Abstract
The emergence of recombinant DNA technology occurred via the appropriation of known tools and procedures in novel ways that had broad applications for analyzing and modifying gene structure and organization of complex genomes. Although revolutionary in their impact, the tools and procedures per se were not revolutionary. Rather, the novel ways in which they were applied was what transformed biology.
FREEMAN Dyson contrasts what he called the “Kuhnian” and “Galisonian” views of the origins of scientific revolutions in a review of Peter Galison's book, Einstein's Clocks, Poincare's Maps: Empires of Time (Dyson 2003). In The Structure of Scientific Revolutions, Thomas Kuhn proposes that revolutionary breakthroughs in science are triggered primarily by ideas that, by their novelty, transform or replace the prevailing paradigm (Kuhn 1962). By contrast, Galison (2003) attributes such breakthroughs to new tools that, by their nature, make possible new approaches to formerly intractable problems. Galison also acknowledges that the application of existing tools in novel ways often provides the means to explore what was previously impossible. As Alfred Hershey has been quoted saying, “There is nothing more satisfying to me than developing a method. Ideas come and go, but a method lasts” (Stahl 1998).
The Galison view is exemplified by the genetic revolution in biotechnology, which relied on both the discovery of new tools and the use of existing tools in new ways. The key methodological advances were: (i) the discovery of enzymes that modify DNA molecules in ways that enable them to be joined together in new combinations; (ii) the demonstration that DNA molecules can be cloned, propagated, and expressed in bacteria; (iii) the development of methods for chemically synthesizing and sequencing DNA molecules; and (iv) the development of the polymerase chain reaction method for amplifying DNA in vitro.
Although the emergence of recombinant DNA technology was transformational in its impact, the tools and procedures that were the keys to its development largely emerged as enhancements and extensions of existing knowledge, i.e., they were evolutionary, not revolutionary, in nature. What was novel was the numerous ways in which many investigators applied these technologies for analyzing and modifying gene structure and the organization of complex genomes. Especially striking was the rapidity with which the new technologies took hold and dominated research into many different biological problems. Today, recombinant DNA technology has altered the ways both questions are formulated and solutions are sought. Scientists now routinely isolate genes from any organism on our planet, alive or dead. The construction of new variants of genes, chromosomes, and viruses has become standard practice in research laboratories. Only science fiction one-half century ago, the introduction of new genes into microbes, plants, and animals, including humans, is a common occurrence. The tools of recombinant DNA greatly expedite sequencing of the genomes of humans and numerous other species. Along with these advances have come astonishing improvements in medical diagnoses, prognoses, and therapies. In addition, many commercial opportunities have been realized, with the United States being the world leader in the biotechnology industry. Equally profound is the influence these developments have had on many related fields. Even a cursory look at journals in such diverse fields as chemistry, evolutionary biology, paleontology, anthropology, linguistics, psychology, medicine, plant science, and, even, forensics, information theory, and computer science shows the pervasive influence of this new technology. This essay traces the conceptual and experimental origins of the recombinant DNA technology.
Background:
During the 1960s, enormous progress was made in understanding the structure of genes and the mechanisms of their replication, expression, and regulation in prokaryotes and the viruses that infect them. However, largely unknown at the end of that decade was whether these findings were applicable to eukaryotes, i.e., organisms with an authentic nucleus, and, in particular, mammalian cells. The reason was that the experimental tools available at that time for exploring the molecular and genetic properties of mammalian organisms were woefully inadequate for the task.
One method that had been very powerful in investigations of the molecular biology of the most widely studied microbe, Escherichia coli, was the property of bacteriophage (commonly abbreviated “phage”) to transfer genes from one strain to another, a process referred to as transduction. For example, in the case of “generalized transduction” by phage P1, E. coli cells are infected with phage P1, the viral proteins are synthesized, the viral genome is replicated, and new infectious virus particles are assembled. However, concomitant with virus multiplication, random segments of the infected cell's DNA are also incorporated into newly formed virus particles in place of viral DNA. When such a pseudo-P1 phage “infects” a bacterium, neither virus replication nor cell death occurs. Instead, the bacterial DNA contained within the pseudo-P1 phage enters the bacterium and recombines at low frequency with the cell's chromosome to become a permanent part of that cell's genetic makeup. If the newly acquired bacterial DNA confers a measurable or selectable property, the rare recombinant can be recovered using an appropriate selection condition. In this way, any part of the genome of one E. coli strain can be transferred to the genome of another E. coli strain. Zinder's recollections of his and Lederberg's discovery of bacteriophage-mediated gene transfer in bacteria has been described in an earlier Perspectives article (Zinder 1992).
An alternate way of transferring genes from one E. coli cell to another is exemplified by phage λ-mediated “specialized transduction.” In this system, transduction occurs when the phage DNA integrates into the infected cell's chromosome, and bacterial DNA adjacent to the site of integration is excised and packaged into phage particles along with the viral DNA. The cellular DNA acquired by the phage can then be transferred to new hosts during subsequent rounds of infection. These two modes of virus-mediated transduction are distinctive in that phage P1 can transfer DNA from any region of the bacterial chromosome while phage λ transfers only regions of the bacterial chromosome adjacent to sequence-specific phage λ integration sites (Campbell 2007).
