al. 2006). Genotyping scores (Table S3) were analyzed using the MultiPoint mapping software package (population type: “RIL-selfing”; initial threshold recombination rate, 0.15; final threshold recombination rate, 0.43) (Mester et al. 2003, 2004). Genetic distances were calculated using Kosambi function and then corrected using IRILmap software (Falque et al. 2005; Fu et al. 2006).
TABLE 1.
SNP validation and mapping
| No. of SNP markers |
|||
|---|---|---|---|
| Validation class | Used for genotyping | With sufficient dataa | Mapped |
| Codominantb | 973 (72) | 909 (87) | 888 (87) |
| B73 dominantc | 142 (10) | 140 (13) | 128 (13) |
| Paramorphismsd | 34 (2) | — | — |
| No SNP | 42 (3) | — | — |
| Assay failed | 168 (12) | — | — |
| Total |
1359e (100) |
1049f (100) |
1016f (100) |
Numbers in parentheses are percentages.
SNPs that yielded genotyping scores for >90% (i.e., 268 of 297) IBM RILs.
SNP markers that have calls for both B73 and Mo17 alleles.
SNP markers that have calls for only B73 alleles.
SNP markers that have multiple calls for an inbred line (Emrich et al. 2007b)
Ten of these SNPs are non-unique.
Nine of these SNPs are non-unique.
As expected, on the basis of the multiple generations of inbreeding used to develop the IBM RILs, most of the RILs were heterozygous at <20 of 909 codominant markers. But 6 RILs were heterozygous for >67 of the 909 codominant markers (Table S1). A total of 1016 codominant and dominant SNP markers (Table 1) were mapped using the 291 IBM RILs that remained.
Data quality and validation:
The quality of the Sequenom-based genotyping scores was evaluated using three types of controls. First, SNPs located in MAGIs [maize assembled genomic islands (Fu et al. 2005)] (which average a few kilobases in size) that had previously been mapped using temperature gradient capillary electrophoresis genotyping technology (Hsia et al. 2005) were mapped using Sequenom technology (Table S4). Second, pairs of SNPs from the same MAGI were mapped (Table S5). Third, the same SNP was mapped using different Sequenom PCR primers or extension primers (Table S6).
Quantitative SNP typing and BSA:
Selection of a set of 124 SNP markers for BSA:
A set of eight multiplex Sequenom assays, which in combination detected 232 different SNPs and were originally designed for analysis of allelic variation (Stupar et al. 2007), were used to perform BSA. To perform BSA, it is critical to use SNP markers that are robust and highly quantitative. Several quality control measures were employed to identify SNP markers that provide robust, quantitative data. Assays were removed if they failed to provide product (45 markers), did not detect a polymorphism (33 markers), or were not highly correlated with the input ratio when tested against a series of controls (30 markers). Following these quality control steps, a set of 124 robust markers involving eight multiplex reactions remained. The genetic map position for each of these markers was inferred on the basis of BLAST alignments to sequenced maize BACs and tests of the map position for other markers within the same BAC contig. These 124 SNP markers were used to analyze bulk DNA samples created for each of the EMS-induced mutants listed in Table S7. Reaction conditions were as described above and the data were extracted using the Sequenom's allelotyping process method. The resulting data provided an estimate of the relative frequency of the B73 and Mo17 alleles for each SNP in each mutant pool. The relative enrichment for the B73 allele was calculated for each SNP in each sample by calculating the difference between the measured frequency of the B73 allele and the trimmed mean of the frequency of the B73 allele at that SNP.
Data analysis of BSA using 1016 SNP markers:
BSA demands codominant markers that can clearly distinguish mutant and non-mutant alleles. A codominant SNP marker should yield two allele-specific Sequenom peaks, and the sizes of these peaks (peak areas) are expected to represent the relative frequencies of the corresponding alleles in a pool of individuals. Pools of mutant and non-mutant DNA were prepared by bulking samples from mutant plants and non-mutant plants within a segregating family generated from a self-pollination cross of a plant heterozygous for mutant and non-mutant alleles. Because not all SNP markers will generate codominant peak patterns in the mutant populations, only those markers that met the following criteria were selected for mapping analysis: (1) both peak areas in the non-mutant pool were greater than an arbitrary cutoff value (20–30 arbitrary units) and (2) at least one peak area in the mutant pool was greater than this cutoff. In the mutant pool, markers that are not linked to the mutant gene of interest are expected to segregate for both peaks in a 1:1 allele ratio. In contrast, markers linked to the mutated gene are expected to exhibit deviations from a 1:1 allele ratio. The ratio of the peak areas of the two alleles was used to estimate the allele ratio. For a given SNP, the allele ratio for a mutant pool (mutant ratio) was defined as the ratio of the smaller peak area (designated as allele 1 in the assay) to that of the larger peak area (designated as allele 2). The non-mutant allele ratio was then calculated by dividing the peak area of allele 1 by that of allele 2 but using data from the non-mutant pool.
