Abstract
The DNA-packaging specificities of phages λ and 21 depend on the specific DNA interactions of the small terminase subunits, which have support helix-turn-recognition helix-wing DNA-binding motifs. λ-Terminase with the recognition helix of 21 preferentially packages 21 DNA. This chimeric terminase's ability to package λDNA is reduced ∼20-fold. Phage λ with the chimeric terminase is unable to form plaques, but pseudorevertants are readily obtained. Some pseudorevertants have trans-acting suppressors that change codons of the recognition helix. Some of these codons appear to remove an unfavorable base-pair contact; others appear to create a novel nonspecific DNA contact. Helper-packaging experiments show that these mutant terminases have lost the ability to discriminate between λ and 21 during DNA packaging. Two cis-acting suppressors affect cosB, the small subunit's DNA-binding site. Each changes a cosBλ-specific base pair to a cosB21-specific base pair. These cosB suppressors cause enhanced DNA packaging by 21-specific terminase and reduce packaging by λ-terminase. Both the cognate support helix and turn are required for strong packaging discrimination. The wing does not contribute to cosB specificity. Evolution of packaging specificity is discussed, including a model in which λ- and 21-packaging specificities diverged from a common ancestor phage with broad packaging specificity.
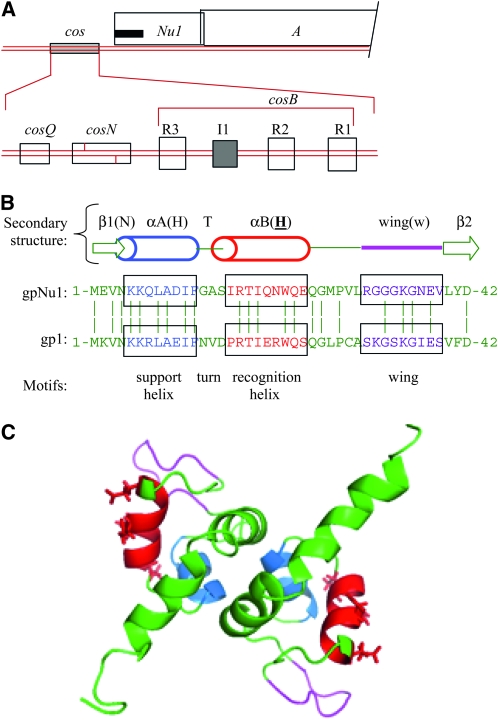
VIRUSES must package viral chromosomes from nucleic acid pools that include host-cell nucleic acids, so specific recognition of the viral nucleic acid is essential during virion assembly. For large DNA viruses, including the tailed double-strand DNA (dsDNA) bacteriophages, the herpesviruses, and the adenoviruses, DNA-packaging proteins recognize specific sequences on the viral chromosomes (reviewed in Baines and Weller 2005 and Ostapchuk and Hearing 2005, respectively). For the dsDNA viruses that produce virion chromosomes by processing concatemeric DNA, a viral terminase enzyme functions in the recognition and cutting of concatemeric DNA and subsequently sponsors DNA translocation. λ-Terminase is a heterooligomer of large and small subunits, gpA and gpNu1, respectively. Cutting of concatemeric DNA is carried out by gpA's endonuclease activity (Becker and Gold 1978; Davidson and Gold 1992; Hwang and Feiss 1996). Three DNA subsites, cosQ, cosN, and cosB, are contained in the ∼200-bp-long cos site and orchestrate DNA packaging through interactions with terminase (Figure 1A; reviewed in Feiss and Catalano 2005). gpA introduces staggered nicks in cosN to generate the 12-bp cohesive ends of mature λDNA molecules. Efficient and accurate nicking of cosN requires anchoring of gpA by gpNu1, which binds to the adjacent cosB subsite (Higgins and Becker 1994b; Hang et al. 2001).
Figure 1.—
The cos and terminase region of the λ-chromosome. (A) (Top) Map of cos and the terminase-encoding Nu1 and A genes. The black bar indicates the location of the winged helix-turn-helix DNA-binding motifs in the N-terminal domain of gpNu1. (Bottom) cos subsites: cosQ is required for termination of DNA packaging; cosN is the site where the large terminase subunit, gpA, introduces staggered nicks to generate the cohesive ends of virion DNA molecules; and cosB contains the gpNu1-binding sites R1, R2, and R3 along with the IHF-binding site I1. (B) (Top) Schematic of gpNu1 residues 1–42, including the support (blue) and recognition (red) α-helixes and the wing loop (magenta). β1 and β2 are short β-strands flanking the DNA-binding elements. (Bottom) Sequences are a comparison of residues of λ's gpNu1 and phage 21's gp1, with conserved resides indicated by vertical lines. Note that the recognition helixes of gpNu1 and gp1 differ by four residues, all likely solvent-exposed (Becker and Murialdo 1990; de Beer et al. 2002). (C) Three-dimensional structure of the winged helix-turn-helix-containing, N-terminal domain of gpNu1 (residues 1–68) (de Beer et al. 2002). Side groups of solvent-exposed residues of the recognition helix are displayed. Color coded as in B.
λ's cosB (cosBλ) is a complex subsite containing three copies of a gpNu1-binding sequence, the R sequence, plus a site, I1, for the integration host factor (IHF), the Escherichia coli DNA-bending protein. The order of sites is cosN–R3–I1–R2–R1. The amino-terminal half of gpNu1 contains a winged helix-turn-helix DNA-binding motif (Figure 1, B and C; Gajiwala and Burley 2000) that interacts with the R sequences. Further, the amino-terminal domain of gpNu1 is a tight dimer (Figure 1C, de Beer et al. 2002). The IHF-induced bend at I1 creates a DNA hairpin in cosB that positions the major grooves of R3 and R2 to face inward, so that the helix-turn-helix motifs of dimeric gpNu1 can be docked into them. The wing loops are positioned to make minor groove contacts with R3 and R2. Thus it is proposed that gpA is positioned to nick cosN by assembly of a bent structure with dimeric gpNu1 bound to R3 and R2 (Becker and Murialdo 1990; de Beer et al. 2002). A variety of studies indicate that the positioning of gpNu1 at R3 is crucial and that the other interactions function to create and/or stabilize the R3–gpNu1 interaction (Cue and Feiss 1993a; Higgins and Becker 1994a; Hang et al. 2001).
DNA packaging initiates when terminase binds and nicks a cos. Following cosN nicking and separation of the cohesive ends, terminase remains bound to the cosB-containing chromosome end (Becker et al. 1977; Yang et al. 1997). The DNA-bound terminase docks on the portal vertex of a prohead, the empty, immature virion head shell. Assembly of the ternary prohead–terminase–DNA complex activates gpA's potent translocation ATPase, and the viral DNA is translocated into the prohead (Yang and Catalano 2003; Dhar and Feiss 2005). Translocation brings the next cos along the concatemer to the portal-docked terminase (Feiss and Widner 1982). The downstream cos is cleaved by terminase, completing packaging of the chromosome. Recognition of the downstream cos requires cosQ and cosN (Cue and Feiss 2001). Following DNA packaging, terminase undocks from the filled head. Attachment of a tail to the DNA-filled head completes virion assembly. The undocked terminase remains bound to and sponsors the packaging of the next chromosome along the concatemer.