It seemed reasonable to consider whether a comparable virus-mediated gene-transfer system exists for mammalian cells. The small DNA viruses, polyoma and SV40, were deemed to be good candidates. It was already known that infection of cultured mouse cells with polyoma virus results in the production of infectious polyoma progeny and virus particles containing exclusively mouse DNA. Importantly, the mouse DNA contained in these polyoma “pseudovirions” is representative of the entire mouse genome. A similar finding was made with the related primate virus, SV40. However, in this case, some virus particles are produced in which host cellular DNA is covalently joined to the viral DNA. Might it be possible, we mused, that polyoma or SV40 could be used to transfer genes from one mammalian cell to another in much the same way that phage transfer genes among bacteria? On the face of it, that seemed unlikely for the following reasons. The amount of bacterial DNA that can be accommodated in a phage P1 particle is ∼2% of the E. coli genome; somewhat less cellular DNA can be transferred by phage λ. By contrast, polyoma and SV40 virions can accommodate only 5–6 kbp of DNA, i.e., roughly one-millionth of a mammalian genome. Thus, the probability of acquiring a specific mammalian gene in a polyoma or SV40 virion particle is at least four orders of magnitude lower than is the probability that a P1 or λ phage particle will contain one or more specific E. coli genes. In addition, the difficulty of picking out a specific, unique segment of mammalian DNA without having on hand a very strong method of selection or detection made the whole notion rather infeasible.
An alternative that seemed worth exploring was whether specific segments of mammalian, or any DNA for that matter, could be recombined with SV40 DNA in vitro. That would bypass the need for the recombinant product to be incorporated into a virus particle. This idea was attractive because mammalian cells have the capacity to take up “naked” DNA such as the SV40 genome, integrating it into the host cell's genome. Thus, any DNA covalently linked to SV40 DNA could become integrated into the chromosomes of a mammalian cell along with the viral DNA. In theory, such cells could be screened or selected for the presence and expression of both the SV40 and foreign DNAs. Thus, the first step toward achieving this game plan involved devising a method for introducing foreign DNA into the SV40 genome.
In early 1971, the American Cancer Society approved a grant application in which Berg proposed to develop the means for transducing foreign DNA into mammalian cells (Berg 1970). In the proposal, he identified SV40 DNA as the vector because it can be taken up by rodent and primate cells, including human ones, where it can replicate to high copy number as an autonomous plasmid or integrate into the host cell's genome. For the recombinant partner, the DNA would, ideally, be one (i) whose integration and possible expression in mammalian cells could be assayed, (ii) that could replicate as an autonomous plasmid in E. coli, and (iii) that has a gene whose expression could provide a way to screen or select E. coli cells containing the DNA.
But first, a method for joining together two DNAs in vitro needed to be developed. The plan was based on the knowledge that the bacteriophage λ genome exists as a linear DNA molecule within its virus particle, yet becomes a circular molecule following infection of its host, E. coli. That property stems from the existence of complementary, single-stranded extensions on the 5′-ends of the linear phage λ DNA enabling the ends to be joined (Hershey et al. 1963). At low DNA concentration, intramolecular base pairing of these complementary single-stranded ends leads primarily to the formation of monomeric circular DNA molecules; at high DNA concentration, intermolecular end-to-end joining leads primarily to the formation of oligomeric DNA molecules. Such complementary ends are referred to as being “cohesive” or “sticky.” Furthermore, these hydrogen-bonded rings can be sealed in vitro by incubation with DNA ligase to create covalently closed circular DNA molecules (Gellert et al. 1968; Wu and Kaiser 1968). Thus, it seemed attractive to consider constructing “artificial” cohesive ends as the strategy for joining together two different DNAs.
Following that strategy required a procedure for constructing short stretches of complementary nucleotides onto the ends of the two molecules to be recombined and to rely on their capacity to base pair in vitro to effect the joining. The enzyme terminal deoxynucleotidyl transferase (TdT) seemed admirably suited for this purpose since it was known to synthesize chains of a single nucleotide onto the 3′-ends of duplex DNA when a single nucleoside triphosphate is provided as the nucleotidyl donor (Kato et al. 1967). Synthesizing short polynucleotide chains of adenylates onto the 3′-ends of one DNA and approximately the same length polynucleotide chains of thymidylates onto the 3′-ends of the other DNA would create the necessary cohesive ends for joining together two DNA molecules. David Jackson4 and Robert Symons5 undertook the task of exploring this approach.
Peter Lobban6 independently conceived the idea of using a series of enzymes to covalently join DNAs together in vitro while fulfilling the Stanford Biochemistry Department's requirement for Ph.D. students to write and defend an original research proposal (Lobban 1969).
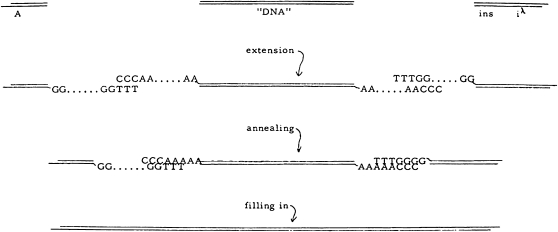
Lobban's stated goal was to create a λ phage-based transduction system by replacing nonessential DNA in the middle of the phage λ genome with “foreign” DNA (Figure 1). He proposed to isolate DNA segments derived from the left and right “arms” of λ phage DNA and then to join the foreign DNA to the two internal ends of these arms. The cohesive ends present on the left and right arms would be left intact to permit the recombinant genome to circularize and replicate. The formation of the recombinant was to be achieved by using TdT to add short polymeric tails to the 3′-ends of the foreign DNA and complementary polymeric tails to the internal 3′-ends of the left and right arms of the λ DNA.