RESULTS
SNP validation and map construction:
A total of 1359 putative maize SNPs (derived from 1290 unique genomic sequence contigs) that we identified previously (Barbazuk et al. 2007) were selected for validation (Table S2). SNPs were detected using Sequenom MassARRAY technology. As shown in Table 1, 72% (973/1359) of the putative SNPs behaved as codominant alleles, and a single, variable allele was detected in B73 and Mo17. Another 10% (142/1359) of the putative SNPs behaved as dominant genetic markers such that the B73 allele was detected, but the Mo17 allele gave no signal. The remaining 244 SNP assays could not be used for mapping (Table 1). For the codominant and dominant SNP assays, we observed a high degree of repeatability, and excellent consistency was observed between Sequenom genotyping and an independent genotyping technology (Table S4, Table S5, Table S6, Table S8).
All of the SNP markers were used to genotype a collection of RILs from the IBM population (materials and methods). A total of 1016 of the dominant and codominant SNPs were successfully mapped, yielding the Iowa State University (ISU) SNP_v1 map (Table 1, Table 2, Figure S1, Table S9).
TABLE 2.
Summary statistics for the 1016 markers on the ISU SNP_v1 Map
| Chromosome | No. of skeleton markersa | No. of muscle markersa | No. of total markers | Length (cM) | Largest gap (cM) | Estimated centromere range (cM) |
|---|---|---|---|---|---|---|
| 1 | 144 | 33 | 177 | 279 | 11.9 | 116.8–122.5 |
| 2 | 80 | 25 | 105 | 213 | 11.2 | 98–98.5 |
| 3 | 105 | 26 | 131 | 230 | 10.9 | 87.2–87.5 |
| 4 | 72 | 20 | 92 | 143 | 11 | 29–29.7 |
| 5 | 71 | 31 | 102 | 142 | 16.4 | 80.7–81.6 |
| 6 | 77 | 25 | 102 | 130 | 10.5 | 12.5–12.9 |
| 7 | 69 | 18 | 87 | 140 | 17.7 | 34–34.5 |
| 8 | 69 | 11 | 80 | 169 | 14.6 | 58.2–60.7 |
| 9 | 50 | 11 | 61 | 145 | 14.3 | 43.2–44.7 |
| 10 | 61 | 18 | 79 | 147 | 19.7 | 60.6–63.6 |
| Total |
798 |
218 |
1016 |
1737 |
— |
— |
Skeleton markers are assigned genetic positions with high certainty; muscle markers are assigned genetic positions relative to skeleton markers, but their orientations relative to those skeleton markers are not specified (Fu et al. 2006).
Accuracy of allele frequency detection via Sequenom assays:
Although the Sequenom MassARRAY platform has the potential to provide quantitative data on the relative frequency of the two alleles (Bansal et al. 2002; Ding and Cantor 2003), many SNPs actually exhibit nonlinear relationships between the input ratio and the detected allele frequency (Stupar et al. 2007). We observed similar deviations from linearity for some SNPs of the codominant category in Table 1 when we analyzed B73 × Mo17 F1 hybrid DNA. To determine the accuracy with which the Sequenom MassARRAY platform calls allelic frequencies, we determined the allele frequency for 50 codominant SNP markers in the B73 × Mo17 F1 hybrid (which is known to contain equal amounts of the two alleles). In addition, B73 and Mo17 genomic DNAs were mixed at 21 different ratios (ranging from 1:100 to 100:1), and the ratios of the B73/Mo17 allele peak areas in these mixed samples were determined. In these titrations, genomic DNAs of B73 and Mo17 served as controls. Four independent replications were performed, and for most (48/50) of the SNP markers the SNP typing was quantitatively repeatable across replicates within the F1 (Figure S2).
