Abstract
Agrobacterium tumefaciens uses a type IV secretion system to deliver a nucleoprotein complex and effector proteins directly into plant cells. The single-stranded DNA-binding protein VirE2, the F-box protein VirF and VirE3 are delivered into host cells via this VirB/D4 encoded translocation system. VirE1 functions as a chaperone of VirE2 by regulating its efficient translation and preventing VirE2-VirE2 aggregation in the bacterial cell. We analyzed whether the VirE1 chaperone is also essential for transport recognition of VirE2 by the VirB/D4 encoded type IV secretion system. In addition, we assayed whether translocation of VirF and VirE3, which also forms part of the virE operon, is affected by the absence of VirE1. We employed the earlier developed CRAFT (Cre recombinase Reporter Assay For Translocation) assay to detect transfer of Cre::Vir fusion proteins from A. tumefaciens into plants, monitored by stable reconstitution of a kanamycin resistance marker, and into yeast, screened by loss of the URA3 gene. We show that the C-terminal 50 amino acids of VirE2 and VirE3 are sufficient to mediate Cre translocation into host cells, confirming earlier indications of a C-terminal transport signal. This transfer was independent of the presence or absence of VirE1. Besides, the translocation efficiency of VirF is not altered in a virE1 mutant. The results unambiguously show that the VirE1 chaperone is not essential for the recognition of the VirE2 transport signal by the transport system and the subsequent translocation across the bacterial envelope into host cells.
Agrobacterium tumefaciens causes crown gall disease on a wide range of plants by genetic transformation of host cells with a piece of its oncogenic plasmid-borne transfer (T)-DNA (for most recent review, see Gelvin, 2003). The expression of the genes located on the T-DNA, which contain plant transcription and translation signals, results in overproduction of the plant hormones auxin and cytokinin and hence increased cell division and the tumor phenotype. A set of accessory virulence or effector proteins (encoded by the vir region on the tumor inducing [Ti] plasmid) is also transported into the transformed cells to confer full virulence. The host range is not confined to the plant kingdom, because A. tumefaciens can also transform yeast (Bundock et al., 1995), fungi (de Groot et al., 1998), and mammalian cells (Kunik et al., 2000).
The translocated proteins of A. tumefaciens described to date include VirD2, VirE2, VirE3, and VirF. In the bacterium, VirD2 introduces a nick at the border sequences surrounding the T-DNA and by replacement synthesis a single-stranded (ss) DNA copy of the bottom strand is released. VirD2 acts as a pilot protein as it remains covalently attached to the 5′ end of the T-strand during transport of the T-DNA into the host cell nucleus (Ward and Barnes, 1988). The ssDNA-binding protein VirE2 binds cooperatively to the T-strand and thereby protects it from degradation in the host cell (Rossi et al., 1996). Both VirD2 and VirE2 contain nuclear localization signals that facilitate transport of the T-complex into the nucleus (Zupan et al., 1996; Citovsky et al., 1997; Ziemienowicz et al., 2001). VirF and VirE3 are only essential for full tumor formation on a subset of host plants, such as Nicotiana glauca and tomato (Lycopersicon esculentum; Melchers et al., 1990; Schrammeijer, 2001). VirF interacts via its N-terminally located F-box domain with plant orthologs of the yeast SKP1 protein (Schrammeijer et al., 2001), which form part of a class of E3 ubiquitin ligases and are involved in the proteolytic degradation of target proteins by the proteasome. F-box proteins determine the specificity of the enzyme complex by interacting with target proteins. However, the identity of the target proteins of VirF remain yet undetermined. A virE3 mutation aggravates the attenuated virulence of a virF mutation; however, its role in the infection process is unclear.
A. tumefaciens uses a type IV secretion system (TFSS) to transport T-DNA/VirD2 and effector proteins into host cells. TFSSs are widely distributed among gram-negative pathogenic bacteria of human and animal cells, such as Brucella spp., Bartonella spp., Helicobacter pylori, and Legionella pneumophila. These bacteria can subvert specific host cells, such as macrophages, and modulate the hosts defense response by hijacking signal transduction cascades, vesicle traffic, and cytoskeletal functions, to be able to multiply and trigger a pathogenic response (Boschiroli et al., 2002; Nagai et al., 2002; Dehio, 2003). Effector proteins involved in the infection process have so far been identified in H. pylori (CagA), L. pneumophila (DotA, RalF, LidA), and Bordetella pertussis (pertussis toxin) (Conover et al., 2003; for review, see Nagai and Roy, 2003). The TFSS is a specialized protein complex, in A. tumefaciens assembled from the 11 VirB and VirD4 subunits, spanning the inner and outer membrane of the bacterium (Christie and Vogel, 2000; Christie, 2001; Baron et al., 2002). It is assumed that this protein complex translocates substrates after attachment and establishing contact with the host cell via the extended pilus structure. Although the structure of the complex is becoming clearer, it is not known how the effector proteins and the nucleoprotein complex are recognized by and transported through the TFSS.
The family of TFSS also contains evolutionary related members involved in conjugative DNA transfer of broad host range plasmids such as RP4 and R388. A. tumefaciens T-DNA transfer to plants by the VirB/D4 system shows similarity to the process of bacterial DNA conjugation, because the VirB/D4 pore can also mobilize the incQ plasmid RSF1010 into recipient bacterial (Beijersbergen et al., 1992) and plants cells (Buchanon-Wollaston et al., 1987). Plasmid conjugation systems are composed of two components; the mating pair formation system (Mpf) for contact between donor and recipient cell, and the DNA transfer and replication system (Dtr) or relaxosome. In conjugation, as in T-DNA transfer, a specific endonuclease nicks the plasmid at specific recognition sequences, the origin of transfer (oriT), in a so-called relaxosome complex. A coupling protein such as the TraG protein of plasmid RP4 and VirD4 of the A. tumefaciens virulence system is thought to form the interface between the relaxosome and the transport complex (Pansegrau and Lanka, 1996; Cabezón et al., 1997; Hamilton et al., 2000; Szpirer et al., 2000; Schröder et al., 2002). Structural analysis of the coupling protein TrwB of plasmid R388 showed that it is indeed a basic integral inner membrane protein with a hexameric ring structure (Gomis-Rüth et al., 2001, 2002).
Originally TFSSs were delineated as DNA transfer systems. However, monomeric proteins in addition are substrates for translocation through TFSSs. The A. tumefaciens virulence proteins VirE2, VirE3, and VirF can still be translocated into host cells in the absence of T-DNA (Vergunst et al., 2000; Schrammeijer et al., 2003), showing that these proteins do not travel as part of the DNA complex. The finding that virulence protein translocation has the same requirement of the VirD4 coupling protein and an intact VirB pore as the T-DNA suggest that the T-DNA is transported by virtue of the VirD2 protein that is covalently attached to the 5′ end, and that TFSSs are actually dedicated protein translocation systems.
An interesting issue concerns the state of the effector proteins during translocation. Twin arginine translocation systems mediate transfer of folded proteins (Berks et al., 2000), whereas Sec-dependent (Mori and Ito, 2001) and type III secretion systems (TTSS), for instance, translocate proteins in an (partly) unfolded state (Delahay and Frankel, 2002). The prototype of the TTSS family of transporters is the Yersinia Ysc (Yop secretion) system and defines another specialized protein translocation pathway among plant and mammalian pathogenic Gram-negative bacteria. These systems also directly inject effector proteins into host cells, using a N-terminal secretion signal in the effector proteins. Translocation of some effectors requires the help of chaperone proteins. Different roles have thus far been assigned to chaperones involved in secretion of bacterial TTSS proteins (for review, see Page and Parsot, 2002; Feldman and Cornelis, 2003). They have been implied to act as (a) “bodyguards” to prevent premature interaction of their binding partner with other proteins or among themselves, preventing aggregation and degradation of effector proteins, (b) maintaining their substrates in a secretion competent state, (c) playing a role in the secretion of their cognate substrate (Birtalan et al., 2002), and (d) playing a role in the hierarchy of secretion of effectors (Boyd et al., 2000).