Figure 1.—
“Steps in the creation of transducing genomes (digestion with λ exonuclease not shown),” the procedure originally proposed by Lobban for inserting “foreign DNA” into the left and right arms of phage λ DNA in vitro. Reproduced from Figure 3 of Lobban (1969).
In his proposal and Ph.D. thesis (Lobban 1972, Lobban) foresaw the prospect of inserting any foreign DNA, including from mammalian cells, into the phage DNA. He suggested that such an approach might enable specific mammalian genes to be identified and their mRNA and protein products to be detected and recovered in E. coli. He speculated that there would be many uses for such transducing phage, including “genetic engineering” (Lobban 1969, 1972). However, rather than directly pursuing the construction of a λ phage transducing virus as proposed, Lobban decided it would be better to focus initially on developing an in vitro DNA joining protocol to form circular dimers of phage P22 DNA from P22 DNA monomers (Lobban 1972; Lobban and Kaiser 1973). His reasoning was that the latter was a better model system for working out the detail methodology since P22 phage DNA naturally has blunt ends and is circularly permuted and, therefore, would be unable to dimerize without the addition of (dA)n and (dT)n tails.
During this period, Lobban and Jackson were in close communication, freely sharing enzymes and their findings while they worked on their respective projects. Unbeknownst to them, Jensen et al. (1971) were also attempting to join together two DNAs in vitro by synthesizing complementary tails with TdT followed by incubation with DNA ligase in the presence of DNA polymerase I; in this case, they used phage T7 DNA as the two templates. Clearly, the idea of joining together DNAs by generating cohesive ends with TdT was a logical extension of facts already known to many biochemists at this time.
A suitable DNA for linking to SV40 DNA was developed during the winter of 1971 through the collaborative efforts of D. Berg7 et al.(1974). This DNA, called λdvgal 120, contains both the genes from phage λ necessary for replication as an autonomous plasmid in E. coli and an intact gal operon, i.e., the three genes from E. coli needed for metabolizing galactose. At the time, Mertz also showed that purified λdvgal 120 DNA could be reestablished as an autonomously replicating plasmid in E. coli using a procedure originally developed by Mandel and Higa (1970) for transformation of linear phage DNAs. Thus, both the mammalian and bacterial cloning DNAs were in hand, along with methods for reintroducing them into their host cells.
Several kinds of experiments could potentially be explored with an SV40-λdvgal 120 recombinant DNA. One was to determine whether the E. coli gal operon is expressed in mammalian cells and, if so, to study its expression and regulation in that environment. The other objective was to determine whether the SV40-λdvgal 120 plasmid DNA could replicate autonomously in E. coli. The latter would provide a way (i) to produce large quantities of SV40 DNA and, possibly, its encoded proteins, and (ii) to generate mutants of SV40 in vitro or in vivo that could be propagated in E. coli and their phenotypes assessed by introduction into mammalian cells.
Creating recombinant DNA in vitro:
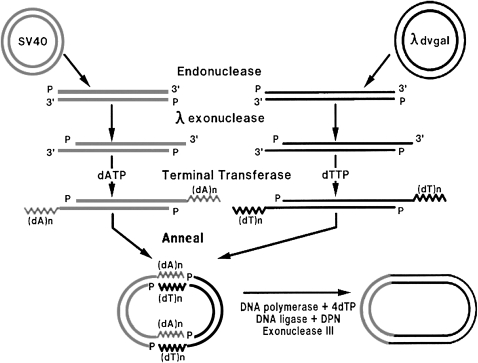
Both SV40 and λdvgal 120 exist naturally as circular DNA molecules. Thus, as a first step, methods were needed to cleave each of them once to produce full-length linear molecules. This task was achieved in two ways. One procedure relied on the fact that circular DNAs can be cleaved to linear molecules by incubation with pancreatic DNase I in the presence of the divalent cation Mn2+, a condition that limits the reaction to one or two double-stranded cleavages per molecule (Melgar and Goldthwait 1968). The second procedure grew out of the seminal findings of Kelly and Smith (1970) and Danna and Nathans (1971) that some restriction endonucleases can be used to quantitatively cleave DNAs at unique sites. By testing several DNA restriction enzymes, John Morrow8 found one, EcoRI endonuclease, an enzyme from E. coli discovered by Herbert Boyer,9 that cleaved both SV40 (Morrow and Berg 1972) and λdvgal 120 (D. Berg et al. 1974) DNA once at unique sites. The latter method was chosen for our studies because it generated much higher yields of linear DNAs that were both unit length and devoid of single-strand nicks.