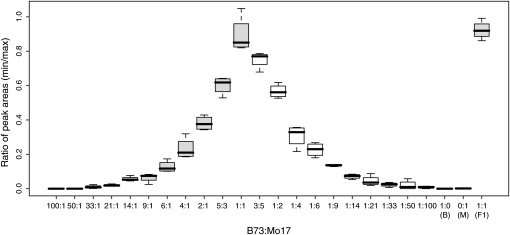
It was expected that B73- and Mo17-derived peak areas from F1 DNA (Figure S2) would be at a 1:1 ratio. On the basis of the results of a two-sample t-test, this was the case for ∼58% (29/50; P-values >0.05; Table S11) of the tested codominant SNP markers. These 29 markers also exhibited high correlations between the ratios of the B73 and Mo17 peak areas and the input allele ratio across a wide range of ratios (see an example in Figure 1), indicating that SNP typing via Sequenom MassARRAY is reasonably quantitative for these markers.
Figure 1.—
Quantitative behavior of a codominant SNP marker. SNP typing of MAGI_19354_W5 was conducted on different concentrations of B73 and Mo17 genomic DNAs (x-axis). DNAs from B73, Mo17, and their F1 hybrid were used as controls. Four independent replications were performed. For B73:Mo17 template ratios from 100:1 to 1:1 (left half of the chart), B73 (B) and F1, the ratios of peak areas of the Mo17/B73 SNP alleles are plotted as solid boxes. For B73:Mo17 template ratios from 3:5 to 1:100 (right half of the chart) and Mo17 (M), the ratios of peak areas of the B73/Mo17 SNP alleles are plotted as open boxes. Horizontal bars in the boxes designate the median of the four replications; the top edge of the box indicates the 75th percentile of the data set and the bottom edge indicates the 25th percentile; the ends of the whiskers indicate the minimum and maximum data values.
In contrast, the other 21 codominant SNP markers exhibited significant deviations from a 1:1 ratio of the peak areas for the B73 and Mo17 alleles in the F1 (P-values <0.05; Table S11). The vast majority of these SNP markers (19/21) have higher-than-expected peak areas for B73 alleles. Because all primers and extension primers were designed on the basis of B73 sequences, we hypothesized that DNA sequence polymorphisms in Mo17 haplotypes could account for this difference by affecting the binding of primers and/or extension primers. It was possible to identify Mo17 genomic sequence reads generated by the Department of Energy (DOE)'s Joint Genome Institute for 24 of the 37 SNPs (of 50) that were surveyed (Table S12). Polymorphisms could be detected at the primer binding sites for 90% (9/10) of the SNP markers that yielded significantly larger peak areas for B73 than Mo17 alleles in the F1. In contrast, 0% of the 13 SNP markers that yielded approximately equal peak areas for B73 and Mo17 alleles in the F1 exhibited polymorphisms at the primer binding sites (Table S12). Hence, we conclude that polymorphisms within PCR primer and/or extension primer binding sites are often responsible for the lack of codominance observed for some polymorphic SNP markers. One SNP marker yielded significantly larger peak areas for the Mo17 than the B73 allele in the F1, but it does not exhibit polymorphisms within primer binding sites. Since copy-number variations (CNVs) are common in maize (Springer et al. 2009), we hypothesize that this SNP marker might exhibit a CNV in the Mo17 haplotype.
SNP-based BSA of maize mutants in a mixed B73- and Mo17-derived genetic background:
The quantitative nature of the Sequenom platform provides the potential to map mutants via BSA (Michelmore et al. 1991; Korol et al. 2007; Lambreghts et al. 2009). A series of 40 recessive mutants (Table S7 and Figure S3) generated via EMS mutagenesis (Till et al. 2004) of B73 was used to demonstrate the utility of combining Sequenom-based quantitative SNP detection with BSA. F2 mapping populations were generated by crossing each mutant (in a B73 genetic background) to Mo17 and then self-pollinating the resulting F1's. Leaf tissue was collected from mutant plants within each of the resulting F2 families. A single BSA sample that contained DNA from 12 to 94 different mutant individuals was produced for each of the mutants (Table S7). Quantitative allelotype data were produced for the 40 mutants using 124 selected SNPs (materials and methods).