The VirE1 protein of A. tumefaciens has the characteristics reminiscent of a chaperone molecule. It has been shown that VirE1 plays a prominent role in the export process of VirE2 by preventing aggregation, enhancing stability, and therefore likely maintaining VirE2 in a (unfolded) transport competent state (Deng et al., 1999; Sundberg and Ream, 1999; Zhao et al., 2001). Dumas et al. (2001) suggested that VirE2 might form a transmembrane channel in the plant cytoplasmic membrane to facilitate transport of ssDNA. Another role of the VirE1 chaperone might thus be to prevent VirE2 from interacting with membranes in the bacterial cell.
We were interested to resolve the question of whether VirE1 plays an unambiguous role also in the VirE2 substrate recognition by the translocation machinery. Therefore, we analyzed transport of the A. tumefaciens effector protein VirE2 in the presence or absence of its chaperone VirE1. We made use of the Cre recombinase as a reporter protein, attached to the N terminus of the effector protein (or deletions thereof) and expressed those in A. tumefaciens, followed by monitoring of Cre activity in host cells as a permanent selectable change in the host genome.
First, we delineated the transport signal in VirE2 and VirE3, because it was suggested before that this may be present in the carboxy terminus (Vergunst et al., 2000; Simone et al., 2001), as experimentally shown for VirF. Our data show that a transport signal is present in the C-terminal 50 amino acids of VirE2 and VirE3, because these peptides were sufficient to mediate translocation of the Cre reporter protein into Arabidopsis and Saccharomyces cerevisiae. Transport is detected with similar efficiency in the presence or absence of VirE1. We show for the first time that VirE1, which is essential for preventing VirE2 from premature protein interactions and thus indirectly for export of VirE2, is not essential for the recognition of the translocation signal of VirE2 by the transport machinery and the subsequent translocation of VirE2 into host cells.
RESULTS
Experimental Strategy
A. tumefaciens is used worldwide as a very efficient tool for the introduction of desired genes into plants. In addition, A. tumefaciens translocates the effector proteins VirE2, VirE3, and VirF from A. tumefaciens into host cells. Translocation was detected by using the Cre reporter assay for translocation (CRAfT) in which the Cre recombinase protein is fused to transport signals of these effector proteins (Vergunst et al., 2000; Schrammeijer et al., 2003). Detection of the fusion proteins in host cells was accomplished by using a transgenic Arabidopsis line (3043; Vergunst et al., 2000) harboring a reporter-detection construct inserted in the genome, in which Cre-mediated loss of a lox-flanked bialaphos resistance (bar) gene resulted in reconstitution of a kanamycin resistance marker.
Deletion studies of VirF have shown that the C-terminal 37 amino acids are sufficient for efficient translocation by the VirB/D4 transport system. Data from Vergunst et al. (2000) and Simone et al. (2001) suggested that also in VirE2 the transport domain is present in the C terminus of the protein. An RPR motif, which could be part of the transport signal, is evident in the outermost C-terminal regions of VirF, VirE2, and VirE3. To demarcate the translocation signals also in VirE2 and VirE3, Arabidopsis line 3043 was used to examine whether the Cre protein would be mobilized after fusion to a C-terminal portion of the proteins. Next, we studied the role of the chaperone VirE1 in translocation of its cognate substrate VirE2. To corroborate the data in another system, we analyzed protein transport in an analogous CRAfT assay in S. cerevisiae, based on selection for excision of a lox-flanked URA3 gene on medium containing fluoro orotic acid (FOA).
Localization of a Transport Signal in the C-Terminal 50 Amino Acids of VirE2 and VirE3
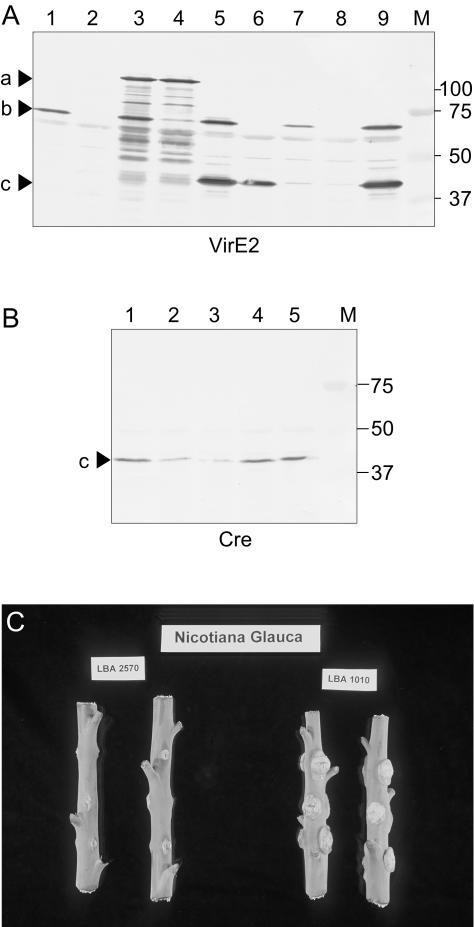
To narrow down the transport signal, we made in frame fusions between the cre open reading frame and the 3′ region of the virE2 (pSDM3210) and virE3 (pSDM3211) coding sequences, resulting in Cre fusions with the C-terminal 50 amino acids of both effector proteins. The fusion genes were contained on a broad host range non-mobilizable plasmid, pRL662 (see Table I for strains and plasmids used in this study), and electroporated into LBA1100 (C58 with an octopine pTiB6 plasmid lacking T-DNA, occ genes and tra region, but containing a wild-type vir region [Beijersbergen et al., 1992]). The fusions were expressed from the virF promoter and contained an additional SV40 NLS, located at the N terminus of the fusion protein, to improve nuclear entry into the host cell. Expression of Cre::VirE2_50C as well as Cre::VirE3_50C fusion proteins was confirmed by immunoblot analysis using VirE2 and Cre antibodies for detection (Fig. 1, A and B).
Table I.
Strains and plasmids used
| Relevant Characteristics | Reference | |
|---|---|---|
| Strains | ||
| LBA1100 | C58 containing pTiB6Δ (T-DNA, occ, tra), Rifr, Spcr | Beijersbergen et al. (1992) |
| LBA1010 | C58 containing oncogenic pTiB6, Rifr | Koekman et al. (1982) |
| LBA2570 | Precise virE1 deletion in LBA1010, Rifr | This study |
| LBA2571 | Precise virE1 deletion in LBA1100, Rifr, Spcr | This study |
| Plasmids | ||
| pRL662 | Broad host range plasmid derived from pBBR1-MCS2, in which Kmr, mob and oriT were replaced by a Gmr marker. | Vergunst et al. (2000) |
| pSDM3147 | pvirE-cre in pRL662, Gmr | Vergunst et al. (2000) |
| Schrammeijer et al. (2003) | ||
| pSDM3129 | pvirE-virE1-cre::virE2 in pRL662, Gmr | Vergunst et al. (2000) |
| Schrammeijer et al. (2003) | ||
| pSDM3155 | pvirF-NLS::cre::virFΔ42N in pRL662, Gmr | Vergunst et al. (2000) |
| Schrammeijer et al. (2003) | ||
| pSDM3210 | pvirF-NLS::cre::virE2_50C in pRL662, Gmr | This study |
| pSDM3211 | pvirF-NLS::cre::VirE3_50C in pRL662, Gmr | This study |
| pSDM3507 | pvirF-NLS::cre::virE3 in pRL662, Gmr | Schrammeijer et al. (2003) |
Figure 1.