The interactions between the recognition helix of gpNu1 and an R sequence are typical for helix-turn-helix proteins, as shown by genetic studies of chimeras between λ and its relative, phage 21, as follows: λ and 21 have similarly organized cos sites; the cosB of 21 also has the R3–I1–R2–R1 structure. Nevertheless, the two phages have distinct packaging specificities. Base-pair differences in the R sequences account for packaging specificity (Becker and Murialdo 1990; Smith and Feiss 1993). cosN and cosQ are interchangeable between λ and 21 (Feiss et al. 1981). The consensus R sequences are 5′-CGTTTCCtTTCT-3′ for cosBλ and 5′-CaTGTCGGncCT-3′ for cosB21, where capitalized residues are conserved in all three R sequences of both phages; underlined and capitalized are two residues conserved in all three R sequences of both phages, but which differ between cosBλ and cosB21 (Becker and Murialdo 1990). These two conserved but phage-specific base pairs are likely to be of major importance for specificity. Similarly, the recognition helixes of the helix-turn-helix motifs of the small subunits of λ (gpNu1) and 21 (gp1) terminases differ in four amino acid residues that account for packaging specificity (Figure 1; Becker and Murialdo 1990).
In earlier work (de Beer et al. 2002), we showed that modifying λ-terminase by replacing the gpNu1 recognition helix with that of 21's gp1 created a terminase (gpNu1hy1 terminase) that was specific for the cosB of phage 21 (designated cosB21). That is, λ cosB21 Nu1hy1 was viable, but λ cosBλ Nu1hy1 was inviable due to the specificity mismatch between cosBλ and the cosB21-specific recognition helix of the chimeric small terminase subunit, gpNu1hy1. The Nu1hy1 terminase packages cosB21 chromosomes ∼10-fold more efficiently than it does cosBλ chromosomes. This 10-fold discrimination between cosB21 and cosBλ chromosomes is much weaker than the >104-fold discrimination shown by wild-type λ and 21 terminases (de Beer et al. 2002). Because of the modest discrimination of Nu1hy1 terminase, the yield of λ cosBλ Nu1hy1 is only slightly below the yield required for plaque formation. Lysates of λ cosBλ Nu1hy1 contain plaque-forming pseudorevertants at a level expected for single mutations. A number of these pseudorevertants were sequenced and found to contain mutations in cosBλ or in the Nu1hy1 gene. Here we report on in vivo packaging studies on the effects of these Nu1hy1 and cosBλ suppressor mutations on packaging specificity.
MATERIALS AND METHODS
Media:
Luria broth (LB), Luria agar (LA), tryptone broth (TB), tryptone agar (TA), and tryptone soft agar (TBSA) were prepared as described (Arber et al. 1983), except that TB, TA, and TBSA were supplemented with 0.01 m MgSO4. When required, kanamycin and/or ampicillin were added to final concentrations of 50 and 100 μg/ml, respectively.
λ sequence designations:
Base-pair positions on the λ-chromosome follow the standard numbering system (Daniels et al. 1983), where the first base pair of the left cohesive end is designated base pair 1, and numbering proceeds 5′−3′ along that strand. Base-pair coordinates refer to positions on the 48,502-bp λ+-chromosome.
Phages, bacteria, and plasmids:
Phages and lysogens used in helper-packaging experiments are described in the results. The construction of derivatives of λ-P1 was described earlier (Frackman et al. 1985; de Beer et al. 2002). Other phages used were derivatives of λ+ and were constructed by standard genetic crosses. MF532 is an E. coli K12 recA1 strain carrying a λ imm434 prophage (Miller and Feiss 1988; Cue and Feiss 1993b). MF532 is deleted for a segment extending rightward from a site in the λ A gene, through the b region and attR, into the E. coli bio operon. Dilysogens were produced by spotting a second (imm21) phage on a lawn of MF532, followed by streaking out and testing candidates for the presence of the second prophage. Strains in which multiple prophages of the second phage are inserted are not common and can be identified by looking at the immunity of the prophage chromosomes packaged by the helper phage. If a strain has only one packageable prophage, the packaged prophage will have the imm434 marker, but if multiple imm21 prophages are present, both imm21 and imm434 virions will be produced in helper-packaging experiments. MF532 derivatives were examined for the presence of imm21-packaged prophages in lysates; these strains were discarded.
Derivatives of the dilysogens, carrying a third prophage, the λ−P1 helper prophage, as a plasmid were isolated as KnR transductants following infection by the appropriate λ−P1 phage. Plating bacteria were C600, an E. coli K12 tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1 strain (Campbell 1961), and C600(λ+). Two constructs—one a Nu1 gene with the codons for the phage 21 support and recognition helixes (H21 Tλ H21 wλ) and another with the turn and recognition helix codons from 21(Hλ T21 H21 wλ)—were purchased from a commercial supplier (Blue Heron) and crossed into λ−P1.
Helper-packaging experiments:
Overnight cultures of helper-containing dilysogens were grown in LB plus kanamycin at 30° without shaking. Aliquots (0.25 ml) of overnight cultures were added to 5 ml of fresh LB, and cultures were grown with aeration at 31°. After 2.5 hr, the cell density was ∼5 × 107 cells/ml. Cell density was determined by direct counting, using a light microscope and a Petroff–Hauser chamber. Helper phages were induced by shifting cultures to 42° (without shaking) for 15 min, and helper phage growth was continued by shaking at 37° for 70 min. Lysis was completed by adding CHCl3, and the lysates were clarified by centrifugation. Phage yields were determined by plating dilutions on TA plates at 37°, using C600 as the plating bacterium. On C600, virions containing helper-packaged prophage chromosomes formed turbid plaques, and the helper phages, with the thermolabile cI857 repressor, formed clear plaques. For lysates containing low levels of helper-packaged prophages, C600(λ+) was used as the plating bacterium, so that the λ-prophage's immunity repressor blocked growth of the helper phage.
RESULTS
Experimental design and genotypic and gene product nomenclature:
To ask about the packaging specificity of terminases in pseudorevertants of λ, in vivo helper-packaging experiments were done as follows: When a phage, the “helper,” grows lytically on a host cell containing a packageable passive prophage inserted in the bacterial chromosome, both helper phage and passive prophage chromosomes are packaged into infectious virions. The yields of both types of phages, which are genetically distinguishable, can be enumerated by titering and expressed as the number of plaque-forming units per cell.