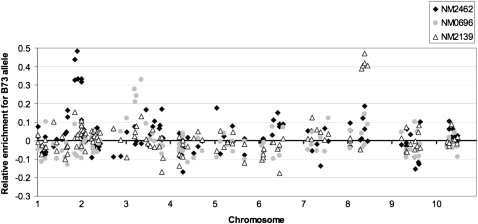
Because the mutations were induced in inbred B73 plants, the mutant allele occurred in coupling with B73 alleles of genetic markers. Consequently, genetic markers that were linked to the mutation were enriched for the B73 allele in BSA samples. The quantitative SNP data were analyzed to identify the genomic locations of the genes that produced the mutant phenotypes (see Figure 2 for three examples of genomic scans). It was possible to determine the genomic locations for 37 of the 40 analyzed mutants by assessing the relative enrichment of the B73 allele for each marker in the mutant pool relative to the chromosomal position of that marker (in centimorgans). Map positions were inferred by visual inspection of the data to identify regions containing multiple SNPs that exhibited enrichment for the B73 allele (see an example in Figure 2). The failure to identify a map position for 3 of the 40 mutants may be due to the fact that these mutants are localized in regions of the genome with relatively few markers in the N = 124 SNP set or due to mutant-specific reasons.
Figure 2.—
Bulked segregant mapping using quantitative SNP typing in B73- and Mo17-derived genetic background. Mapping data for 124 quantitative codominant SNP markers are presented for three different phenotypic mutations. The genetic location of each SNP marker is displayed on the x-axis in which the 10 maize chromosomes have been concatenated. The y-axis displays the relative enrichments for the B73 allele for each of the SNP markers determined by subtracting the trimmed mean value for this SNP in all samples from the value determined for the bulk DNA sample of the event being mapped. Enrichment for the B73 allele (to a maximum expected value of 0.5) indicates linkage of the SNP marker to the mutation causing the mutant phenotype. The numbers of mutant seedlings pooled to establish the BSA bulk were 94, 71, and 74 for events NM2462, NM0696, and NM2139, respectively.
The map positions predicted by BSA for 22 of the mutants were validated using an independent mapping strategy. We selected insertion-deletion polymorphisms markers (Fu et al. 2006) located near the predicted locations of the mutants and tested for linkage by genotyping individual mutant DNA samples (Table S7). The predicted map locations were validated for 20 of the 22 tested mutants.
We tested the effects of pooling size on the ability to identify the genomic location for a mutation. A pool containing ∼20 mutant individuals was sufficient to identify a region of enrichment for the B73 allele with relatively little noise. However, pools of 5–10 mutant individuals exhibit relatively high levels of variation at many SNPs due to sampling variation (data not shown).
BSA of mutants in genetic backgrounds that are not derived from B73 and/or Mo17:
BSA requires access to multiple quantitatively codominant markers distributed across the genome. The experiments reported above demonstrate that mutants can be mapped by quantitative SNP-typing pooled DNA samples from mutants using as few as 124 markers. To determine the potential of the quantitative codominant SNP markers from the ISU SNP_v1 map to conduct BSA in other genetic backgrounds, we genotyped these markers in the inbred parents of the NAM population, which sample the genetic diversity of maize (Yu et al. 2008). The number of markers that are polymorphic between each pair of the 27 inbreds was computed (Table S10). Approximately 50% of the codominant markers are polymorphic between B73 (or Mo17) and any of the 25 parents of the NAM population. In addition, the number of codominant SNPs that are polymorphic between any pair of inbreds included in this analysis is greater than the number of SNPs used for BSA in the experiments described above (N = 124). It must be remembered, however, that ∼50% of the markers that exhibit codominance between B73 and Mo17 are not quantitatively codominant. This fraction is expected to vary on the basis of the frequency of SNPs between a pair of haplotypes. Even so, we predict that the set of 1016 SNPs contains sufficient markers to conduct BSA in a wide variety of genetic backgrounds.