On the basis of a finding by Lobban, Jackson and Symons pared back the 5′-ends of the duplex linear DNAs with a λ phage-encoded 5′-exonuclease to improve the TdT-catalyzed addition of nucleotides at the 3′-ends. Accordingly, they digested EcoRI-cleaved SV40 and λdvgal 120 DNAs with λ 5′-exonuclease to create 3′-extensions and then added 50–100 adenylate nucleotides to the 3′-ends of the SV40 DNA and 50–100 thymidylate nucleotides to the 3′-ends of the λdvgal 120 DNA (Figure 2). Specific annealing conditions led to the formation of noncovalently associated chimeric circular DNA molecules. Because the (dA)n and (dT)n tails had only approximately similar lengths, there were gaps at the (dA)n:(dT)n joints. These gaps were filled in using E. coli DNA polymerase I in the presence of the four deoxynucleoside triphosphates and exonuclease III, and the joints were covalently sealed using E. coli DNA ligase I. Exo III was included in the final reaction mixture because Lobban had found that the enzyme's presence greatly increased his yield of covalently closed circular P22 dimers. Jackson proved he had succeeded in constructing covalently closed SV40-λdvgal 120 chimeric DNA molecules in vitro by separating them from the unreacted linear DNAs by CsCl-ethidium bromide equilibrium centrifugation and documenting their existence and size by electron microscopy (Jackson et al. 1972).
Figure 2.—
Method used by Jackson et al. (1972) for constructing SV40-λdvgal 120 recombinant DNA in vitro.
Thus, by the spring of 1972, the first chimeric recombinant DNA had been produced by sequentially using six enzymes with previously known properties: EcoRI endonuclease provided by Boyer and the others provided by colleagues in the Stanford Biochemistry Department. Undoubtedly, the ready availability of all of the above-mentioned enzymes and the expertise in their use was a very important contributor to the venture's success. Noteworthy is the fact that none of the individual procedures, manipulations, and reagents used to construct this recombinant DNA was novel; the novelty lay in the specific way in which they were used in combination. The procedure outlined above worked well with two relatively pure DNAs. However, the complexity of the products is problematic with mixtures of DNAs. Indeed, when David Hogness10 and his colleagues used the (dA)n:(dT)n joining procedure to recombine random-sized fragments of Drosophila DNA with a bacterial plasmid, they ended up with a complex mixture of inseparable recombinants (Wensink et al. 1974). To overcome that problem, a method was needed to enrich or, preferably, completely separate recombinants one from another.
The plan to construct SV40-λdvgal 120 recombinant DNAs and to propagate them in E. coli became public in July, 1971, while Mertz was taking a course on animal cells and viruses at the Cold Spring Harbor Laboratory. Upon hearing her description of this project, Robert Pollack, the course's instructor, expressed concern about it. His anxiety, soon repeated by others, centered on the facts that: (i) SV40 can promote oncogenic transformation of human cells in culture and produce tumors in rodents; and (ii) E. coli, the presumptive carrier of the recombinant plasmid, is a natural inhabitant of the human intestinal tract. Most of the scenarios imagined the inadvertent or intentional release of E. coli carrying the SV40 DNA, with the attendant potential to spread a cancer-causing gene within the human population. Our initial reaction was that those fears were overblown and that procedures could be designed to mitigate against those risks. While some experienced tumor virologists and bacteriologists were also dismissive of the fears of the potential hazards, others thought the likelihood of something amiss happening were quite small, but not absolutely zero. Although there was little reason to believe that the SV40-λdvgal 120 recombinant DNA itself posed a risk to human health, we, nevertheless, agreed after considerable hesitation to defer the introduction of this chimeric DNA into E. coli until better assessments regarding its safety were developed.
Prompted by concerns relating to the possible oncogenic potential of SV40 in humans, Berg and other prominent scientists convened a meeting to assess the risks of working with tumor viruses and recombinant DNAs that contain them. That meeting, sponsored by the National Institutes of Health and the National Science Foundation, was held in January, 1973 at the Asilomar Conference Center in Pacific Grove, California. Although no well-documented problems arising from working with these agents were uncovered, several recommendations were made for scientists working with them (Hellman et al. 1973). These recommendations included to periodically monitor researchers who work with tumor viruses for infection, to prohibit pipetting by mouth, and to use laminar flow hoods during all manipulations involving potentially infectious material.
Shortly thereafter, another important breakthrough occurred. In the spring of 1972, Mertz discovered an unexpected property of the EcoRI endonuclease. She had repeatedly observed that EcoRI-cleaved linear SV40 DNA is approximately one-tenth as infectious as circular SV40 DNA in monkey cells; the recovered replicated viral DNA is circular and contains an intact EcoRI site. Although Kelly and Smith (1970) had shown that the restriction endonuclease they had characterized from Haemophilus influenza cleaves DNA leaving blunt ends, Mertz hypothesized that EcoRI-cut SV40 DNA contained cohesive ends and that it could form circles by annealing of these ends in the same way that linear phage λ DNA forms circles. Using electron microscopy, she showed that incubation of EcoRI-cut linear SV40 DNA with E. coli DNA ligase I at 15° results in the efficient reformation of covalently closed circular DNA molecules. Then, working in collaboration with Ronald Davis,11 Mertz determined that, although less than 1% of EcoRI-cut SV40 DNA molecules are circular when spread in 50% formamide at room temperature, more than half of them are circular when incubated and spread at 3°. Thus, the ends created by cleavage with EcoRI endonuclease are cohesive. The Tm for the circular-to-linear molecule transition is 6°. Mertz and Davis (1972) also found that at least 18 of the 19 fragments of various lengths produced by EcoRI cleavage of an ∼74-kbp plasmid, F8 (P17), can form intramolecular circles when incubated and spread for electron microscopy at 3°. Thus, they concluded that all ends created by EcoRI cleavage are probably identical, cohesive, and can be joined together with DNA ligase.