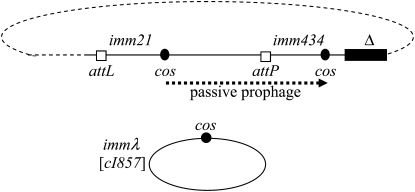
The normal substrate for λDNA packaging is concatemeric DNA. Packaging is initiated at a randomly chosen cos and proceeds in a cosB-to-cosQ direction to the next cos along the concatemer. Thus a packageable λ-chromosome is bounded by two cos sites. Constructing a packageable prophage chromosome bounded by two cos sites requires constructing a tandem double prophage; i.e., two prophages inserted in tandem in the bacterial chromosome. To do this, we started with the prophage deletion strain MF532, which contains a deleted prophage containing phage DNA from the left attachment site, attL, through cos, but is deleted for the late genes and attR (Figure 2; Cue and Feiss 1993b). When MF532 is lysogenized by a second prophage, the incoming prophage inserts at attL, forming a structure with a packageable chromosome where packaging can initiate at the cos of the second prophage and terminate at the cos of the deleted prophage. Because packaging initiates at the cos of the second prophage, second prophages with different cos markers, such as cosλ or cos21, enable us to ask specifically about the initiation effects of cos alleles. To maintain genetic stability, MF532 contains the null recA1 mutation. To keep the prophages from being induced during helper phage growth, the prophage repressor immunities differed from the immλ of the helper: imm434 for the deleted prophage and imm21 for the added prophage.
Figure 2.—
Genetic structure of strains used for helper packaging. The small oval represents the λ-P1 helper prophage, and the large oval indicates the E. coli chromosome with the tandem double-prophage structure. imm21, imm434, and immλ indicate the immunity segments imm21, imm434, and immλ, respectively. Solid dots indicate cos sites. The solid line indicates the deletion of the λ imm434 prophage of MF532. Addition of the second prophage (λ imm21) generates the tandem prophage structure. The dotted arrow indicates the passive prophage that can be packaged by the helper phage. Passive prophage packaging proceeds left to right, and the specificity of the left cos, which is derived from the imm21 prophage, can be varied, i.e., can have λ or 21 specificity. Shifting growing cells to 42° induces the immλ (cI857) helper phage to carry out lytic growth.
The helper phage background was λ-P1:5R cI857 nin5 Δbet-gam∷kan (Sternberg and Austin 1983; Pal and Chattoraj 1988). In λ-P1:5R, the normal site-specific recombination genes have been substituted by a phage P1 DNA segment encoding the plasmid replication and partitioning system. As a consequence, λ-P1:5R prophage is a plasmid. The Δbet-gam∷kan marker, an insert of a kanamycin-resistance cassette, enables selection of λ-P1:5R lysogens with kanamycin. The Δbet-gam∷kan marker inactivates λ's bet and gam genes, resulting in the inability to produce concatemeric DNA in recA mutant cells, so helper phage yields on MF532 strains are reduced to a few phages per cell (Furth and Wickner 1983). The cI857 mutation renders the immunity repressor thermolabile; a λ-P1:5R cI857 nin5 Δbet-gam∷kan can be induced by shifting a growing culture to 42°. Thermo-inactivation of the cI857 repressor does not result in induction of the imm434 and imm21 prophages in the MF532 constructs. Hereafter, λ-P1:5R cI857 nin5 Δbet-gam∷kan will simply be referred to as λ-P1 or the λ-P1 helper. Derivatives of λ-P1 used as helpers include hybrid phages with chimeric small terminase subunit genes with various combinations of the λ and 21 segments specifying features of the N termini of λ's gpNu1 and 21's gp1 (Figure 1, Table 1). Hybrid Nu1 alleles and gene products will be indicated by superscripts: e.g., the Nu1hy1 gene encodes gpNu1hy1. The four N-terminal segments relevant to DNA binding are as follows, with identifying designations in parentheses: Residues 5–12 form the support α-helix (H), residues 13–15 compose the turn (T), residues 16–24 are the recognition helix (H), and residues 31–39 form the wing loop (w). The virus specificity for each of these segments will be given in parentheses: gpNu1hy1 contains the four amino acids of the 21-specific recognition helix, so the Nu1hy1 small subunit is indicated as Hλ Tλ H21 wλ. Additional hybrid phages are listed in Table 1.
TABLE 1.
Chimeric Nu1 alleles used in this study
| Allele (motif segment origin)a | Remarks | Reference |
|---|---|---|
| Nu1hy51 (H21 T21H21 w21) | Codons 1–103 derived from small terminase subunit gene 1 of phage 21. | Frackman et al.(1985) |
| Nu1hy1 (Hλ TλH21 w λ) | Produces gpNu1 with the four amino acids of the recognition helix replaced with those of 21's gp1. | de Beer et al. (2002) |
| Nu1hy2 (H21 T21H21 w λ) | Codons 1–24 derived from phage 21's 1 gene. Small subunit residues derived from phage 21's gp1 include the amino terminus and the helix-turn-helix motif. | de Beer et al. (2002) |
| Nu1hy3 (H21 TλH21 w λ) | Small subunit differs from gpNu1 by two amino acids of the support helix and four amino acids of the recognition helix being replaced with those of 21's gp1. | This work |
|
Nu1hy4 (Hλ T21H21 w λ) |
Small subunit differs from gpNu1 in four amino acids of the recognition helix and three amino acids of the turn being replaced with those of 21's gp1. |
This work |
Origins of gpNu1 segments of the winged helix-turn-helix DNA-binding motif, as described in materials and methods.
Pseudorevertants of λ cosBλ Nu1hy1 included five types with changes affecting residues in or near the small terminase subunit's DNA-binding motifs (de Beer et al. 2002). Two additional types contained changes in cosB R sequences. We wondered if the packaging specificities of these pseudorevertant terminases had switched back, so that they had reacquired λ specificity while losing 21 specificity. Alternatively, the terminases might have acquired broadened specificity, so that the pseudorevertant terminases might package both λ and 21 chromosomes. To ask about the packaging specificity of the pseudorevertants' terminases, derivatives of MF532 were constructed so that passive prophage packaging was initiated at cosλ or cos21.
Packaging specificity of chimeric terminases:
As preliminary controls, packaging specificities were determined for four helper phages, each with a different Nu1 gene. The phages were (1) λ-P1 cosBλ Nu1+, (2) λ-P1 cosB21 Nu1hy51 (H21 T21 H21 w21), (3) λ cosB21 Nu1hy2 (H21 T21 H21 wλ), and (4) the parent of the pseudorevertants λ cosB21Nu1hy1 (Hλ Tλ H21 wλ). λ-P1 cosB21 Nu1hy51 is a chimera in which the first 89 codons of the small terminase subunit are derived from 21 gene 1 (Frackman et al. 1985). Thirty-eight of these 89 codons specify amino acids different from those of λ's gpNu1. The λ-P1 cosB21 Nu1hy51 phage's gp1-derived segment includes the entire helix-turn-helix, the wing, and additional residues. The λ cosB21 Nu1hy2 phage's small subunit gene has the codons specifying the 10 phage 21-specific amino acids found in small subunit residues 1–24, which include the helix-turn-helix segment. (The 10 phage 21-specific residues in the Nu1hy2 chimera also include Lys at residue 2.) The λ cosB21 Nu1hy1 chimera produces gpNu1hy1 terminase that contains the recognition helix from 21, with just four amino acid differences from λ's gpNu1 (Figure 1). For viability, the cosB of each helper phage was cognate, i.e., derived from the same phage as the small subunit recognition helix.