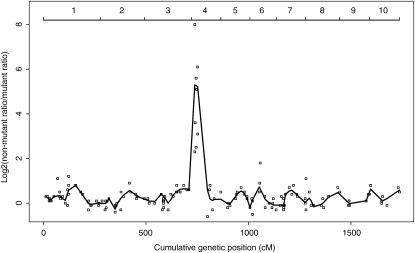
To test this prediction, the 1016 SNPs were used in combination with BSA to map nine additional recessive mutants, each of which affects the biosynthesis or accumulation of cuticular waxes (Schnable et al. 1994). Because the genetic backgrounds of these F2 families are more complex than those of the B73 × Mo17 F2 families used in the previously described BSA mapping experiments, not all markers from the ISU SNP_v1 map are polymorphic in a given F2 family. In addition, due to the presence of uncharacterized polymorphisms at primer binding sites in non-B73 and non-Mo17 alleles, we would expect that some of the markers that exhibited codominance between the B73 and Mo17 alleles might fail to exhibit codominance in F2 families that included novel alleles. DNAs of both mutant and non-mutant tissue pools from individual F2 families were extracted and analyzed. Non-mutant DNA pools were used to identify SNP markers that exhibited codominant behavior in a given F2 family. Markers to be used for BSA must exhibit quantitative codominance. A filtering procedure was developed to identify those codominant markers that provide reasonably quantitative allele frequencies and that could therefore be used for BSA (see materials and methods). Locally weighted polynomial regression (LOWESS) (Cleveland 1979) was used to visualize the map positions of mutants (Figure 3). Markers that exhibited a pronounced peak were deemed to be close to the affected gene. Eight of the nine mutants were successfully mapped in this manner. Among these eight mutants, the map positions of seven were consistent with prior mapping results obtained using other technologies (Table 3).
Figure 3.—
BSA mapping of gl7. Allele ratios of each of 150 quantitative codominant markers were calculated by using peak areas obtained for each marker used to SNP-type the gl7 mutant pool and the non-mutant pool (materials and methods). The genetic location of each SNP marker was plotted on the x-axis in which the 10 maize chromosomes have been concatenated. The position of each chromosome is shown at the top. The y-axis represents the log 2 ratio of the non-mutant ratio to the mutant ratio for each of these markers. Mutant ratios equal to 0 were arbitrarily assigned a log 2 ratio value of 8. Locally weighted polynomial regression (LOWESS) with a smoother span (f) equal to 0.05 was performed (solid line).
TABLE 3.
Comparison between BSA mapping results and previous mapping results
| Gene | Allelea | No. of informative markers | No. of mutants in pool | No. of nonmutants in pool | Chromosome location via BSA (cM)b | Independent map locationc | Consistency between both mappings |
|---|---|---|---|---|---|---|---|
| gl3 | gl3-ref | 137 | 21 | 47 | Chr4L: 66–95 | 4L | Yes |
| gl3 | gl3-93-4700-6 | 90 | 16 | 60 | Chr4L: 91 | 4L | Yes |
| gl3 | gl3-94-4700-7 | 166 | 28 | 65 | Chr4L: 78–90 | 4L | Yes |
| gl6 | gl6-ref | 137 | 25 | 45 | Chr3: 85–141 | 3L | Yes |
| gl7 | gl7-ref | 139 | 28 | 111 | Chr4: 16–30 | 4S | Yes |
| gl27 | gl27-ref | 264 | 21 | 66 | Chr1: 118–159 | 1 | Yes |
| gl28 | gl28-ref | 216 | 17 | 72 | NDd | 10 | NA |
| gl32e | gl32-ref | 189 | 23 | 71 | Chr5L: 86–99 | 2L | Nof |
|
gl33 |
gl33-ref |
93 |
17 |
45 |
Chr8: 78–90 |
8 |
Yes |
All mutants are controlled by single recessive alleles. gl3, gl6, gl7, gl27, gl28, gl32, and gl33 all show a glossy phenotype on the juvenile leaves. gl3-ref and gl6-ref alleles were described previously (Schnable et al. 1994). gl3-mu alleles were isolated from Mutator transposon direct tagging. gl7 was located on chromosome 4S (Stinard 1997). gl27, gl28, gl32, and gl33 mutants are either Mutator-induced alleles generated from random-tagging experiments or alleles identified in M2 or M3 families derived from the treatment of pollen with EMS.