To demonstrate directly that the cohesive ends created by EcoRI cleavage could be used to create chimeric DNAs, they also incubated EcoRI-cleaved SV40 DNA and EcoRI-cleaved λdvgal 120 DNA together in equimolar amounts at high DNA concentration with E. coli DNA ligase I at 15°. While the linear DNAs ligated separately had distinctive buoyant densities in CsCl, most of the molecules produced when the two DNAs were ligated in the same reaction mixture had an intermediate buoyant density. Taken together, these experiments definitively established that any two DNA molecules whose ends are created by cleavage with EcoRI endonuclease can be readily joined together by ligation in vitro. Electron microscopic analysis of the lengths of these chimeric DNA molecules indicated that most consisted of circular DNAs containing a mixture of three or more copies of the input DNAs. Thus, the products of this reaction probably included some containing two or more tandem copies of λdvgal 120 DNA covalently linked to one or more copies of SV40 DNA. These chimeric molecules would have been able to replicate in E. coli. However, that supposition was not tested because of our self-imposed moratorium on producing E. coli containing SV40 oncogenes.
Boyer was promptly informed about the discovery that cleavage of DNA with EcoRI endonuclease generates cohesive ends. Together with Joe Hedgpeth12 and Howard Goodman,13 Boyer used this knowledge to determine that the nucleotide sequence of the 5′-extensions generated by cleavage with EcoRI endonuclease is 5′-AATT-3′ (Hedgpeth et al. 1972). This finding agreed well with the Mertz and Davis (1972) estimate of 4 or 6 bases obtained by measuring the Tm for annealing of the ends.
Cloning in bacteria:
Prior to 1972, Stanley Cohen14 had been studying the structure and replication of DNA plasmids such as pSC101 that bear antibiotic resistance genes in bacteria. Aware of the not-yet-published findings of Mertz and Davis (1972) and D. Berg et al. (1974), Cohen realized that these techniques could be quite helpful for his research. In collaboration with Annie Chang15 and Leslie Hsu,15 Cohen showed that EcoRI endonuclease-cleaved pSC101 DNA can be taken up by E. coli where it recircularizes and replicates as an autonomously replicating plasmid (Cohen et al. 1972). Next, Cohen, Chang, Boyer, and Robert Helling16 (1973) relied on the cohesive property of EcoRI endonuclease-generated ends to recombine pSC101 with a segment of DNA from an E. coli plasmid that contained a different antibiotic resistance gene; the new plasmid could be propagated in E. coli where it expressed both antibiotic resistance properties. Chang and Cohen (1974) then constructed a wholly novel interspecies recombinant plasmid by joining together pSC101 and a plasmid DNA originating from the gram-positive bacterium, Staphylococcus aureus. This chimeric plasmid propagated efficiently in gram-negative E. coli, exhibiting the unique antibiotic resistance characteristics of both parental plasmids. Thus, Cohen and his collaborators demonstrated that novel recombinant DNAs created in vitro, including even interspecies ones, can be cloned, propagated, and expressed in E. coli.
The finding that DNAs of different microbial origins can be propagated in E. coli still left unanswered the provocative, key question of whether eukaryotic or, for that matter, any DNA can be cloned in a bacterial host. John Morrow, who was finishing his Ph.D. thesis research in 1973 in Berg's laboratory and was aware of the Mertz and Davis (1972) and Cohen et al. (1972, 1973) discoveries, undertook to answer that question. Knowing about the concerns of introducing potentially biohazardous genes into bacteria, Morrow proposed to Boyer at the June 1973 Gordon Conference on Nucleic Acids that they attempt to propagate Xenopus laevis ribosomal DNA in E. coli. Morrow had already determined that a sample of purified X. laevis ribosomal DNA obtained from Donald Brown, Morrow's prospective postdoctoral mentor, was cleaved by EcoRI endonuclease. With Cohen joining the collaborative effort, pSC101 was chosen as the cloning vector because it contained a readily selectable marker. After ligating the mixture of EcoRI-cleaved pSC101 and X. laevis ribosomal DNAs, they selected and characterized clones expressing the pSC101-encoded antibiotic resistance gene. The outcome was quite clear: ∼20% of the bacterial clones containing pSC101 DNA also contained 18S or 28S X. laevis ribosomal DNA (Morrow et al. 1974). In some instances, RNA complementary to the X. laevis ribosomal DNA could be detected in the cells containing the chimeric plasmid DNAs, although these RNAs probably arose from transcripts initiated within pSC101 sequences. Thus, the Morrow et al. experiment demonstrated that genes from a eukaryotic organism can be cloned and replicated in E. coli.