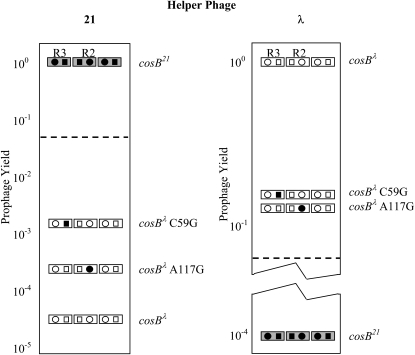
Of these four phages, three showed strong discrimination between chromosomes with either cosBλ or cosB21(Table 2, rows 1–4). The ability of terminase to discriminate between cosBλ and cosB21 can be expressed as a discrimination index: the ratio of the yield of the cognate phage to the yield of the noncognate phage (Table 2). Helpers λ cosBλ Nu1+, λ-P1 cosB21 Nu1hy51, and λ cosB21 Nu1hy2 discriminated strongly with ratios of ∼104. The discrimination indexes for the Nu1hy2 and Nu1hy51 chimeras are about the same as that of a phage with the complete set of 21 head genes (Feiss et al. 1981), and they show that the ten 21-specific residues in gpNu1hy2 are sufficient for strong discrimination. In contrast, Nu1hy1 terminase had a discrimination index of only ∼10 (line 4 in Table 2), consistent with the yields of λ cosBλ Nu1hy1 and λ cosB21 Nu1hy1 determined earlier (de Beer et al. 2002). Thus, gpNu1hy1 terminase has only a modest ability to discriminate against cosBλ chromosomes.
TABLE 2.
Mutant terminases with broad packaging specificity
| Line | Helper | Passive prophage | Prophage yielda (SEM)b | Passive prophage | Prophage yielda (SEM)b | Discrimination indexc |
|---|---|---|---|---|---|---|
| 1 | λ-P1 Nu1+ | λ | 1.02 (0.36) | 21 | 3.24 (1.37) × 10−5 | 3.1 × 104 |
| 2 | λ-P1 Nu1hy51 (H21 T21H21 w21) | λ | 8.02 (0.9) × 10−5 | 21 | 2.62 (0.29) | 3.3 × 104 |
| 3 | λ-P1 Nu1hy2 (H21 T21H21 wλ) | λ | 2.9 (0.55) × 10−6 | 21 | 2.15 (0.175) | 7.4 × 105 |
| 4 | λ-P1 Nu1hy1 (Hλ TλH21 wλ) | λ | 0.27 (0.040) | 21 | 2.86 (0.40) | 1.1 × 101 |
| 5 | λ-P1 Nu1hy1E20D | λ | 0.36 (0.14) | 21 | 0.41 (0.17) | 1.1 |
| 6 | λ-P1 Nu1hy1E20G | λ | 1.03 (0.14) | 21 | 1.90 (0.35) | 1.8 |
| 7 | λ-P1 Nu1hy1Q23R | λ | 0.70 (0.14) | 21 | 1.12 (0.22) | 1.6 |
| 8 | λ-P1 Nu1hy1Q23K | λ | 1.35 (0.18) | 21 | 1.29 (0.21) | 0.95 |
| 9 | λ-P1 Nu1hy1L40I | λ | 0.90 (0.20) | 21 | 2.55 (0.17) | 2.8 |
| 10 | λ-P1 Nu1hy3 (H21 TλH21 wλ) | λ | 0.03 (0.006) | 21 | 1.63 (0.24) | 54 |
| 11 |
λ-P1 Nu1hy4 (Hλ T21H21 wλ) |
λ |
0.15 (0.04) |
21 |
1.72 (0.33) |
11 |
Plaque-forming units/induced lysogen.
Values in parentheses are the standard error of the mean (3 ≤ n ≤ 5).
Discrimination index is the yield of the cognate passive prophage/yield of the noncognate passive prophage.
Specificities of the pseudorevertant terminases with gpNu1 changes:
The five pseudorevertant phages with mutations affecting the Nu1 gene of λ cosBλ Nu1hy1 were used as helpers. All were found to efficiently package cosBλ and cosB21 chromosomes. The λ-P1 Nu1hy1 pseudorevertant phages with changes in the recognition helix have the changes E20G, E20D, Q23K, and Q23R (Table 2, rows 5–8). The discrimination ratios for these four phages varied from 0.95 and 1.84, in contrast to the ratio of 10 for the λ cosBλ Nu1hy1 parent. The discrimination ratio for the fifth phage, with the change L40I, was 2.83 (Table 2, row 9). The L40I change is located just toward the C terminus from the wing loop. In sum, the pseudorevertants' mutant Nu1hy1 alleles encode terminases with broadened specificity, rather than encoding terminases that have acquired λ specificity and lost 21 specificity.
Roles of the support helix and turn in discrimination:
The chimeric terminase of phage λ Nu1hy2 (H21 T21 H21 wλ) strongly discriminated between 21 and λ-chromosomes, but Nu1hy1 terminase (Hλ Tλ H21 wλ) did not. The two terminases differ only by the N terminus (with one differing residue), the support helix (two differing residues), and the turn (three differing residues), which are from 21 in Nu1hy2 terminase and from λ in Nu1hy1 terminase. To ask if the contributions of the support helix and the turn make separable, independent contributions to discrimination, phages carrying the Nu1hy3 (H21 Tλ H21 wλ) and Nu1hy4 (Hλ T21 H21 wλ) alleles were constructed and used as helpers for packaging λ and 21 chromosomes. λ-P1 Nu1hy3 was about fivefold better at discriminating than λ-P1 Nu1hy1 (Table 2, row 10). λ-P1 Nu1hy4 (Hλ T21 H21) was no better at discriminating than λ-P1 Nu1hy1 (Table 2, row 11). In sum, the addition of the gp1 support helix (Nu1hy3) contributes modestly to the ability of the Nu1hy1 terminase to distinguish between 21 and λDNAs. Full discrimination requires the presence of the entire helix-turn-helix motif of phage 21's gp1. Thus, the components of the DNA-binding motif act in concert in cos recognition.