The genetic locations of mutant-associated markers (the allele ratio of the mutant pool <0.3 and the allele ratio of the non-mutant pool >1.2). When unambiguous, centromere positions shown in Table 2 were used to assign chromosome arm positions.
These mutants were independently mapped using the B-A or the wx translocation series (Beckett 1978; Burnham 1982).
Not successfully mapped via BSA.
May be allelic to gl8a on chromosome 5L.
BSA mapping result provides support for hypothesis that gl32 is allelic to gl8a on chromosome 5L.
DISCUSSION
Conversion of putative SNPs to genetic markers:
Over 80% (1115/1359) of putative SNPs identified via comparative next-generation transcriptomic sequencing were successfully converted into informative genetic markers. Few (3%) of the putative SNPs were definitively false-positive SNP calls. Instead, most of the remaining conversion failures (13% of total) were due to Sequenom assay failures. Another 13% (142/1115) of the markers were dominant in that only the B73 allele could be called using Sequenom technology. We assume many of the markers that exhibit dominance do so as a result of polymorphisms that block amplification or extension of the Mo17 allele. The remaining 87% (973/1115) of markers were codominant, in that both B73 and Mo17 alleles could be “called” by Sequenom technology. However, only approximately one-half of these codominant markers were quantitatively codominant and therefore suitable for BSA. Our analyses indicate that this allele specificity is often caused by the presence of polymorphisms that flank a mapped SNP and that therefore interfere with the binding of PCR primers or extension primers in an allele-specific manner.
Recommendations for mapping mutants via quantitative SNP typing:
The use of quantitative, multiplex SNP markers can facilitate the rapid analysis of large numbers of phenotypic mutants. We found that it is critical to use DNA controls to identify quantitative SNP assays and to remove from the analysis those assays that do not provide quantitative allelic ratios. The proportion of codominant assays that were quantitative varied by genetic background, but in F2 families derived from B73 and Mo17 approximately one-half of codominant markers were suitable for BSA. We recommend conducting the analysis BSA on mutant pools that contain at least 20 individuals. In addition, when analyzing non-B73/non-Mo17 mapping populations, it is advisable to include F1 or non-mutant (control) pooled DNA samples to identify (and remove from) polymorphic markers that do not exhibit codominance quantitatively.
In the experiments reported here, SNP markers were assigned to multiplexes without regard to their genetic map positions. But for future SNP designs, we recommend assigning SNP markers from a common chromosome or chromosome arm to a single multiplex. This will allow the efficient use of quantitative SNP typing and BSA for mutants that have already been assigned to a chromosome or chromosome arm via other mapping procedures.
We demonstrate that as few as eight multiplex reactions containing 124 SNPs could be used to identify the map positions of >90% (37/40) of the mutants tested. The mapping can be done using ∼20 mutant F2 individuals. It should be noted that quantitative SNP typing and BSA can also define potential complementation groups. For example, 9 of the 40 mutants tested exhibit reddish coloration of the seedling leaves (Table S7). Three of these mutants map near the same location on chromosome 3, and another 3 mutants map together on chromosome 8. These may reflect two complementation groups, and indeed for two examples, genetic tests have confirmed that these independent mutations affect the same gene (data not shown). Using this type of rapid, low-cost system, it is possible to perform mapping on large classes of mutants and rapidly assign chromosomal positions and potential complementation groups. Although we have so far mapped only qualitative mutants using this procedure, we predict that it will also be useful for mapping QTL (Korol et al. 2007).
Broader applications:
As a consequence of technological improvements in SNP discovery and detection, it is now possible to develop genetic maps even in species for which substantial investments in genomic resources have not been made. Next-generation sequencing technology is used to conduct deep EST sequencing (Emrich et al. 2007a) of the parents of a mapping population. The resulting ESTs are aligned the gene-enriched sequences to identify SNPs (Barbazuk et al. 2007). This approach can be successful even in cases for which reference genomic sequences are not available (Novaes et al. 2008; Buggs et al. 2009). Once identified, SNPs are converted into genetic markers and are used to genotype the mapping population and build a genetic map. These markers can be used to map mutants and QTL in preparation for investigations of biological function and/or breeding. Given the value of SNP-based genetic maps to geneticists and breeders and the ease with which they can now be generated, we advocate the early development of SNP-based genetic maps for the world's important fruit, vegetable, and “orphan” crops.