A and B, Western-blot detection. A, VirE2 antibodies: 1, LBA1100; 2, LBA2571; 3, LBA1100(Cre::VirE2); 4, LBA2571 (Cre::VirE2); 5, LBA1100(Cre::VirE2-50C); 6, LBA2571(Cre:: VirE2-50C); 7, LBA1100(Cre::VirE3-50C); 8, LBA2571(Cre:: VirE3-50C); 9, virD4(Cre::VirE2-50C). B, Cre antibodies: 1, LBA1100(Cre::VirE2-50C); 2, LBA2571(Cre::VirE2-50C); 3, LBA1100(Cre::VirE3-50C); 4, LBA2571(Cre::VirE3-50C); 5, virD4 (Cre::VirE2-50C). M, Molecular weight marker (kD). C, Tumor assay on N. glauca with LBA1010 and LBA2570 (virE1). Triangles in a, Cre::VirE2; in b, VirE2; in c, Cre::VirE2_50C.
Table II (top) summarizes the results of four independent transport assays with Arabidopsis line 3043. Three weeks after cocultivation, the number of kanamycin-resistant (Kmr) calli was calculated per number of root explants, defining the efficiency with which Cre::Vir transport is detected. Transport of the fusion proteins consisting of the 50 C-terminal amino acids of VirE2 and VirE3 fused to the C terminus of Cre was detected in this way. On average, two to four Kmr calli were found per 100 explants, although the overall efficiency varied between experiments. The negative control strain (LBA1100 [pSDM3147]), expressing the native Cre protein, yielded only a single Kmr callus in four experiments due to a very low incidence of homologous recombination at the 34-bp lox sites, as described earlier (Vergunst et al., 2000). As a positive control for transfer, we used the Cre::VirFΔ42N fusion protein, because it is the most efficiently transported protein fusion identified thus far. The transfer efficiency of the positive control was on average 80 calli per 100 root explants, which is slightly higher than data reported earlier (54 calli per 100 explants; Vergunst et al., 2000). Transfer of Cre::VirE2 yielded on average 18 calli per 100 explants, which was also higher than reported before (six calli per 100 explants). This increase in transport efficiency may have been due to the application of an additional 24 h of cocultivation of A. tumefaciens with root tissue in this study. The efficiency with which transport of Cre::VirE2_50C is seen, is about 5- to 13-fold lower than that of Cre:VirE2. This may be because the Cre::virE2_50C fusion adopts a conformation that is not optimal for interaction with the transport system compared with the full-length VirE2 protein. Transport of Cre, mediated by the VirE3 signal, seems equally efficient as transport mediated by the VirE2 signal.
Table II.
Cre::Vir protein translocation from A. tumefaciens into Arabidopsisa
| Total No. Kmr Calli/No. Root Explantsc
|
Transfer Efficiencyd
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strainb | Plasmidb | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 |
| No. kmr Calli/100 Expl | |||||||||
| LBA1100 | Cre | 1/500 | 0/500 | ND | 0/515 | 0.2 ± 0.3 | 0 | - | 0 |
| Cre::VirFΔ42N | 338/350 | 352/600 | 352/580 | 612/515 | 97 ± 3 | 58 ± 2 | 60 ± 4 | 120 ± 18 | |
| Cre::VirE2 | 87/475 | 88/460 | 22/530 | 157/515 | 18 ± 3 | 19 ± 6 | 4 ± 0.3 | 31 ± 2 | |
| Cre::VirE2-50C | ND | 20/480 | 2/600 | 10/280 | - | 4 ± 0.3 | 0.3 ± 0.5 | 4e | |
| Cre::VirE3-50C | ND | 21/540 | 13/580 | 124/525 | - | 4 ± 0.5 | 2 ± 1 | 24 ± 4 | |
| Cre::VirE3 | ND | ND | 0/500 | 10/800 | - | - | - | 1 ± 0.7 | |
| LBA2571 | Cre::VirFΔ42N | 200/345 | 388/620 | ND | 390/530 | 57 ± 16 | 63 ± 18 | - | 74 ± 9 |
| Cre::VirE2 | 42/420 | 42/565 | 14/450 | 157/480 | 10 ± 3 | 7 ± 1 | 3 ± 1 | 33 ± 5 | |
| Cre::VirE2-50C | Inf | 15/600 | 0/520 | 6/310 | - | 2 ± 1 | 0 | 2e | |
| Cre::VirE3-50C | 24/445 | 22/495 | 2/555 | 99/590 | 5 ± 0.9 | 4 ± 0.3 | 0.3 ± 0.01 | 17 ± 2 | |
a Arabidopsis C24, line 3043 contains a lox-substrate for detection of in planta Cre activity, detected as kanamycin resistance. A. tumefaciens was cocultivated with root explants for 3 d. Kmr calli were scored 3 weeks after cocultivation. In four independent experiments, two petri dishes per strain/plasmid combination were assayed.
b A. tumefaciens strain LBA1100 (wild-type vir) or virE1 mutant LBA2571 carrying a non-mobilizable plasmid expressing Cre alone, Cre::VirFΔ42N, Cre::VirE2, Cre::VirE2-50C, Cre::VirE3-50C, or Cre::VirE3, respectively.
c Total no. of calli and root explants per two petri dishes. ND, Not determined.
d Transport efficiency presented as no. of Kmr calli per 100 root explants (± sd).
e Nos. per single petri dish due to infection.
Evidence for transport of a Cre::VirE3 full length fusion protein (LBA1100 [pSDM3507]) into Arabidopsis, however, was only obtained in experiment 4 (Table II) in which overall the efficiencies of transfer of the different fusion proteins were higher than in the other three assays. We could not detect transfer of the Cre::VirE3 fusion protein in five additional independent transformation experiments (data not shown). This result contrasts observations in yeast where translocation of Cre::VirE3 is reproducibly detected (Schrammeijer et al., 2003; see below). This difference may be caused by an interaction of the full-length VirE3 protein with host proteins during its journey to the cell nucleus in plants, but not in yeast, diminishing its ability to initiate recombination at the lox sites in the plant cell nucleus.
With the exception described above, transport experiments to yeast gave similar results to those obtained with Arabidopsis (Table III, top). After cocultivation for 6 d, dilution series of the cocultivation mix were plated on medium containing FOA to select for colonies that had lost the URA3 gene due to recombination at the lox sites. Efficiency of transfer is indicated as number of colonies on FOA per number of surviving yeast colonies in the absence of selection. In yeast, the homologous recombination efficiency is high compared with plants resulting in some positive yeast colonies in the order of 10–6 per output yeast. This background is also seen in cocultivation with A. tumefaciens strains not expressing Cre (data not shown). Translocation of Cre::VirE2_50C and Cre::VirE3_50C into yeast was detected, which corroborates the data obtained in plants. Transfer was detected with about 10-fold lower efficiency than Cre::VirE2, which is 2 orders of magnitude above background (LBA1100[pSDM3147]). Cre::VirFΔ42 transfer was 3 orders of magnitude above background levels, as shown before. As shown earlier, Cre::VirE3 transport into yeast occurred at detectable levels. In addition we tested transfer of Cre::VirE2_50C and Cre::VirE3_50C from a virD4 mutant strain (Fig. 1, A and B). We did not detect transfer (data not shown), indicating that the transport signal in these 50 C-terminal amino acids requires the VirB/VirD4 system for translocation.
Table III.
