Abstract
Purpose
The current classification systems of myelodysplastic syndromes (MDS), including the International Prognostic Scoring System (IPSS), do not fully reflect the molecular heterogeneity of the disease. Molecular characterization may predict clinical outcome and help stratify patients for targeted therapies. Epigenetic therapy using decitabine, a DNA hypomethylating agent, is clinically effective for the treatment of MDS. Therefore, we investigated the association between DNA methylation and clinical outcome in MDS.
Patients and Methods
We screened 24 patients with MDS for promoter CpG island methylation of 24 genes and identified aberrant hypermethylation at 10 genes. We then performed quantitative methylation analyses by bisulfite pyrosequencing of the identified genes in 317 patient samples from three independent studies and assessed relations between methylation and clinical outcome.
Results
In an initial training cohort of 89 patients with MDS, methylation frequencies of individual genes ranged from 7% to 70% and were highly concordant. Therefore, we defined a methylation z score based on all genes for each patient. We found that patients with higher levels of methylation, compared with patients with lower levels, had a shorter median overall survival (12.3 v 17.5 months, respectively; P = .04) and shorter median progression-free survival (6.4 v 14.9 months, respectively; P = .009). This methylation prognostic model was independent of age, sex, and IPSS group. Applied to two validation cohorts (228 patients), this model was confirmed as an independent prognostic predictor for survival. Although methylation at baseline did not correlate with clinical response to decitabine, we observed a significant correlation between reduced methylation over time and clinical responses.
Conclusion
DNA methylation predicts overall and progression-free survival in MDS.
INTRODUCTION
Myelodysplastic syndromes (MDS) are heterogeneous hematopoietic disorders characterized by bone marrow failure, dysplasia of one or more of the myeloid blood cell lineages, and an increased risk of developing acute myeloid leukemia.1 Various models have been proposed to predict patient outcomes, including the International Prognostic Scoring System (IPSS).2 On the basis of the percentage of blasts, karyotype, and number of cytopenias, patients are classified into four IPSS groups (Low, Intermediate-1, Intermediate-2, and High). The median survival declines from almost 6 years in patients with IPSS low risk to less than 1 year in patients with IPSS high risk.2 Despite the usefulness of IPSS in clinical practice, however, outcome varies considerably among MDS patients with identical IPSS scores.3 Although cytogenetic abnormalities in MDS are among the most valuable independent prognostic determinants, their prognostic value is limited by the inherent cytogenetic diversity of these diseases.4 Because of the marked heterogeneity of MDS and the increasing number of emerging therapeutic targets approved for its treatment, molecular characterization of MDS is needed to further improve prognostic prediction and for the selection of patients for targeted therapeutic approaches.
Recently, epigenetic therapy using hypomethylating agents (azacitidine and decitabine) has demonstrated clinical effectiveness; azacitidine and decitabine are now approved by the US Food and Drug Administration for the treatment of patients with MDS.5,6 DNA methylation is a well-established epigenetic mechanism that regulates gene transcription through modification at cytosines in the context of CpG dinucleotides.7 Aberrant DNA methylation at the promoter CpG islands is increasingly recognized as a common event in human cancers and has been associated with silencing of important tumor suppressor genes.8 The relation between prognosis and DNA methylation has been investigated in MDS at single genes9–13 or combinations of multiple genes.14,15 For instance, hypermethylation of p15INK4B was found to be negatively correlated with prognosis, but this association was not statistically significant in multivariate analysis.11,12 Another study of 37 patients with MDS found that the combination of promoter methylation of p15INK4B, HIC1, CDH1 and ERα genes predicts poor prognosis in early-stage MDS but not advanced-stage MDS.14 These studies had a small sample size, and whether DNA methylation could predict responsiveness to hypomethylation agents has not been established. Therefore, in the current study of a large number of adult patients with MDS, we set out to investigate the prognostic significance of DNA methylation measured before treatment, to test the significance of methylation at baseline in predicting clinical responses, and to explore the correlation between modulation of DNA methylation and response to decitabine.
PATIENTS AND METHODS
Patients and Samples
We analyzed tumor samples from 317 patients with MDS. Patients were divided into one training cohort and two validation cohorts. Each of the three cohorts is unique. The training cohort consisted of 89 patients enrolled between July 2001 and February 2003 as part of a multicenter randomized phase III trial comparing decitabine with supportive care; the study design has been described in detail.5 Of the patients studied, 40 received decitabine, and 49 received supportive care alone. For each treatment cycle, decitabine was administered intravenously at a dose of 15 mg/m2/d for 3 days, and each course was repeated every 6 weeks. The first validation cohort comprised 75 patients from a phase II trial comparing three schedules of low-dose decitabine treatment at The University of Texas M. D. Anderson Cancer Center between November 2003 and August 2005.6 All patients received decitabine in one of the following three schedules: 20 mg/m2/d intravenously for 5 days (46 patients); 20 mg/m2/d subcutaneously for 5 days (13 patients); or 10 mg/m2/d intravenously for 10 days (16 patients). Courses of decitabine were administered every 4 weeks. The second validation cohort was an independent study of 153 patients with a diagnosis of MDS referred to The University of Texas M. D. Anderson Cancer Center between December 2001 and April 2007. Of these 153 patients, 40 received either decitabine or azacitidine, and 113 received either other chemotherapy or supportive care. Seventeen patients received decitabine alone, 18 received azacitidine alone, and five received both decitabine and azacitidine. Decitabine 20 mg/m2/d was administered intravenously for 5 days, and azacitidine 75 mg/m2/d was administered subcutaneously for 7 days.