Acknowledgments
We thank Sarah Hargreaves, Hailing Jin, Mitzi Wilkening, Jia-Ling Pik, Peter Hermanson, and Summer St. Pierre for technical assistance; Kai Ying and Cheng-Ting “Eddy” Yeh for assistance with data analyses; Dan Nettleton and Tieming Ji for helpful discussions regarding the analysis of BSA data; An-Ping Hsia for the useful comments; and Dan Rokhsar of the DOE's Joint Genome Institute for sharing unpublished whole-genome shotgun sequences of the Mo17 genome. This project was supported in part by a competitive grant from the National Science Foundation Plant Genome Program (DBI-0321711) to P.S.S.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107557/DC1.
References
- Alleman, M., L. Sidorenko, K. McGinnis, V. Seshadri, J. E. Dorweiler et al., 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295–298. [DOI] [PubMed] [Google Scholar]
- Bansal, A., D. van den Boom, S. Kammerer, C. Honisch, G. Adam et al., 2002. Association testing by DNA pooling: an effective initial screen. Proc. Natl. Acad. Sci. USA 99 16871–16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk, W. B., S. J. Emrich, H. D. Chen, L. Li and P. S. Schnable, 2007. SNP discovery via 454 transcriptome sequencing. Plant J. 51 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, J. B., 1978. B-A translocations in maize. J. Hered. 69 27. [Google Scholar]
- Briggs, W. H., M. D. McMullen, B. S. Gaut and J. Doebley, 2007. Linkage mapping of domestication loci in a large maize teosinte backcross resource. Genetics 177 1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs, R. J., A. N. Doust, J. A. Tate, J. Koh, K. Soltis et al., 2009. Gene loss and silencing in Tragopogon miscellus (Asteraceae): comparison of natural and synthetic allotetraploids. Heredity 103 73–81. [DOI] [PubMed] [Google Scholar]
- Burnham, C. R., 1982. The location of genes to chromosome by the use of chromosomal interchanges, pp. 65–70 in Maize for Biological Research: A Special Publication of the Plant Molecular Biology Association, edited by W. F. Sheridan. Charlottesville, VA.
- Cleveland, W. S., 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74 829–836. [Google Scholar]
- Cookson, W., L. Liang, G. Abecasis, M. Moffatt and M. Lathrop, 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. R., F. Cui, M. L. Packila, J. Li, D. A. Ashlock et al., 2002. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, C., and C. R. Cantor, 2003. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc. Natl. Acad. Sci. USA 100 3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich, S. J., W. B. Barbazuk, L. Li and P. S. Schnable, 2007. a Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 17 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich, S. J., L. Li, T. J. Wen, M. D. Yandeau-Nelson, Y. Fu et al., 2007. b Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falque, M., L. Decousset, D. Dervins, A. M. Jacob, J. Joets et al., 2005. Linkage mapping of 1454 new maize candidate gene loci. Genetics 170 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., S. J. Emrich, L. Guo, T. J. Wen, D. A. Ashlock et al., 2005. Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc. Natl. Acad. Sci. USA 102 12282–12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., T. J. Wen, Y. I. Ronin, H. D. Chen, L. Guo et al., 2006. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, S., and L. Ziaugra, 2004. SNP genotyping using Sequenom MassARRAY 7K platform. Curr. Protoc. Hum. Genet Chapter 2 Unit 2.12. [DOI] [PubMed]
- Glazier, A. M., J. H. Nadeau and T. J. Aitman, 2002. Finding genes that underlie complex traits. Science 298 2345–2349. [DOI] [PubMed] [Google Scholar]
- Gunderson, K. L., F. J. Steemers, G. Lee, L. G. Mendoza and M. S. Chee, 2005. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 37 549–554. [DOI] [PubMed] [Google Scholar]
- Hsia, A. P., T. J. Wen, H. D. Chen, Z. Liu, M. D. Yandeau-Nelson et al., 2005. Temperature gradient capillary electrophoresis (TGCE): –a tool for the high-throughput discovery and mapping of SNPs and IDPs. Theor. Appl. Genet. 111 218–225. [DOI] [PubMed] [Google Scholar]
- Hui, L., T. DelMonte and K. Ranade, 2008. Genotyping using the TaqMan assay. Curr. Protoc. Hum. Genet. Chapter 2: Unit 2.10. [DOI] [PubMed]