The profound implication of this experiment was that DNA from any organism on the planet could probably be cloned and propagated in E. coli. This experiment also provided a prototype for many subsequent ones aimed at cloning specific genes. By 1976, Davis and his colleagues demonstrated functional expression of a protein-coding gene from yeast (Struhl 2008). Eventually, cloning served as the archetypical approach used to sequence entire genomes. It also paved the way toward creating E. coli containing recombinant plasmids in which genes encoding proteins or RNAs are linked to regulatory sequences, thereby enabling the expression of their products.
Patenting and start of biotechnology industry:
None of the members of the Berg, Kaiser, or Davis groups ever considered patenting the reagents or procedures that were used for recombining DNA in vitro. Neither had the scientists who discovered TdT, DNA polymerases, DNA ligases, exonucleases, and restriction enzymes ever sought patents for their efforts. Indeed, few, if any, of the discoveries, reagents, and methods that constitute the foundations of molecular biology were ever patented. While some academic institutions such as the University of Wisconsin–Madison had a long history of patenting inventions in the biological and biochemical sciences (e.g., vitamins, antibiotics), the sociology among most U. S. life scientists prior to the 1970s was to eschew patents, believing that they would restrict the free flow of information and reagents and impede the pace of discovery. However, that reticence disappeared in November, 1974 when Stanford University and the University of California at San Francisco jointly filed a United States patent application citing their respective faculty members, Stanley Cohen and Herbert Boyer, as the sole inventors of the recombinant DNA technology. Their claims to commercial ownership of the techniques for cloning all possible DNAs, in all possible vectors, joined in all possible ways, in all possible organisms were dubious, presumptuous, and hubristic. Nevertheless, these claims, only slightly modified, were eventually approved in 1980 by the U. S. Patent Office (Cohen and Boyer 1980). By employing what proved to be very wise terms regarding licensing and royalties, the two universities collectively garnered nearly $300 million in revenues during the life of this and two other related patents. Following university practices, Cohen, Boyer, and their respective university departments each received shares of the income from the “Cohen-Boyer patents,” while the institutions' shares were used to support universitywide research and education. In retrospect, Stanford's and UCSF's action set in motion an escalating cascade of patent claims by universities covering their faculties' respective discoveries that continues to this day. The emergence of the biotechnology industry followed naturally from the encouragement of academic scientists to patent their research discoveries and to explore their newly discovered entrepreneurial instincts. The early successes of Genentech, Biogen, and Amgen owe much to those encouragements. The events leading to the approval of the Cohen-Boyer patents and the founding of the biotechnology industry are described in detail by Hughes (2001) and Yi (2008).
Development of regulatory guidelines:
Boyer's presentation of the Cohen et al. (1973) experiments, resulting in the creation of plasmids with novel combinations of antibiotic resistance genes, triggered concerns about the safety of such recombinants among the participants attending the June 1973 Gordon Conference on Nucleic Acids (Singer and Söll 1973). In response to those concerns, the U. S. National Academy of Sciences (NAS) asked Berg to convene a committee of scientists who were familiar with and likely to use the new tools in their own research. That committee was asked to examine the scientific prospects and potential risks of what came to be known as recombinant DNA. Just before the committee met, news of the Morrow et al. (1974) experiment became known. Even though this experiment involved the cloning of a DNA segment generally accepted as being quite innocuous, its success was viewed as having “opened the door” to cloning DNAs from any biological source, including viruses, toxin-coding genes, and mammalian oncogenes. At the spring 1974 meeting of the NAS committee, the participants acknowledged that recombinant DNA technology had great promise for advancing basic and applied biology, but agreed there was insufficient information and data to determine the magnitude, if any, of the risks (P. Berg et al. 1974). In light of the uncertainty, the committee recommended that certain types of DNA cloning experiments be deferred until a conference of experts could be convened to assess the nature of the benefits and risks associated with such research.
The International Conference on Recombinant DNA was convened in February of 1975 at the Asilomar Conference Center in Pacific Grove, California. After considerable debate, the conference recommended that the moratorium on the previously deferred experiments be lifted and replaced with guidelines governing such research (Berg et al. 1975). In the summer of 1976, the National Institutes of Health issued its first set of Guidelines for Research Involving Recombinant DNA. These guidelines and analogous ones from other international jurisdictions along with their updates have been adhered to throughout the world. In the over three decades since adoption of these various regulations for conducting recombinant DNA research, many millions of experiments have been performed without reported incident. No documented hazard to public health has ever been attributable to the applications of recombinant DNA technology. Moreover, the concern that moving DNA among species would breach customary breeding barriers with profound effects on natural evolutionary processes has substantially diminished as research has revealed such exchanges occur in nature as well. Table 1 summarizes the chronology as we know it of the events described in this essay.
TABLE 1.