Packaging specificity of pseudorevertants with mutant cos sites:
Two pseudorevertants of λ cosBλ Nu1hy1 contained changes in cosB. Interestingly, both cosB mutations, C59G in R3 and A117C in R2, changed a base pair that was conserved in all three cosBλ R sequences to the analogous but different base pair conserved in all three cosB21 R sequences (de Beer et al. 2002). Because of the importance of the R sequences in recognition by terminase, we tested the effects of the two R-sequence mutations on recognition by the λ and 21 packaging systems. Dilysogens were constructed in which the initial (left) cosBλ had either the R3 C59G or the R2 A117C mutation, and the ability of λ or 21 helpers to package chromosomes with these mutations was determined (Table 3, Figure 3). For the 21 helper, λ-P1 Nu1hy51, with the complete DNA-binding motif of 21, was used. When helper λ-P1 Nu1hy51 provided 21-specific terminase, the cosBλ C59G and cosBλ A117C chromosomes were packaged 45- and 9-fold more efficiently, respectively, than the cosBλ positive control prophage. These results indicate that both the C59G and A117C changes improve cosB recognition by 21 terminase. The C59G and A117C mutations also reduced λ-packaging by 6- and 10-fold, respectively, as expected for base pairs important for terminase recognition. Thus, the cosB mutations C59G and A117C change the specificity of cosB away from that of λ and toward that of 21, rather than broadening cosB's specificity.
TABLE 3.
cosB mutations that enable packaging by 21-specific terminases reduce packaging by λ-specific terminase
| Line | Helpera | Initial cosb | Prophage yieldcd (SEM) | Helper | Prophage initial cos | Prophage yieldcd (SEM) | Discrimination indexe |
|---|---|---|---|---|---|---|---|
| 1 | λ-P1 Nu1hy1 (Hλ TλH21 wλ) | cosBλ | 0.20 (0.046) | λ-P1 Nu1hy1 (Hλ TλH21 wλ) | cosB21 | 2.87 (0.36) | 14.4 |
| 2 | λ-P1 Nu1+ | cos+Bλ | 0.84 (0.15) | λ-P1 Nu1hy51 (H21 T21H21 w21) | cosBλ | 4.55 (0.12) × 10−5 | 1.8 × 104 |
| 3 | λ-P1 Nu1+ | cosBλC59G | 0.13 (0.016) | λ-P1 Nu1hy51 (H21 T21H21 w21) | cosBλC59G | 2.05 (0.26) × 10−3 | 63 |
| 4 |
λ-P1 Nu1+ |
cosBλA117C |
0.09 (0.015) |
λ-P1 Nu1hy51 (H21 T21H21 w21) |
cosBλA117C |
4.1 (1.7) × 10−4 |
219 |
The genetic background for helper phages was λ-P1:5R cI857 nin5 Δbet-gam∷kan. Helper phage yields varied from 0.58 to 3.03.
The initial cos is the left cos at which DNA packaging initiates (Figure 2).
Plaque-forming units/induced lysogen.
Values in parentheses are the standard error of the mean (3 ≤ n ≤ 5).
Discrimination index is the yield of cognate prophage/yield of noncognate passive prophage.
Figure 3.—
The effects of the cosBλ mutations on packaging efficiency by λ and 21 helpers. Packaging efficiencies are normalized by setting the cognate packaging efficiencies to 1.0. cosB structures are indicated by rectangles: open rectangles indicate cosBλ and shaded rectangles represent cosB21. Dots and squares indicate two base-pair positions that (1) are conserved in all three R sequences of λ and 21 and (2) differ between λ and 21. Dots represent base pair 56 in R3, base pair 117 in R2, and base pair 158 in R1. Open circles represent the λ-specific TA base pair, and solid dots the 21-specific GC base pair. Note that R2 is oriented opposite to R3 and R1, so the R2 base pair is an AT base pair in λ and a CG in 21. Squares represent base pair 59 in R3, base pair 114 in R2, and base pair 161 in R1. Open squares indicate the λ-specific CG base pair, and solid squares the 21-specific GC base pair. Again, because of the opposite orientation of R2, base pair 114 is GC in λ and CG in 21. The dashed lines indicate the approximate relative yield (5% of wild type) required for plaque formation.
cosB suppressors do not have trans effects, and Nu1 suppressors do not have cis effects:
Pseudorevertants of λ-P1 Nu1hy1 with suppressor mutations in the Nu1hy1 gene showed trans effects: the mutant terminases had broadened specificity (Table 2). To see if the Nu1hy1E20G suppressor had a cis-specific effect on packaging, we asked if the Nu1hy1E20G change affected packaging by helper phages λ-P1 Nu1+ and Nu1hy2. A strain was constructed that contained a tandem double prophage with the packaging initiation cos of λ Nu1hy1E20G. As expected, the results (Table 4, rows 1 and 2) showed that the Nu1hy1E20G allele did not alter packaging by λ and 21 helpers when compared with packaging of a Nu1+ prophage (Table 1, rows 1 and 3).
TABLE 4.
cosB suppressors do not have trans effects, and Nu1 suppressors do not have cis effects
| Line | Helpera | Prophage | Helper yieldb | Prophage yieldb |
|---|---|---|---|---|
| 1 | λ-P1 Nu1+ | cosBλNu1hy1E20G | 3.22 (0.74) | 0.49 (0.21) |
| 2 | λ-P1Nu1hy2 | cosBλNu1hy1E20G | 1.10 (0.21) | 4.55 (1.2) × 10−5 |
| 3 | λ-P1Nu1hy1 cosBλC59G | cosBλ | 2.55 (0.43) | 0.51 (0.12) |
| 4 | λ-P1Nu1hy1 cosBλC59G | cosB21 | 1.79 (0.39) | 2.14 (0.44) |
| 5 | λ-P1Nu1hy1 cosBλA117C | cosBλ | 1.25 (0.33) | 0.31 (0.06) |
| 6 |
λ-P1Nu1hy1 cosBλA117C |
cosB21 |
1.6 (0.35) |
2.75 (0.28) |
The genetic background for helper phages was λ-P1:5R cI857 nin5 Δbet-gam∷kan. Tandem double prophages were constructed by lysogenizing MF532 as described in materials and methods.
The genetic background for helper phages was λ-P1:5R cI857 nin5 Δbet-gam∷kan.
Values in parentheses are the standard error of the mean (3 ≤ n ≤ 5).
Similarly, helper phages λ-P1 cosBC59G Nu1hy1 and λ-P1 cosBA117C Nu1hy1 packaged λ and 21 prophages with about the same discrimination as λ-P1 Nu1hy1 (compare Table 4, rows 3–6, with Table 1, row 4), verifying that the cosB suppressors do not affect terminase specificity.