- Korol, A., Z. Frenkel, L. Cohen, E. Lipkin and M. Soller, 2007. Fractioned DNA pooling: a new cost-effective strategy for fine mapping of quantitative trait loci. Genetics 176 2611–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korstanje, R., and B. Paigen, 2002. From QTL to gene: the harvest begins. Nat. Genet. 31 235–236. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L., 2008. The road to genome-wide association studies. Nat. Rev. Genet. 9 314–318. [DOI] [PubMed] [Google Scholar]
- Lambreghts, R., M. Shi, W. J. Belden, D. Decaprio, D. Park et al., 2009. A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., J. Ruan and R. Durbin, 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Y. Li, X. Fang, H. Yang, J. Wang et al., 2009. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth, G. T., I. Korf, M. D. Yandell, R. T. Yeh, Z. Gu et al., 1999. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 23 452–456. [DOI] [PubMed] [Google Scholar]
- Menzel, S., C. Garner, I. Gut, F. Matsuda, M. Yamaguchi et al., 2007. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 39 1197–1199. [DOI] [PubMed] [Google Scholar]
- Mester, D., Y. Ronin, D. Minkov, E. Nevo and A. Korol, 2003. Constructing large-scale genetic maps using an evolutionary strategy algorithm. Genetics 165 2269–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester, D. I., Y. I. Ronin, E. Nevo and A. B. Korol, 2004. Fast and high precision algorithms for optimization in large-scale genomic problems. Comput. Biol. Chem. 28 281–290. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novaes, E., D. R. Drost, W. G. Farmerie, G. J. Pappas, Jr., D. Grattapaglia et al., 2008. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics 9 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeth, P., M. Beaulieu, C. Park, D. Kosman, G. Mistro et al., 2005. iPLEX™ assay: increased plexing efficiency and flexibility for MassARRAY® system through single base primer extension with mass-modified terminators. http://www.agrf.org.au/assets/files/PDF%20Documents/Sequenom%20iPlex.pdf.
- Prigge, V., A. E. Melchinger, B. S. Dhillon and M. Frisch, 2009. Efficiency gain of marker-assisted backcrossing by sequentially increasing marker densities over generations. Theor. Appl. Genet. 119 23–32. [DOI] [PubMed] [Google Scholar]
- Salvi, S., G. Sponza, M. Morgante, D. Tomes, X. Niu et al., 2007. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P. S., P. S. Stinard, T. J. Wen, S. Heinen, D. Weber et al., 1994. The genetics of cuticular wax biosynthesis. Maydica 39 279–287. [Google Scholar]
- Shifman, S., J. T. Bell, R. R. Copley, M. S. Taylor, R. W. Williams et al., 2006. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 4 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X. J., W. Huang, M. Shi, M. Z. Zhu and H. X. Lin, 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 623–630. [DOI] [PubMed] [Google Scholar]
- Springer, N. M., K. Ying, Y. Fu, T. Ji, C. Yeh et al., 2009. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 5(11) e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinard, P. S., 1997. gl7 and v17 map to the short arm of chromosome. Maize Genet. Coop. Newsl. 71 83. [Google Scholar]
- Stupar, R. M., and N. M. Springer, 2006. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., P. J. Hermanson and N. M. Springer, 2007. Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. 145 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B. J., S. H. Reynolds, C. Weil, N. Springer, C. Burtner et al., 2004. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Y. Yu, J. Zhao, Y. Shi, Y. Song et al., 2008. Population structure and linkage disequilibrium of a mini core set of maize inbred lines in China. Theor. Appl. Genet. 117 1141–1153. [DOI] [PubMed] [Google Scholar]
- Weckx, S., J. Del-Favero, R. Rademakers, L. Claes, M. Cruts et al., 2005. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 15 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., J. B. Holland, M. D. McMullen and E. S. Buckler, 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., D. A. Wheeler, I. Yakub, S. Wei, R. Sood et al., 2005. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput. Biol. 1 e53. [DOI] [PMC free article] [PubMed] [Google Scholar]