Cre:vir protein translocation from A. tumefaciens to S. cerevisiaea
| Frequency ura Excision per Output Yeastc
|
|||||
|---|---|---|---|---|---|
| Strainb | Plasmidb | 1 | 2 | 3 | 4 |
| LBA1100 | Cre | 3.2 × 10-6 | 2.9 × 10-6 | 2.0 × 10-5 | 8.5 × 10-6 |
| Cre::VirFΔ42N | 1.2 × 10-2 | 3.5 × 10-3 | 1.7 × 10-2 | 1.0 × 10-2 | |
| Cre::VirE2 | 5.6 × 10-4 | 7.6 × 10-5 | ND | 3.5 × 10-4 | |
| Cre::VirE2-50C | ND | 1.0 × 10-5 | 9.6 × 10-5 | 2.7 × 10-5 | |
| Cre::VirE3-50C | 2 × 10-4 | 3.2 × 10-5 | 2.0 × 10-4 | ND | |
| Cre::VirE3 | ND | 1.1 × 10-4 | 1.9 × 10-4 | ND | |
| LBA2571 | Cre::VirFΔ42N | 3.3 10-2 | ND | ND | 4.0 × 10-3 |
| Cre::VirE2 | 1.5 × 10-3 | ND | ND | 4.1 × 10-4 | |
| Cre::VirE2-50C | ND | ND | 1.2 × 10-4 | 8.8 × 10-5 | |
| Cre::VirE3-50C | ND | ND | 2.0 × 10-4 | ND | |
| Cre::VirE3 | ND | ND | 1.1 × 10-4 | ND | |
a The recipient yeast strain contains a floxed URA3 gene at chromosome V. A. tumefaciens and yeast were cocultivated for 6 d at 22°C and plated on medium containing 0.1% FOA.
b A. tumefaciens strain LBA1100 (wild-type vir) or virE1 mutant LBA2571 carrying a non-mobilizable plasmid expressing Cre alone, Cre::VirFΔ42N, Cre::VirE2, Cre::VirE2-50C, Cre::VirE3-50C, or Cre::VirE3, respectively.
c After cocultivation of A. tumefaciens with yeast, 100 μL of cells was plated on medium containing 0.1% FOA. In 4 independent experiments, the no. of Ura- yeast colonies was determined after 4 d at 30°C. Also, the output no. of yeast was determined. The Cre excision efficiency (frequency URA3 excision per output yeast) is determined by the no. of yeast colonies on medium containing FOA per no. of surviving yeast colonies (output yeast). ND, Not determined.
Summarizing, the 50 C-terminal amino acids of both VirE2 and VirE3 are sufficient for translocation into plant and yeast cells and thus must contain a translocation signal recognized by the VirB/D4 transport system.
Construction of an A. tumefaciens virE1 Deletion Strain
To analyze the effect of virE1 (coding for the VirE2 chaperone protein) on translocation of VirE2, we constructed a precise in frame deletion of virE1 by using the marker exchange-eviction mutagenesis method as described by Ried and Collmer (1987). This resulted in a double crossover event at the virE operon of the A. tumefaciens Ti-plasmid of both wild-type A. tumefaciens (LBA1010; Table I) and its disarmed derivative LBA1100. The virE1 deletion derivative of LBA1010 was named LBA2570, and the LBA1100-derived deletion, LBA2571.
It was previously shown that VirE1 is essential for tumor formation (McBride and Knauf, 1988; Sundberg et al., 1996) and for a VirE2 helper strain to complement tumor formation of a virE2 mutant strain in extracellular complementation experiments. We confirmed loss of virulence of LBA2570 in tumor assays on N. glauca plants (Fig. 1C). Besides, LBA2571 could not complement the virE2 transposon insertion mutant LBA1514 (Hooykaas et al., 1984) in extracellular complementation experiments (data not shown).
VirE2 Is Unstable in the Absence of VirE1 in Cis
The cre::virE2 fusion gene in pSDM3129 was cloned in its original context, expressed from the virE promoter in a transcriptional unit with virE1, based on earlier findings that VirE2 is unstable in the absence of VirE1 (Dombek and Ream, 1997; Deng et al., 1999). When we analyzed expression levels of VirE2 after vir gene induction in medium containing acetosyringone, we observed instability of VirE2 in the virE1 deletion mutant LBA2571 (Fig. 1A, LBA1100 versus LBA2571). Western-blot analysis of LBA1100(pSDM3129) and LBA2571(pSDM3129) also showed decreased levels of VirE2 in the absence of VirE1 in cis (Fig. 1A, lanes 3 and 4). In contrast, in the same strains, the Cre::VirE2 fusion protein (99 kD), cotranslationally expressed with VirE1 from pSDM3129, was detected in similar amounts. From these data, we can conclude that the VirE1 protein that is expressed from plasmid pSDM3129 is unable to stabilize VirE2, expressed from the virE operon (in the absence of VirE1 in cis) to wild-type levels. This result is in line with the findings of Zhou and Christie (1999) who found that expression of virE2 from a virE promoter sequence in the absence of coexpression in cis of the virE1 gene resulted in accumulation of low levels of VirE2.
Attempts to transform a derivative of plasmid pSDM3129, with a precise deletion of virE1, into LBA1100 or LBA2571 resulted in very low A. tumefaciens transformation efficiencies. A few resistant colonies obtained were analyzed by restriction analysis. These strains mainly carried recombined plasmid molecules (data not shown), possibly due to a toxic effect of aggregation of the Cre::VirE2 fusion protein in the absence of VirE1 in cis, resulting in selection of clones that had lost the correct coding sequence. Therefore, we could not analyze transport of Cre::VirE2 in the absence of VirE1. Instead, we used the Cre::VirE2_50C fusion protein in further studies. Presence of the Cre::VirE2_50C fusion protein is detected in LBA1100 (pSDM3210) and in LBA2571(pSDM3210) (Fig. 1, A and B), indicating that VirE1 is not needed in cis for its stability. Expression levels of the Cre fusion proteins with VirF and VirE3 were similar in the virE1 mutant background (LBA2571) and in the control LBA1100 (data not shown).
Translocation of VirE2 Does Not Require Its Chaperone VirE1
We performed transfer experiments with Arabidopsis line 3043 and A. tumefaciens strain LBA2571, carrying the plasmids listed in Table I. As shown in Table II (lower panel), the absence of VirE1 had no significant effect on the transport efficiency of the fusion proteins analyzed. Both the 50 C-terminal amino acids of VirE2 and VirE3, as well as Cre::VirFΔ42N, seemed as efficient in directing Cre transfer into plant cells from an A. tumefaciens strain carrying wild-type vir genes as from a virE1 deletion strain. The number of kanamycin-resistant calli may seem low, but is significantly above background. Besides, preliminary data (not shown) indicate that the use of a different detector plant line, based on induction of green fluorescent protein (GFP) expression rather than kanamycin resistance, results in higher efficiency of detection. The increase in sensitivity of the GFP assay may be due to the fact that in this assay, Cre activity can also be detected in cells that are not competent for regeneration, which is needed to select kanamycin-resistant calli.
The results show that VirE1 is not necessary to accomplish the interaction between the VirE2 transport domain with the TFSS and the subsequent translocation of its cognate substrate VirE2.
In yeast (Table III, bottom), comparable results were found to those obtained in plants. Transport of Cre::VirE2_50C was detected at similar efficiency from the virE1 mutant as from the wild-type control LBA1100. No differences in translocation efficiency of VirF and VirE3 were seen in the absence or presence of VirE1.
In conclusion, the transport signal of VirE2 is recognized by the VirB/D4 transport system in the absence of VirE1, and this suggests a chaperone-independent translocation for VirE2. Furthermore, the data show that VirE1 is not necessary for translocation of VirF and VirE3 into host cells.