DISCUSSION
Mutations that broaden the specificity of gpNu1Hy1 terminase:
Helper-packaging experiments reported here show five variants of gpNu1hy1 terminase, with the changes E20D, E20G, Q23K, Q23R, and L40I, have broadened specificity so that λ- and 21-specific chromosomes are packaged with roughly equal efficiency. Reasonable conjectures can be made about how these five changes might broaden the specificity of gpNu1Hy1 terminase. For example, the changes Q23K and Q23R are changes to basic residues. These basic residues could strengthen the interactions of terminase with cosBλ and cosB21 through nonspecific salt bridges with DNA backbone phosphates. All 14 pseudorevertants with changes in residue 23 were changes to arginine and lysine (de Beer et al. 2002). No isolate containing any of the other codons that could be generated by a single base-pair mutation, namely those for Glu, Leu, Pro, and His, was recovered. Interestingly, residue 23 is a glutamine in the recognition helixes of both λ and 21, and so residue 23 is not involved in specificity; rather, the changes to basic residues appear to add a new contact. The preferential improvement of the packaging of cosBλ chromosomes by the Nu1hy1Q23K/R suppressors is likely because the packaging of cosB21 chromosomes by λ-P1 Nu1hy1 is relatively efficient and not much improved by the suppressors, while inefficient packaging of cosBλ chromosomes is much improved by the Nu1hy1Q23K/R suppressors.
In the five pseudorevertant isolates of λ-P1 cosBλ Nu1hy1 (H21 T21 H21 wλ) with changes affecting gpNu1hy1 residue 20, the suppressors cause the changes E20D and E20G. These changes appear to remove a clash between E20 of gpNu1hy1 terminase and the R sequences of cosλ. That is, it is unlikely that residues that are so structurally different as Asp and Gly would create contacts with R-sequence base pairs. Rather, the shorter side chain of aspartic acid, and the absence of the acidic group in glycine, may lessen and abolish, respectively, a clash due to the acidic group of Glu. This argument would be strengthened by examination of more pseudorevertants of λ-P1 cosBλ Nu1hy1 or by direct examination of the behavior of the other changes at 20; Lys, Gln, Val, and Ala are the additional changes that can be obtained by single base-pair mutations. Also, it is an open question as to whether E20 makes a specific contact with the R sequences of cosBλ. If so, loss of the contact caused by the E20G change seems not to be critical for efficient packaging, because the pseudorevertant helper λ-P1 cosBλ Nu1hy1E20G packages λDNA (yield = 1.03) as efficiently as the λ-P1 Nu1+ helper (yield = 1.02; Table 2, rows 6 and 1, respectively). The phage with the E20D change shows a general reduction in prophage packaging, perhaps because the clash is not fully relieved by the Glu-to-Asp substitution. Consistently, λ-P1 Nu1hy1E20D forms tiny plaques and has a yield, at 23 phages/cell, only slightly higher than the 20 phages/cell of the parent phage, λ-P1 Nu1hy1 (de Beer et al. 2002).
The Nu1 mutation giving the L40I change was recovered previously as a suppressor of a cosB defect caused by three point mutations, as follows: A C-to-T transition mutation in a base pair conserved in all three R sequences of cosBλ (at base pair 58 in R3, base pair 115 in R2, and base pair 160 in R1; Figure 1) is lethal (Cue and Feiss 1992b, 1993b). In vitro studies showed that the R-sequence mutations had mild effects on cos cleavage and strong effects on DNA packaging (Cue and Feiss 1993a). Among pseudorevertants of the cosB triple mutant were variants with suppressor mutations in Nu1, causing the changes L40F and L40I (Cue and Feiss 1992a). gpNu1 L40F and L40I terminases behave similarly in suppression studies and presumably have the same mechanism, with L40F being a somewhat stronger suppressor. The gpNu1 L40F terminase showed no increased ability to carry out cos cleavage of the mutant DNA, but rather showed increased efficiency of DNA packaging of the mutant DNA (Cai et al. 1997). These results suggest that the R-sequence mutations lower the stability of the terminase–DNA complex, complex I, that forms following cos cleavage, and that the L40F and L40I compensate by enhancing the efficiency of the assembly, and/or the stability, of complex I formed on the R mutant DNA. Residue 40 is adjacent to the wing (residues 31–39), so the L40I change may alter wing–R-sequence interaction(s).
Genetic observations indicate that the L40F and L40I changes enhance terminase's interactions with cos, as follows: Wild-type λ-terminase's yield (∼100 phages/cell) is reduced to ∼30% in cells lacking IHF (Feiss et al. 1988; Granston et al. 1988). The reduction is likely due to reduced formation or stability of complex I. In contrast, λ cos+ Nu1L40F and λ cos+ Nu1L40I phages produce normal yields on IHF− cells but have yields reduced by ∼50% in IHF+ cells (Cue and Feiss 1992a). The reduced yield in the presence of IHF suggests that complex I is perhaps too stable and that progression to the next step of DNA packaging is impeded. Using these ideas for this work, we speculate that the L40I mutation enhances the assembly and/or stability of the complex I formed on cosBλ DNA by gpNu1hy1 terminase, so that packaging efficiency is increased to a level near to that of λ cosB21 DNA. It is important to note that the quaternary structure of terminase is complex. Terminase protomers with the subunit ratio of (gpNu1)2:gpA1 assemble into a higher oligomeric form, a tetramer of the protomers, that has distinctly different properties, including independence from IHF for cos cleavage and DNA packaging (Maluf et al. 2005, 2006). It is not known what the assembly state of wild-type terminase is during development in vivo or whether mutant terminases such as gpNu1 L40I have altered assembly behavior.
cosB mutations that change specificity:
The cosB mutations found in pseudorevertants of λ Nu1hy1, C59G in R3, and A117C in R2 each change a λ base pair to a 21 base pair. The helper-packaging experiments reported here show that each of these mutations enhances packaging by 21 and reduces packaging by λ (Table 3, Figure 3). The C59G mutation enhances packaging by 21-specific phages significantly more efficiently than the A117C mutation. The A117C change alters the base pair corresponding to R3 base pair 56. It is not clear whether the relatively stronger effects of C59G are due to the greater importance of R3 and/or to the greater importance of base pair 59. Overall, it is clear that both positions are important for recognition by the 21 recognition helix. The nature of the specific contacts of the small terminase subunits with the R sequences is unclear; although the gpNu1 dimer modeling shows that the DNA-binding motifs fit well into the major grooves of R3 and R2 (de Beer et al. 2002), much more genetic and structural information is required to define the specific contacts.
Note that these experiments used the λ Nu1hy51 helper phage and that gpNu1hy51 terminase discriminates strongly. The C59G and A117C changes do not raise the level of packaging up to the level required for plaque formation by λ Nu1hy51. Because of the relatively weak discrimination by the gpNu1hy1 terminase, the mutations do allow plaque formation by λ cosλ Nu1hy1. In the model that the two positions (56 and 59 in R3) are the most important for small subunit recognition, each of the mutant cos sites has only a single base-pair change and the other 5 base-pair positions remain mismatched with the helix-turn-helix motif. Packaging reductions by the mutations are similar, with the reduction by A117C being slightly greater.
Weisberg and colleagues used chimeras and mutants to analyze the specificity determinants of site-specific recombination for the integrases of phages λ and HK022 (Yagil et al. 1989, 1995). The λ and HK022 integrases have ∼70% sequence identity. Replacing two amino acids of Intλ with the corresponding IntHK residues generated a broad specificity integrase that sponsored efficient λ and HK recombination. Codon randomization studies indicated that one of these mutations (E319R) increased integrase's catalytic activity with HK022 and that the other (N99D) removed an unfavorable interaction between that residue and the HK022 DNA target. Three additional replacements reduced λ-recombination so that the integrase had activity and specificity approximating that of wild-type HK022 integrase. The three additional changes were argued to reduce λ-recombination by affecting λ-specific integrase–DNA contacts.