Chronology of main events relating to development of methods for constructing and cloning recombinant DNAs
| Yeara | Event |
|---|---|
| 1969–1970 | P. Berg (1970) and Lobban (1969) independently conceive ideas for generating recombinant DNAs in vitro and using them for cloning, propagating, and expressing genes across species. |
| 1971 | D. Berg et al. (1974) isolate the first plasmid bacterial cloning vector, λdvgal 120. |
| 1971 | Concern regarding potential biohazards of cloning first raised by Robert Pollack. |
| 1971–1972 | Jackson et al. (1972) and Lobban and Kaiser (Lobban 1972; Lobban and Kaiser 1973) concurrently and collaboratively develop the terminal transferase tailing method for joining together DNAs in vitro. |
| 1972 | Jackson et al. (1972) create first chimeric DNA in vitro. |
| 1972 | Mertz and Davis (1972) discover that cleavage with EcoRI generates cohesive ends. They use EcoRI plus DNA ligase to generate SV40-λdvgal 120 chimeric DNAs in vitro. |
| 1972–1973 | Cohen et al. (1972) isolate the drug-selectable bacterial cloning vector, pSC101. They use it to construct, clone, and express bacterial intra- (1973) and interspecies (1974) recombinant DNAs. |
| 1973 | Morrow et al. (1974) clone and propagate ribosomal DNA genes from a eukaryote in E. coli. |
| 1973–1976 | Renewed concerns regarding potential biohazards of cloning recombinant DNAs (Singer and Söll 1973; P. Berg et al. 1974, 1975) lead to NIH Guidelines. |
| 1974–1975 | Filing of initial Stanford University/University of California, San Francisco (UCSF) (Cohen/Boyer) patent applications relating to recombinant DNA. |
| 1976 | Boyer and Robert Swanson cofound Genentech, the first biotechnology company. |
| 1980 |
Stanford/UCSF (Cohen/Boyer) patent issued by U. S. Patent Office. |
Year(s) in which event occurred.
Impacts of recombinant DNA technology:
The most far-reaching consequence of the emergence of the recombinant DNA technology has been the great strides made in understanding fundamental life processes and the ability to investigate problems that had previously been unapproachable. Emerging from myriad investigations has been the appreciation that nothing in the man-made world rivals the complexity and diversity of this earth's organisms. No man-made information system invented to date comes anywhere close to containing the amount of information encoded in their genomes or encompassing the complexity of the intricate machinery for their functioning. We have learned enough to reveal how much we do not know and to acknowledge that nature's secrets are not beyond our capabilities of discovery.
The advances made possible by recombinant DNA technology have profound implications for the future of medicine for they have placed us at the threshold of new methods of diagnosis, prevention, and treatment of numerous human diseases. Hormones, vaccines, therapeutic agents, and diagnostic tools developed using recombinant DNA methods are already greatly enhancing medical practice. Although the production and consumption of genetically engineered food are realities, the benefits have yet to be fully realized. Nevertheless, recombinant DNA technologies will, undoubtedly, play roles in the future in increasing the supply of both food and energy needed by the world's growing human population.
Acknowledgments
We thank Douglas Berg, William Dove, David Jackson, A. Dale Kaiser, Peter Lobban, John Morrow, Maxine Singer, and Adam Wilkins for their suggestions for improving this article and Peter Lobban for permission to reproduce Figure 1. Much of the work described here was funded in large part by grants to Paul Berg from the National Institutes of Health and the American Cancer Society.
This article is dedicated to Arthur Kornberg, who fostered a group of colleagues that made this work possible.
David Jackson was a postdoctoral fellow in P. Berg's laboratory.
Robert Symons was a visiting professor in P. Berg's laboratory.
Peter Lobban was a graduate student in A. D. Kaiser's laboratory in the Biochemistry Department at Stanford University.
Douglas Berg was a postdoctoral fellow in A. D. Kaiser's laboratory.
John Morrow was a graduate student in P. Berg's laboratory.
Herbert Boyer was an associate professor in the Department of Microbiology at University of California, San Francisco (UCSF).
David Hogness was a professor in the Biochemistry Department at Stanford University.
Ronald Davis was an assistant professor in the Biochemistry Department at Stanford University.
Joe Hedgpeth was a postdoctoral fellow in Boyer's laboratory.
Howard Goodman was an associate professor in the Department of Biochemistry and Biophysics at UCSF.
Stanley Cohen was an assistant professor in the Department of Medicine at Stanford University.
Annie Chang and Leslie Hsu were technician and graduate student, respectively, in Cohen's laboratory.
Robert Helling was a postdoctoral fellow in Boyer's laboratory.
References
- Berg, D. E., D. A. Jackson and J. E. Mertz, 1974. Isolation of a λdv plasmid carrying the bacterial gal operon. J. Virol. 14 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, P., 1970. Viral oncogenesis and other problems of regulation. American Cancer Society grant application, submitted November, 1970.
- Berg, P., D. Baltimore, H.W. Boyer, S.N. Cohen, R.W. Davis et al., 1974. Letter: potential biohazards of recombinant DNA molecules. Science 185 303. [PubMed] [Google Scholar]
- Berg, P., D. Baltimore, S. Brenner, R. O. Roblin and M. F. Singer, 1975. Summary statement of the Asilomar conference on recombinant DNA molecules. Proc. Natl. Acad. Sci. USA 72 1981–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, A., 2007. Phage integration and chromosome structure. A personal history. Annu. Rev. Genet. 41 1–11. [DOI] [PubMed] [Google Scholar]
- Chang, A. C., and S. N. Cohen, 1974. Genome construction between bacterial species in vitro: replication and expression of Staphylococcus plasmid genes in Escherichia coli. Proc. Natl. Acad. Sci. USA 71 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. N., and H. W. Boyer, 1980. Process for Producing Biologically Functional Molecular Chimeras, U. S. Patent 4,237,224, initially filed November 4, 1974, issued December 2, 1980.