DISCUSSION
The Role of Chaperones in Secretion
Molecular chaperones are involved in many biological processes by stabilizing their target proteins, maintaining their functional properties, or preventing premature interactions with other proteins. Also in secretion of bacterial proteins, chaperones play an important role. In TTSS, a specialized protein secretion pathway of Gram-negative bacteria, which directly injects effector proteins in host cells via a N-terminal secretion signal, the role of chaperones has been investigated in great detail (for review, see Page and Parsot, 2002; Feldman and Cornelis, 2003). Most chaperones identified associate with one effector protein. In the majority of cases, the genes encoding the chaperone are located directly upstream of the coding sequence of the substrate protein. A common feature of these chaperones is that they function as dimers. From crystallographic data of several chaperones and their substrates, including the Yersinia SycE/YopE, the enterohemorrhagic Escherichia coli CesT, and the Salmonella enterica SigD chaperone (Birtalan and Ghosh, 2001; Luo et al., 2001), it has become clear that these chaperones bind to a unique region of the substrate and that the effector proteins are not globally unfolded. This is different from structural data on Salmonella sp. SicP/SptP (Stebbins and Galan, 2001), which suggest that the chaperone is essential for maintaining the substrate in an unfolded translocation competent state. It has been difficult to study the role of chaperones in secretion of their cognate substrate, due to instability in the absence of the chaperone. YopH, YscM proteins, and YopN seem not to be secreted in the absence of their chaperone (Day and Plano, 1998; Cambronne et al., 2000), and thus a chaperone-dependent mechanism for secretion was proposed. For YopE, the first 15 amino acids were sufficient for secretion (but not translocation) of hybrid proteins in culture, whereas the first 50 amino acids (including both the transport signal as well as the SycE chaperone-binding site) were needed for translocation into the host cytosol (Woestyn et al., 1996) suggesting a specific role for chaperones in translocation. Boyd et al. (2000) showed, however, that the YopE transport signal was sufficient to transport a CyaA protein from a mutant Yersinia, lacking all other effectors, but not from wild-type Yersinia cells. This suggested that the chaperone and chaperone-binding site are not required for secretion or translocation itself, but pointed to a role in competition between the effectors for transport. Birtalan et al. (2002) recently presented biochemical and structural data, which suggested that the chaperone/substrate complex SycE/YopE functions as a general three-dimensional translocation signal that is recognized by the TTSS.
The A. tumefaciens TFSS system mediates translocation of a T-DNA nucleoprotein complex and effector proteins into host cells. Only one of these transported substrates, VirE2, is accompanied by a chaperone molecule called VirE1 (Deng et al., 1999; Sundberg and Ream, 1999; Zhao et al., 2001). VirE1 has the structural characteristics of a molecular chaperone (Wattiau et al., 1996), being a small, acidic protein with a dimerization domain. Besides, the gene encoding the chaperone is located in the same operon as the gene encoding its cognate substrate. VirE2 is unstable in the absence of VirE1 (Deng et al., 1999; Zhao et al., 2001; this study). VirE1 has been implicated in posttranslational stabilization of VirE2 and maintaining VirE2 in a translocation competent state, by preventing premature interaction of VirE2 with T-DNA in the bacterial cell, membrane insertion, and/or VirE2/VirE2 interaction (Deng et al., 1999; Dumas et al., 2001; Zhao et al., 2001). The precise role of VirE1 as a chaperone in the secretion process of VirE2 has not been investigated previously. Our results show that VirE1 is not necessary for recognition of its cognate substrate VirE2 by the VirB/D4 transport system.
Presence of a C-Terminal Transport Signal in VirE2 and VirE3
It was previously demonstrated that A. tumefaciens translocates at least three effector proteins into plant cells: VirE2, VirE3, and VirF. In this study, we showed that as with VirF, the 50 C-terminal amino acids of VirE2 and VirE3 were sufficient to mediate translocation of Cre recombinase into host cells. These data unambiguously show that the A. tumefaciens virulence system translocates proteins, which have a transport signal located in the C-terminal region. This finding is not unexpected because the results described by Vergunst et al. (2000) and Simone et al. (2001) have already pointed to the presence of a transport domain in the C terminus of VirE2. Our earlier observation that an RPR motif, present in the C-terminal 37 amino acids of VirF as well as in the C termini of VirE2 and VirE3, may form part of the transport signal was strengthened by the experiments described here. In addition, we did not detect transfer from a VirD4 mutant, indicating that the transport signal in these 50 C-terminal amino acids requires the VirB/VirD4 system for translocation.
The VirE1 Chaperone Is Not Essential for VirE2 Translocation
It was shown earlier that a Cre::VirE2 protein fusion is translocated from A. tumefaciens into Arabidopsis (Vergunst et al., 2000) and yeast cells (Schrammeijer et al., 2003). This fusion protein was expressed in a similar context as the native virE operon, which is driven by the virE promoter and cotranscribed with the virE1 gene. Likely, this construction with virE1 in cis was essential to express and stabilize fusion proteins between Cre and full-length VirE2, as found for native VirE2 (Fig. 1A; Deng et al., 1999). In fact, we were unable to obtain A. tumefaciens strains carrying plasmids expressing Cre::VirE2 in the absence of virE1 in cis from the virE or virF promoter (data not shown), probably due to the role that virE1 plays to stabilize Cre::VirE2 as it does VirE2 (Zhao et al., 2001). Therefore, we were unable to assay Cre::VirE2 protein transport in the absence of VirE1. However, we found that a fusion protein between the C-terminal 50 amino acids of VirE2, containing the translocation signal and the Cre recombinase, was translocated both into Arabidopsis and yeast in the absence of VirE1. This result shows unambiguously that VirE1 does not contain essential information for VirE2 to proceed through the translocation pore and its subsequent translocation directly into host cells. The data suggest that VirE1 is not involved in acting in a complex together with VirE2 as a three-dimensional secretion signal, as suggested for the SycE/YopE complex (Birtalan et al., 2002).
The efficiency of translocation of a full-length Cre::VirE2 fusion is significantly higher (5- to 13-fold in plants; 10-fold in yeast) compared with the Cre::VirE2_50C fusion. Deviation from the natural binding conformation in the Cre::VirE2_50C fusion may be the reason for this, resulting in less optimal interaction with components of the transport complex.
The ssDNA-binding site as well as one of the VirE1-binding domains is located in the C-terminal half of VirE2 extending from residues 288 to 495. Insertions in these domains as well as deletions abolished both ssDNA-binding activity and VirE1 binding (Dombek and Ream, 1997; Sundberg and Ream, 1999). The Cre::VirE2_50C protein fusion does not contain the VirE1-binding domain. Thus, the observation that Cre::VirE2_50C is translocated, even from an A. tumefaciens strain expressing VirE1 in trans from the native virE operon, already suggests that VirE1 binding is not a driving factor for signal sequence recognition. However, structural data from a VirE1/VirE2 complex as well as the C-terminal transport signal may shed more light on the exact role of VirE1 in determining transport efficiency of VirE2.
The data indirectly suggest that VirE1 is not a translocated effector itself. We have tested transport using the CRAfT assay of a Cre::VirE1 fusion protein, but we were unable to detect translocation into host cells (data not shown). The role of VirE1 therefore seems restricted to stabilization of VirE2 by preventing premature interactions in the bacterial cell before translocation into host cells. This role, together with the finding that the VirE2 protein can be translocated in the absence of T-DNA, favors the model that cooperative binding of the T-strand by VirE2 takes place in the plant cell and not in the bacterial cell.