Effects of the support helix, turn, and wing on specificity:
Chimeric gpNu1hy2 and gpNu1hy51 terminases contain the entire 21 helix-turn-helix motif and discriminate strongly against λ-chromosomes, and the gpNu1hy1 chimera does not. We asked whether strong discrimination might be obtained if phage 21's support helix or turn was present along with 21's recognition helix. The gpNu1hy3 (H21 Tλ H21) chimera showed a modest fivefold increase in specificity. The gpNu1hy4 (Hλ T21H21) chimera did not discriminate any better than the Hλ Tλ H21 chimera did. We conclude that the turn can enhance discrimination to a modest degree, presumably by altering the positioning of the recognition helix in the major grooves of the R sequences. It is clear, however, that for strong discrimination against λ-chromosomes, the entire helix-turn-helix motif must be from 21.
The two chimeras with the complete helix-turn-helix motif of 21 differ in the origin of the wing: λ-P1 Nu1hy51 has w21, and the Nu1hy2 phage has wλ. These phages have similar discrimination ratios, indicating that the DNA interactions of the wing are not phage-specific. At the tip of each wing loop is residue 35, a lysine in both cases. Fitting the recognition helixes into the major grooves of R3 and R2 places the wing lysine in a position to make electrostatic interactions with either backbone phosphates or base pairs in the minor groove (de Beer et al. 2002). Terminase with gpNu1 K35D has reduced DNA affinity and reduced the ability to distinguish between cosB and nonspecific DNA, indicating that lysine-35 plays a role in DNA binding and in specific binding of cosB (Hwang and Feiss 1997). Our results indicate that the DNA contacts made by the wing, whether sequence-specific or not, do not contribute significantly to the specificity difference between cosBλ and cosB21.
How did λ and 21 specificities evolve?
λ and 21 clearly have closely related chromosome recognition determinants, but each is highly phage-specific. It is obviously advantageous for viruses to have differing specificities. When a λ-like phage infects a cell carrying a heteroimmune prophage with the same packaging specificity, the infecting phage can initiate packaging at the prophage cos. DNA packaging would proceed into bacterial DNA adjacent to the prophage, a nonproductive packaging event leading to loss of the prohead and terminase engaged in packaging (Little and Gottesman 1971). Clearly the fitness of such a virus is increased if the virus possesses a DNA-packaging specificity that differs from the specificity of a heteroimmune prophage found at a significant frequency in host cells.
One scenario for how a new packaging specificity might arise is that a phage with a new specificity could arise as a variant of an existing phage through a series of mutations that alters the specific interactions between R sequences and small subunit helixes. One imagines that such a series of changes might produce an intermediate phage with a nonspecific terminase, which would allow for genetic drift in the R sequences to produce new sequences. The new R sequences could participate in specific interactions while a parallel series of changes would confer specificity on the small subunit for the new R sequence.
A problem with this scenario is that all of the nonspecific terminase variants that we have generated here have reduced yields (de Beer et al. 2002) and hence are clearly less fit than the specific parental phages, λ and 21. Less-fit variants would not effectively compete with the parental phages. The prophage state provides an opportunity for genetic drift to occur in the absence of selective pressure for robust lytic growth and might provide a way for specificity changes involving less-fit intermediates (Dove 1971). Of course there is a myriad of potential mutational paths by which a broad-specificity intermediate could arise, and paths involving neutral mutations may exist. Here we have looked at only a few particular examples. The yield of λ-P1 Nu1hy51, the equivalent of phage 21 with respect to packaging specificity, was 120 phages/induced lysogenic cell, that of λ-P1 cosB21 Nu1hy1 was 70, and those for the phages with broadened specificity ranged from 23 to 106 (de Beer et al. 2002). The reduced yields of the pseudorevertants could be due to many factors that include folding defects or difficulty in distinguishing between cos and non-cos DNA sequences.
A second scenario is that λ and 21 DNA-packaging specificities have evolved from an ancestor phage with relatively nonspecific cos–terminase interactions. Packaging by the ancestor phage would have been less efficient, but as the founder organism, more efficient and fit competitors simply would not exist. λ and 21 would result from gradual acquisitions of greater specificity and hence would have replaced their common ancestor. Given the strongly conserved cosB structures of λ and 21, the ancestor phage's cosB would likely be similarly structured, i.e., with three R sequences and an IHF site.
Acknowledgments
We thank the editor and reviewers for helpful comments. M.F. thanks Ben Hall, Walt Fangman, and Allan Campbell for genetics lessons. This work was supported by National Institutes of Health grant GM-51611 and National Science Foundation (NSF) grant 0717620. M.S. was a student in the Department of Microbiology Summer Research Program and was supported by NSF-Research Experience for Undergraduates grant DBI-0097361.
References
- Arber, W., L. Enquist, B. Hohn, N. E. Murray and K. Murray, 1983. Experimental methods for use with lambda, pp. 433–366 in Lambda II, edited by R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Baines, J. D., and S. K. Weller, 2005. Cleavage and packaging of herpes simplex virus 1 DNA, pp. 135–150 inViral Genome Packaging Machines: Genetics, Structure and Mechanism, edited by C. E. Catalano. Landes Bioscience, Georgetown, TX.