- Cohen, S. N., A. C. Chang and L. Hsu, 1972. Non-chromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69 2110–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. N., A. C. Chang, H. W. Boyer and R. B. Helling, 1973. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 70 3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna, K., and D. Nathans, 1971. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenza. Proc. Natl. Acad. Sci. USA 68 2913–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, F., 2003. Clockwork science. The New York Review of Books 50(17): 6 (http://www.nybooks.com/articles/16739). [Google Scholar]
- Galison, P., 2003. Einstein's Clocks, Poincaré's Maps: Empires of Time. W. W. Norton, New York.
- Gellert, M., J. W. Little, C. K. Oshinsky and S. B. Zimmerman, 1968. Joining of DNA strands by DNA ligase of E. coli. Cold Spring Harbor Symp. Quant. Biol. 33 21–26. [DOI] [PubMed] [Google Scholar]
- Hedgpeth, J., H. M. Goodman and H. W. Boyer, 1972. DNA nucleotide sequence restricted by the RI endonuclease. Proc. Natl. Acad. Sci. USA 69 3448–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman, A., M. N. Oxman and R. Pollack (Editors), 1973. Biohazards in Biological Research. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hershey, A. D., E. Burgi and L. Ingraham, 1963. Cohesion of DNA molecules isolated from phage lambda. Proc. Natl. Acad. Sci. USA 49 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, S, 2001. Making dollars out of DNA: the first major patent in biotechnology and the commercialization of molecular biology, 1974–1980. Isis 92 541–575 (http://www.jstor.org/stable/3080733). [DOI] [PubMed] [Google Scholar]
- Jackson, D. A., R. H. Symons and P. Berg, 1972. Biochemical method for inserting new genetic information into DNA of simian virus 40: circular DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc. Natl. Acad. Sci. USA 69 2904–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R. H., R. J. Wodzinski and M. H. Rogoff, 1971. Enzymatic addition of cohesive ends to T7 DNA. Biochem. Biophys. Res. Comm. 43 384–392. [DOI] [PubMed] [Google Scholar]
- Kato, K.-I., J. M. Goncalves, G. E. Houts and F. J. Bollum, 1967. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J. Biol. Chem. 242 2780–2789. [PubMed] [Google Scholar]
- Kelly, Jr., T. J., and H. O. Smith, 1970. A restriction enzyme from Hemophilus influenzae. II. J. Mol. Biol. 51 393–409. [DOI] [PubMed] [Google Scholar]
- Kuhn, T., 1962. The Structure of Scientific Revolutions. University of Chicago Press, Chicago.
- Lobban, P. E., 1969. The generation of transducing phage in vitro. Essay for third Ph.D. examination, Stanford University, November 6, 1969.
- Lobban, P. E., 1972. An Enzymatic Method for End-to-End Joining of DNA Molecules. Ph.D. Thesis, Stanford University, Stanford, CA.
- Lobban, P. E., and A. D. Kaiser, 1973. Enzymatic end-to-end joining of DNA molecules. J. Mol. Biol. 78 453–471. [DOI] [PubMed] [Google Scholar]
- Mandel, M., and A. Higa, 1970. Calcium dependent bacteriophage DNA infection. J. Mol. Biol. 53 159–162. [DOI] [PubMed] [Google Scholar]
- Melgar, E., and D. A. Goldthwait, 1968. Deoxyribonucleic acid nucleases. II. The effects of metals on the mechanism of action of deoxyribonuclease I. J. Biol. Chem. 243 4409–4416. [PubMed] [Google Scholar]
- Mertz, J. E., and R. W. Davis, 1972. Cleavage of DNA by RI restriction endonuclease generates cohesive ends. Proc. Natl. Acad. Sci. USA 69 3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, J. F., and P. Berg, 1972. Cleavage of simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc. Natl. Acad. Sci. USA 69 3365–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, J. F., S. N. Cohen, A. C. Chang, H. W. Boyer, H. M. Goodman et al., 1974. Replication and transcription of eukaryotic DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 71 1743–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health, 1976. Guidelines for Research Involving Recombinant DNA. Bethesda, MD. (http://oba.od.nih.gov/rdna/nih_guidelines_oba.html).
- Singer, M., and D. Söll, 1973. Guidelines for DNA hybrid molecules. Science 181 1114. [DOI] [PubMed] [Google Scholar]
- Stahl, F. W., 1998. Hershey. Genetics 149 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, K., 2008. The hisB463 mutation and expression of a eukaryotic protein in Escherichia coli. Genetics 180 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink, P. C., D. F. Finnegan, J. E. Donelson and D. S. Hogness, 1974. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell 3 315–325. [DOI] [PubMed] [Google Scholar]
- Wu, R., and A. D. Kaiser, 1968. Structure and base sequence in the cohesive ends of phage lambda DNA. J. Mol. Biol. 35 523–537. [DOI] [PubMed] [Google Scholar]
- Yi, D., 2008. Cancer, viruses, and mass migration: Paul Berg's venture into eukaryotic biology and the advent of recombinant DNA research and technology, 1967–1980. J. Hist. Biol. 41 589–636. [DOI] [PubMed] [Google Scholar]
- Zinder, N. D., 1992. Forty years ago: the discovery of bacterial transduction. Genetics 132 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]