We showed that the efficiency of translocation of both VirF and VirE3 effectors is not altered in a virE1 mutant. This is expected, because VirE1 has not been implied as a chaperone for either of these effector proteins. Most chaperones are encoded by genes located in close vicinity to the genes encoding their cognate substrate. The virE3 gene forms part of the virE operon (F. Garcia and P.J.J. Hooykaas, unpublished data) but is located downstream of virE2, and VirF is expressed from a distal single gene. Besides, expression and translocation of Cre::VirE3 and Cre::VirF is unaltered in the absence of VirE1. Therefore, VirE1 does not play a role in translocation of the VirF or VirE3 effectors.
Detection of Protein Translocation Using the CRAfT System
As described above, we detected translocation of Cre by the 50 C-terminal amino acids of VirE3 into Arabidopsis and yeast. Previously, we described translocation of a Cre::VirE3 fusion protein, harboring the full-length VirE3 protein into yeast (Schrammeijer et al., 2003; Table III). However, here, we had great difficulty in detecting translocation of Cre::VirE3 into Arabidopsis (Table II). Only one of seven experiments was successful. This may be due to binding interactions of the full-length protein with host plant but not yeast proteins. Many parameters may determine the efficiency with which translocation is detected in the Cre reporter assay. Not only the number of protein molecules, but also the interaction with other host proteins, stability of the protein, or efficiency in nuclear uptake contribute to this. This difference in detection of translocation efficiency is not without precedent. Previously, we detected a difference in transfer efficiency between Cre fusions with full-length VirF or an N-terminally truncated protein (Cre::VirFΔ42N) to yeast and plants. Translocation of the Cre::VirFΔ42N fusion was detected with a five times higher efficiency in the plant cell nucleus, either by improved stability or the lack of interaction via the N-terminal part of the protein that contains the F-box. When using the CRAfT assay to find new effector proteins, it may therefore be prudent to use not only the plant but also the yeast assay. In fact, use of the yeast system may be advantageous, due to the absence of host (plant)-specific interactors that may sequester the effector proteins.
Concluding Remarks
Our data suggest that chaperones do not play an important role in the recognition of the effector proteins of the A. tumefaciens virulence system by the transport complex. Also, there seems to be no specific role in giving priority to transport of the effectors, because the translocation efficiency of individual effectors is not altered by the absence of other effector proteins (Vergunst et al., 2000). Nevertheless, transport of Cre::VirE2 is detected with a higher efficiency than the Cre::VirE2_50C fusion protein, which lacks the VirE1-binding site. It may be that VirE1 still has a role in VirE2 translocation by keeping it in a (partially) unfolded conformation. Recently, Feldman et al. (2002) described this for the TTSS, in which the SycE chaperone prevented folding of DHFR. Using such a chaperone-binding site together with the transport signal the DHFR protein could be kept in a partially unfolded state to allow translocation. Such a strategy may also be used in A tumefaciens. We have found that GFP cannot be translocated through the TFSS (our unpublished data). Efforts to detect translocation of a Cre::GFP::virF-37C fusion protein using the CRAfT assay were unsuccessful (data not shown). GFP has a barrel structure and is a strongly folded protein and may therefore jam the transport pore. Addition of a chaperone-binding site and the cognate chaperone coding sequence might improve the transfer of such hybrid proteins also in TFSS.
We defined the translocation signal now to the C-terminal 50 amino acids of three different A. tumefaciens effector proteins, VirF, VirE2, and VirE3. A C-terminal signal may be present in effector proteins of other bacterial pathogens of humans using a TFSS. Currently, we are defining the transport signal in more detail. Further studies to the exact functioning of the VirE1 chaperone and the translocation signal will shed more light on the interaction of the effectors with the transport system. Such data may be applied in the development of antimicrobial strategies directed against the TFSS of important mammalian pathogens.
MATERIALS AND METHODS
Recombinant DNA Techniques
Standard cloning techniques were carried out according to Sambrook et al. (1989). Restriction enzymes were purchased from New England Biolabs (Beverly, MA). Bacteria were grown in standard Luria culture medium (10 g L–1 bacto-tryptone, 5 g L–1 yeast extract, and 8 g L–1 NaCl, pH 7), with antibiotics. Kanamycin, rifampicin, carbenicillin, spectinomycin, and gentamycin were purchased from Duchefa (Duchefa Biochemie BV, Maarlem, The Netherlands) and used at a concentration of 100 mg L–1 kanamycin, 20 mg L–1 rifampicin, 250 mg L–1 spectinomycin, and 40 mg L–1 gentamycin for Agrobacterium tumefaciens. For selection of Escherichia coli, 100 mg L–1 carbenicillin, 10 mg L–1 gentamycin, or 25 mg L–1 kanamycin was used.
PCR reactions were performed in a PelkinElmer Life Sciences (Boston) apparatus using cloned Pfu polymerase from Stratagene (La Jolla, CA) for amplification. Primers were purchased from Sigma-Genosys Ltd (Pampisford, UK). Sequencing was carried out at BaseClear (Leiden, The Netherlands).
Bacterial Strains
A. tumefaciens strain LBA1100 (C58C1 with a disarmed octopine-type pTiB6 plasmid [Beijersbergen et al., 1992]) was used for protein transport experiments to plant and yeast cells. A precise virE1 deletion mutant was constructed from LBA1100 using the marker exchange-eviction mutagenesis method as described by Ried and Collmer (1987). To this end, two primers were designed to amplify a 5′ fragment of virE2 introducing an NdeI site (underlined) at the ATG start codon. The oligonucleotide primers virE2/3 (5′-TGTATTCATATGGATCTTTCTGGCAATGAG) and virE2/4 (5′-CTTAGTTTATAATTCCGGTC) were used for amplification of the 5′ sequence of virE2 using pBlueE1-E2 (an XhoI/SmaI fragment of pRAL3248 [Melchers et al., 1990] cloned in pBluescriptIISK–) as template DNA. The PCR fragment was subjected to digestion (NdeI-BglII) and used to replace an NdeI-BglII fragment in pBlueE1-E2, in which the NdeI site is located at the virE1 ATG start codon. The new plasmid was named pSDM3213. Thus, replacement resulted in precise deletion of the virE1 gene in pBluevirE1-virE2. A BglII/SacI fragment, containing about 400 bp of flanking DNA sequences of virE1 on either side was cloned in pSDM3005 (pSDM3600). Plasmid pSDM3005 was the result of cloning a PstI fragment from pUM24 (Ried and Collmer, 1987) into the PstI site of pBGS19 (Spratt et al., 1986). pSDM3005 contains the Bacillus subtilis sacR/B gene and a kanamycin resistance marker and does not replicate in A. tumefaciens to allow easy selection for double crossover events in A. tumefaciens. The sequence of pSDM3600 was confirmed by sequence analysis, and the plasmid was electroporated into LBA1100 (den Dulk-Ras and Hooykaas, 1995) and LBA1010 (C58 with pTiB6; Hooykaas et al., 1984), and Kmr colonies were tested for Suc sensitivity (sucrs) in LC medium (lacking NaCL, but with 6% [w/v] Suc), indicating a single crossover event with the flanking regions of virE1. Kmr/sucrs colonies were grown overnight in LC medium (lacking NaCl) to allow a second excisional recombination event, resulting in precise deletion of virE1 from the vir region. The overnight bacterial culture was plated on medium containing 6% (w/v) Suc. Kms/sucrr colonies were subjected to PCR and Southern-blot analysis to show precise deletion of virE1 both in LBA1100 (named LBA2571) and in LBA1010 (named LBA2570).