- Becker, A., and M. Gold, 1978. Enzymatic breakage of the cohesive end site of phage lambda DNA: terminase (ter) reaction. Proc. Natl. Acad. Sci. USA 75 4199–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, A., and H. Murialdo, 1990. Bacteriophage lambda DNA: the beginning of the end. J. Bacteriol. 172 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, A., H. Murialdo and M. Gold, 1977. Early events in the in vitro packaging of bacteriophage DNA. Virology 78 291–305. [DOI] [PubMed] [Google Scholar]
- Cai, Z. H., Y. Hwang, D. Cue, C. Catalano and M. Feiss, 1997. Mutations in Nu1, the gene encoding the small subunit of bacteriophage lambda terminase, suppress the post-cleavage DNA packaging defect of cosB mutations. J. Bacteriol. 179 2479–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, A., 1961. Sensitive mutants of bacteriophage lambda. Virology 14 22–32. [DOI] [PubMed] [Google Scholar]
- Cue, D., and M. Feiss, 1992. a Genetic analysis of mutations affecting terminase, the bacteriophage lambda DNA packaging enzyme, that suppress mutations in cosB, the terminase binding site. J. Mol. Biol. 228 72–87. [DOI] [PubMed] [Google Scholar]
- Cue, D., and M. Feiss, 1992. b Genetic analysis of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda. J. Mol. Biol. 228 58–71. [DOI] [PubMed] [Google Scholar]
- Cue, D., and M. Feiss, 1993. a The role of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda, in the nicking reaction. J. Mol. Biol. 234 594–609. [DOI] [PubMed] [Google Scholar]
- Cue, D., and M. Feiss, 1993. b A site required for termination of packaging of the phage lambda chromosome. Proc. Natl. Acad. Sci. USA 90 9290–9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue, D., and M. Feiss, 2001. Bacteriophage lambda DNA packaging: DNA site requirements for termination and processivity. J. Mol. Biol. 311 233–240. [DOI] [PubMed] [Google Scholar]
- Daniels, D., J. Schroeder, W. Szybalski, F. Sanger, A. Coulsen et al., 1983. Completed annotated lambda sequence, pp. 519–676 in Lambda II, edited by R. W. Hendrix, J. W. Roberts, F. W. Stahl and R. A. Weisberg. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Davidson, A., and M. Gold, 1992. Mutations abolishing the endonuclease activity of bacteriophage lambda terminase lie in two distinct regions of the A gene, one of which may encode a leucine zipper DNA binding domain. Virology 161 305–315. [DOI] [PubMed] [Google Scholar]
- de Beer, T., J. Fang, M. Ortega, Q. Yang, L. Maes et al., 2002. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell 9 981–991. [DOI] [PubMed] [Google Scholar]
- Dhar, A., and M. Feiss, 2005. Bacteriophage lambda terminase: alterations of the high-affinity ATPase affect viral DNA packaging. J. Mol. Biol. 347 71–80. [DOI] [PubMed] [Google Scholar]
- Dove, W., 1971. Biological inferences, pp. 297–312 in The Bacteriophage Lambda, edited by A. Hershey. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Feiss, M., and C. E. Catalano, 2005 Bacteriophage lambda terminase and the mechanism of viral DNA packaging, pp. 5–39 in Viral Genome Packaging Machines: Genetics, Structure and Mechanism, edited by C. E. Catalano. Landes Bioscience, Georgetown, TX.
- Feiss, M., and W. Widner, 1982. Bacteriophage lambda DNA packaging: scanning for the terminal cohesive end site during packaging. Proc. Natl. Acad. Sci. USA 79 3498–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiss, M., R. Fisher, D. A. Siegele and W. Widner, 1981. Bacteriophage lambda and 21 packaging specificities, pp. 213–222 in Bacteriophage Assembly, edited by M. S. DuBow. Alan R. Liss, New York. [PubMed]
- Feiss, M., S. Fogarty and S. Christiansen, 1988. Bacteriophage λ DNA packaging: a mutant terminase that is independent of integration host factor. Mol. Gen. Genet. 212 142–148. [DOI] [PubMed] [Google Scholar]
- Frackman, S., D. A. Siegele and M. Feiss, 1985. The terminase of bacteriophage lambda: functional domains for cosB binding and multimer assembly. J. Mol. Biol. 183 225–238. [DOI] [PubMed] [Google Scholar]
- Furth, M., and S. Wickner, 1983. Lambda DNA replication, pp. 145–174 in Lambda II, edited by R. Hendrix, J. Roberts, F. Stahl and R. Weisberg. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Gajiwala, K., and S. Burley, 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10 110–116. [DOI] [PubMed] [Google Scholar]
- Granston, A. E., E. M. Alessi, L. J. Eades and D.I. Friedman, 1988. A point mutation in the Nu1 gene of bacteriophage λ facilitates phage growth in Escherichia coli with himA and gyrB mutations. Mol. Gen. Genet. 212 149–156. [DOI] [PubMed] [Google Scholar]
- Hang, J., C. Catalano and M. Feiss, 2001. The functional asymmetry of cosN, the nicking site for bacteriophage lambda DNA packaging, is dependent on the terminase binding site, cosB. Biochemistry 40 13370–13377. [DOI] [PubMed] [Google Scholar]
- Higgins, R. R., and A. Becker, 1994. a The lambda terminase enzyme measures the point of its endonucleolytic attack 47 +/− 2 bp away from its site of specific DNA binding, the R site. EMBO J. 13 6162–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, R. R., and A. Becker, 1994. b Chromosome end formation in phage lambda, catalyzed by terminase, is controlled by two DNA elements of cos, cosN and R3, and by ATP. EMBO J. 13 6152–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, Y., and M. Feiss, 1996. Mutations affecting the high affinity ATPase center of gpA, the large subunit of bacteriophage lambda terminase, inactivate the endonuclease activity of terminase. J. Mol. Biol. 261 524–535. [DOI] [PubMed] [Google Scholar]
- Hwang, Y., and M. Feiss, 1997. Mutations affecting lysine-35 of gpNu1, the small subunit of bacteriophage lambda terminase, alter the strength and specificity of holoterminase interactions with DNA. Virology 231 218–230. [DOI] [PubMed] [Google Scholar]
- Little, J. W., and M. Gottesman, 1971. Defective lambda particles whose DNA carries only a single cohesive end, pp. 371–394 in The Bacteriophage Lambda, edited by A. D. Hershey. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Maluf, N., Q. Yang and C. Catalano, 2005. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J. Mol. Biol. 347 523–542. [DOI] [PubMed] [Google Scholar]
- Maluf, N., H. Gaussier, E. Bogner, M. Feiss and C. Catalano, 2006. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry 45 15259–15268. [DOI] [PubMed] [Google Scholar]
- Miller, G., and M. Feiss, 1988. The bacteriophage lambda cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol. Gen. Genet. 212 157–165. [DOI] [PubMed] [Google Scholar]
- Ostapchuk, P., and P. Hearing, 2005. Control of adenovirus packaging. J. Cell Biochem. 96 25–35. [DOI] [PubMed] [Google Scholar]
- Pal, S. K., and D. K. Chattoraj, 1988. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J. Bacteriol. 172 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. P., and M. Feiss, 1993. Sites and gene products involved in lambdoid phage DNA packaging. J. Bacteriol. 175 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, N., and S. Austin, 1983. Isolation and characterization of P1 minireplicons. J. Bacteriol. 153 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil, E., S. Dolev, J. Oberto, N. Kislev, N. Ramaiah et al., 1989. Determinants of site-specific recombination in the lambdoid coliphage HK022: an evolutionary change in specificity. J. Mol. Biol. 207 695–717. [DOI] [PubMed] [Google Scholar]
- Yagil, E., L. Dorgai and R. A. Weisberg, 1995. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J. Mol. Biol. 252 163–177. [DOI] [PubMed] [Google Scholar]
- Yang, Q., and C. E. Catalano, 2003. Biochemical characterization of bacteriophage lambda genome packaging in vitro. Virology 305 276–289. [DOI] [PubMed] [Google Scholar]
- Yang, Q., A. Hanagan and C. E. Catalano, 1997. Assembly of a nucleoprotein complex required for DNA packaging by bacteriophage lambda. Biochemistry 36 2744–2752. [DOI] [PubMed] [Google Scholar]