Plasmid Constructions
The plasmids used in this study are summarized in Table I. Construction of the plasmids pSDM3147 (pvirE1-cre), pSDM3155 (pvirF-NLS:cre:virFΔ42N), pSDM3129 (pvirE1-NLS::cre::virE2), and pSDM3507 (pvirF-NLS::cre::virE3) was described earlier (Vergunst et al., 2000; Schrammeijer et al., 2003).
For protein transport studies, we made in frame fusions between the 3′ region of virE2 and virE3 encoding the C-terminal 50 amino acids and the cre coding region. Oligonucleotide primers virE2/7 (5′-ccgctcgagTAAGGCTGCCAGCCGATGC [XhoI underlined]) and VirE2/6 (5′-gctctagagTCAAAAGCTGTTGACGC [XbaI underlined, stop codon bold, coding sequence uppercase]) were used for amplification of the 150 3′ bases of the virE2 coding region. SalI/XbaI-digested PCR product was cloned into SalI/XbaI digested pSDM3197 (Schrammeijer et al., 2003). This cloning constructed a NLS::cre::virE2_50C translational fusion expressed from the virF promoter sequence. Sequencing was performed to determine the accuracy of the PCR reaction.
Oligonucleotide primers VirE3/3 (5′-acgcgtcgacttGATTACCATTTGTCAGCTTCG [SalI underlined]) and virE3/4 (5′-gctctagaTTAGAAACCTCTGGAGGTGG [XbaI underlined, stop codon bold]) were used for amplification of the 150 3′ bases of virE3 coding sequence. SalI/XbaI-digested PCR product was cloned into SalI/XbaI digested pSDM3197 (Schrammeijer et al., 2003). This cloning constructed a NLS::cre::virE3_50C translational fusion expressed from the virF promoter. Sequence reaction was performed to confirm the precision of the PCR reaction.
Tumor Assays
Six-week-old Nicotiana glauca plants, grown at 25°C under 16 h of illumination and 60% humidity, were infected with A. tumefaciens as follows: Bacteria were inoculated form fresh LC plates with antibiotics into 10 mL of liquid LC (with antibiotics) and grown overnight at 28°C. One milliliter of bacterial suspension with an OD600 of 1 was concentrated (2 min, 9,000 rpm), and the pellet was resuspended in 0.9% (w/v) NaCl solution. With a toothpick, a hole was made in the stem between two internodes of the young N. glauca plants, three per plant. Twenty microliters of bacterial suspension was pipetted into the hole. Plants were scored for tumors 2 weeks after infection.
Plant Line and Transport Assay
Arabidopsis ecotype C24, transformed with pSDM3043, was described previously (Vergunst et al., 2000). Plants are resistant to phosphinothricin (10 mg L–1 at seedling level) and sensitive to kanamycin. Homozygous plants were used in transport assays, essentially as described by Vergunst et al. (2000). Seedlings were sterilized and cultured in liquid B5 medium, at 80 rpm, as described by Vergunst et al. (1998). Ten days after initiation of the cultures, roots were collected, spread on agar plates containing callus induction medium, and precultured for 3 d. A. tumefaciens was inoculated 1 d before cocultivation from fresh LC plates into 10 mL of liquid LC with antibiotics and grown overnight at 28°C. The roots were collected in 20 mL of liquid B5 medium containing overnight-grown A. tumefaciens cells at a final OD of 0.1. Two minutes after adding the root explants, the roots were cut in small explants of about 5 mm in length. The explants were dried on filter paper and spread on CIM-containing acetosyringone (100 μm). Roots were cocultivated for 3 d at 25°C, at 2,000 lux. Explants were collected and washed in B5 medium. Finally, the root explants were dried briefly on filter paper and plated on shoot induction medium medium containing 50 mg L–1 kanamycin and 100 mg L–1 timentin. Three weeks later, the number of kanamycin-resistant calli was determined using a stereo microscope.
Yeast Strain and Transport Assay
Yeast strain LBY2 contains a lox-URA3-lox construct to detect Cre activity. Loss of the URA3 gene will lead to the ability of the yeast cells to grow on medium containing 0.1% (w/v) FOA (Apollo Scientific, Ltd., Derbyshire, UK). Construction of LBY2 and the cocultivation procedure with A. tumefaciens were described in great detail (Schrammeijer et al., 2003). Calculations of the frequency of FOA-resistant colonies per output yeast were also performed as described by Schrammeijer et al. (2003).
Western-Blot Analysis
Expression level of the fusion genes in A. tumefaciens, upon induction of the virulence system with acetosyringone (purchased from Aldrich-Chemie, Sigma-Aldrich Chemie BV (Zwyndrecht, The Netherlands) was determined using western-blot analysis. A. tumefaciens strains were grown overnight in minimal medium (Hooykaas et al., 1979). A 1:10 dilution in induction medium was followed by overnight growth at 28°C. Induced A. tumefaciens cells were collected (1 mL of an OD 1) and dissolved in 150-μL sample buffer.
An amount of 12 μL of the bacterial suspension was boiled for 10 min and cooled on ice. Samples were loaded on a denaturing 10% (w/v) polyacrylamide (36.5:1 [w/w] acryl:bisacryl) gel for electrophoresis. Proteins were transferred to Immobilon-P membranes (Millipore, Bedford, MA) by using a semidry blotting apparatus (2117 Multiphor II electrophoresis unit from LKB [Uppsala]). The blotting procedure and buffers were essentially prepared as described in the Immun-blot goat-anti-rabbit-AP and goat-anti-mouse-AP assay kits instruction manual (Bio-Rad Laboratories, Hercules, CA). The blot was washed in Tris-buffered saline (TBS) and blocked with 3% (w/v) gelatin in TBS. The antibodies used for protein detection were used as a 1:2,000 (VirE2) and 1:500 dilution (Cre, purchased from Eurogentec, Seraing, Belgium). After incubation for 16 h, blots were washed with TBS with 0.05% Tween, followed by a 1-h incubation with goat-anti-rabbit-IgG-AP (1:7,500 [v/v] dilution, Promega, Madison, WI) for detection with VirE2 antibodies and incubation with goat-anti-mouse-IgG-AP for detection with Cre antibodies. After washing the blots with TBS with 0.05% Tween and TBS, they were incubated in 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium staining solution. Staining was stopped by washing for 10 min with water. The marker we used was obtained from Bio-Rad (dual color prestained precision plus protein standards).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank B. Schrammeijer for preparing pSDM3005, F. Garcia-Rodriguez for helpful discussions and critically reading the manuscript, W. de Winter for technical assistance, and P. Hock for preparation of Figure 1.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.029223.
This work was supported by the European Union (EU Framework 5-Quality of Life and Management of Living Resources; grant no. QLRT–2000–01200).
References
- Baron C, Callaghan DO, Lanka E (2002) Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol Microbiol 43: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Beijersbergen A, den Dulk-Ras A, Schilperoort RA, Hooykaas PJJ (1992) Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256: 1324–1327 [DOI] [PubMed] [Google Scholar]
- Berks BC, Sargent F, Palmer T (2000) The Tat protein export pathway. Mol Microbiol 35: 260–274 [DOI] [PubMed] [Google Scholar]
- Birtalan S, Ghosh P (2001) Structure of the Yersinia type III secretory system chaperone SycE. Nat Struct Biol 8: 974–978 [DOI] [PubMed] [Google Scholar]
- Birtalan SC, Phillips RM, Ghosh P (2002) Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell 9: 971–980 [DOI] [PubMed] [Google Scholar]
- Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Lavigne J, Liautard JP, Ramuz M et al. (2002) Type IV secretion and Brucella virulence. Vet Microbiol 90: 341–348 [DOI] [PubMed] [Google Scholar]
- Boyd AP, Lambermont I, Cornelis GR (2000) Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J Bacteriol 182: 4811–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanon-Wollaston V, Passiatore JE, Cannon F (1987) The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 328: 172–175 [Google Scholar]
- Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJJ (1995) Transkingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J 14: 3206–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezón E, Sastre JI, de la Cruz F (1997) Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254: 400–406 [DOI] [PubMed] [Google Scholar]
- Cambronne ED, Cheng LW, Schneewind O (2000) LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol Microbiol 37: 263–273 [DOI] [PubMed] [Google Scholar]
- Christie PJ (2001) Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol 40: 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Vogel JP (2000) Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol 8: 354–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Guralnick B, Simon MN, Wall JS (1997) The molecular structure of Agrobacterium VirE2-single-stranded DNA complexes involved in nuclear import. J Mol Biol 271: 718–727 [DOI] [PubMed] [Google Scholar]
- Conover GM, Derré I, Vogel JP, Isberg RR (2003) The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol 48: 305–321 [DOI] [PubMed] [Google Scholar]
- Day JB, Plano GV (1998) A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol Microbiol 30: 777–788 [DOI] [PubMed] [Google Scholar]
- de Groot MJA, Bundock P, Hooykaas PJJ, Beijersbergen AGM (1998) Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nature Biotechnol 16: 839–842 [DOI] [PubMed] [Google Scholar]
- Dehio C (2003) Recent progress in understanding Bartonella-induced vascular proliferation. Curr Opin Microbiol 6: 61–65 [DOI] [PubMed] [Google Scholar]
- Delahay RM, Frankel G (2002) Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol Microbiol 45: 905–916 [DOI] [PubMed] [Google Scholar]
- den Dulk-Ras A, Hooykaas PJ (1995) Electroporation of Agrobacterium tumefaciens: plant cell electroporation and electrofusion protocols. 55: 63–72 [DOI] [PubMed] [Google Scholar]
- Deng WY, Chen LS, Peng WT, Liang XY, Sekiguchi S, Gordon MP, Comai L, Nester EW (1999) VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol Microbiol 31: 1795–1807 [DOI] [PubMed] [Google Scholar]
- Dombek P, Ream W (1997) Functional domains of Agrobacterium tumefaciens single-stranded DNA-binding protein virE2. J Bacteriol 179: 1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas F, Duckely M, Pelczar P, Van-Gelder P, Hohn B (2001) An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc Natl Acad Sci USA 98: 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Cornelis GR (2003) The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol Lett 219: 151–158 [DOI] [PubMed] [Google Scholar]
- Feldman MF, Muller S, Wuest E, Cornelis GR (2002) Syc E allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol Micro 46: 1183–1197 [DOI] [PubMed] [Google Scholar]
- Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67: 16–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth F, Moncalian G, de la Cruz F, Coll M (2002) Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein: detailed structural features and mapping of the active site cleft. J Biol Chem 277: 7556–7566 [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth F, Moncalian G, Perez LR, Gonzalez A, Cabezón E, de laCruz F, Coll M (2001) The bacterial conjugation protein TrwB resembles ring helicases and F-1-ATPase. Nature 409: 637–641 [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, von-Bodman SB, Lanka E, Ream W, Farrand SK (2000) TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol 182: 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooykaas PJJ, Hofker M, den Dulk-Ras A, Schilperoort RA (1984) A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid 11: 195–205 [DOI] [PubMed] [Google Scholar]
- Hooykaas PJJ, Roobol C, Schilperoort RA (1979) Regulation of the transfer of Ti-plasmids of Agrobacterium tumefaciens. J Gen Microbiol 110: 99–109 [Google Scholar]
- Koekman BP, Hooykaas PJJ, Schilperoort RA (1982) A functional map of the replicator region of the octopine Ti plasmid. Plasmid 7: 119–132 [DOI] [PubMed] [Google Scholar]
- Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V (2000) Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci USA 98: 1871–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Bertero MG, Frey EA, Pfuetzner RA, Wenk MR, Creagh L, Marcus SL, Lim D, Sicheri F, Kay C et al. (2001) Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat Struct Biol 8: 1031–1036 [DOI] [PubMed] [Google Scholar]
- McBride KE, Knauf VC (1988) Genetic analysis of the virE operon of the Agrobacterium Ti plasmid pTiA6. J Bacteriol 170: 1430–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers LS, Maroney MJ, den Dulk-Ras A, Thompson DV, van Vuuren HA, Schilperoort RA, Hooykaas PJ (1990) Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol Biol 14: 249–259 [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K (2001) The sec protein-translocation pathway. Trends Microbiol 9: 494–500 [DOI] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu XJ, Kahn RA, Roy CR (2002) A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295: 679–682 [DOI] [PubMed] [Google Scholar]
- Nagai H, Roy CR (2003) Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol 5: 373–383 [DOI] [PubMed] [Google Scholar]
- Page AL, Parsot C (2002) Chaperones of the type III secretion pathway: jacks of all trades. Mol Microbiol 46: 1–11 [DOI] [PubMed] [Google Scholar]
- Pansegrau W, Lanka E (1996) Mechanisms of initiation and termination reactions in conjugative DNA processing. J Biol Chem 271: 13068–13076 [DOI] [PubMed] [Google Scholar]
- Ried JL, Collmer A (1987) An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57: 239–246 [DOI] [PubMed] [Google Scholar]
- Rossi L, Hohn B, Tinland B (1996) Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 93: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schrammeijer B (2001) Functional analysis of the virulence protein VirF from Agrobacterium tumefaciens. PhD thesis. Leiden University, Leiden, The Netherlands
- Schrammeijer B, den-Dulk-Ras A, Vergunst AC, Jurado Jacome E, Hooykaas PJJ (2003) Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res 31: 860–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuïnk AJG, Crosby WL, Hooykaas PJJ (2001) Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol 11: 258–262 [DOI] [PubMed] [Google Scholar]
- Schröder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E (2002) TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol 184: 2767–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone M, McCullen CA, Stahl LE, Binns AN (2001) The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol 41: 1283–1293 [DOI] [PubMed] [Google Scholar]
- Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broome-Smith JK (1986) Kanamycin resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41: 337–342 [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE (2001) Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414: 77–81 [DOI] [PubMed] [Google Scholar]
- Sundberg C, Meek L, Carroll K, Das A, Ream W (1996) VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J Bacteriol 178: 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg CD, Ream W (1999) The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J Bacteriol 181: 6850–6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer CY, Faelen M, Couturier M (2000) Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol Microbiol 37: 1283–1292 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, De Waal EC, Hooykaas PJJ (1998) Root transformation by Agrobacterium tumefaciens. In J Martinez-Zapater, J Salinas, eds, Arabidopsis Protocols. Meth Mol Biol 82: 227–244 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den-Dulk-Ras A, de-Vlaam CMT, Regensburg TT, Hooykaas PJJ (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290: 979–982 [DOI] [PubMed] [Google Scholar]
- Ward EW, Barnes WM (1988) VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242: 927–930 [Google Scholar]
- Wattiau P, Woestyn S, Corenelis GR (1996) Customized secretion chaperones in pathogenic bacteria. Mol Microbiol 20: 255–262 [DOI] [PubMed] [Google Scholar]
- Woestyn S, Sory MP, Boland A, Leguenne O, Cornelis GR (1996) The cytosolic SycE and sycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Micro 20: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Zhao ZM, Sagulenko E, Ding ZY, Christie PJ (2001) Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol 183: 3855–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XR, Christie PJ (1999) Mutagenesis of the Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol 181: 4342–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Merkle T, Schoumacher F, Hohn B, Rossi L (2001) Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13: 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan JR, Citovsky V, Zambryski P (1996) Agrobacterium virE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc Natl Acad Sci USA 93: 2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]