We obtained bone marrow or (if bone marrow was not available) peripheral blood before treatment (at baseline) for all patients. In addition, we obtained bone marrow at 1, 2, and 4 months after treatment from 34 patients in the training cohort. Among them, 20 patients received supportive care, and 14 received decitabine. All patients were enrolled onto protocols approved by the institutional review boards of the involved institutions and provided written informed consent.
Quantitative DNA Methylation Analyses by Bisulfite Pyrosequencing
We used bisulfite pyrosequencing to quantitatively assess DNA methylation.16 Details of the method, including primer sequences and conditions used for methylation determination, are described in the Data Supplement.
Statistical Analysis
Details of statistical analysis used in this study are described in the Data Supplement. All methylation data were generated without knowledge of the clinical status of the samples and analyzed as continuous values. To evaluate the effect of the overall genes, methylation for each gene among the patients was standardized by the z score method.17
Standard statistical methods for survival analysis were used. These included log-rank tests to determine univariate differences between groups, Kaplan-Meier tests to calculate and generate survival curves, and multivariate Cox proportional hazards models to assess whether methylation z score or any known risk factor, such as age, sex, treatment, MDS type, prior treatment, cytogenetic category, and IPSS group, was independently associated with survival. All P values are based on two-tailed tests, and P ≤ .05 was considered statistically significant.
RESULTS
Baseline Patient Characteristics and Follow-Up
Of the 170 patients enrolled onto the phase III trial, we excluded 81 patients from the training cohort because no specimen was available. We compared patient clinicopathologic characteristics between the patients with tissue available and those without and found similar distributions for patient age; sex; hemoglobin, WBC, platelet, and marrow blast counts; cytogenetic categories; prior malignancy; prior treatment for MDS; and IPSS status. However, compared with patients who were excluded, there were slightly more patients with chronic myelomonocytic leukemia among the patients with tissue available (13 of 89 studied patients v one of the other 81 patients; P = .001). In the first validation cohort, we excluded 20 patients as a result of sample unavailability. The data for patients included and for those excluded had similar distributions with regard to demographic and clinicopathologic characteristics. Table 1 lists the clinical and demographic characteristics of the patients in the three cohorts. Patients in the training cohort and those in the validation cohorts had similar characteristics, except that compared with the other two cohorts, the second validation cohort included a broad spectrum of patients with more low-risk MDS (Table 1).
Table 1.
Baseline Patient Demographics Clinical and Hematologic Characteristics
| Demographic or Characteristic | Training Cohort (n = 89) |
First Validation Cohort (n = 75) |
Second Validation (n = 153) |
|||
|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | |
| Age, years | ||||||
| Median | 69 | 66 | 66 | |||
| Range | 30-85 | 44-89 | 23-91 | |||
| < 45 | 3 | 3 | 1 | 1 | 8 | 5 |
| 45-65 | 31 | 35 | 34 | 45 | 66 | 43 |
| > 65 | 55 | 62 | 40 | 53 | 79 | 52 |
| Female sex | 28 | 31 | 18 | 24 | 51 | 33 |
| Hemoglobin level, g/dL | ||||||
| Median | 9 | 9.5 | 9.8 | |||
| Range | 5.9-14.1 | 4.5-13.7 | 6.8-14.8 | |||
| < 10 | 60 | 67 | 52 | 69 | 82 | 56 |
| Missing data | 4 | 5 | 0 | 0 | 0 | 0 |
| Absolute neutrophil count, × 109/L | ||||||
| Median | 1.1 | 1 | 1.7 | |||
| Range | 0.1-18.7 | 0.1-61.6 | 0.1-31.4 | |||
| < 1.8 | 57 | 64 | 51 | 68 | 80 | 52 |
| Missing data | 7 | 8 | 1 | 1 | 0 | 0 |
| Platelet count, × 109/L | ||||||
| Median | 38 | 48 | 79 | |||
| Range | 7-480 | 3-416 | 3-1,040 | |||
| Platelet count | ||||||
| < 100 | 65 | 73 | 53 | 71 | 91 | 59 |
| Missing data | 5 | 6 | 0 | 0 | 0 | 0 |
| Marrow blast cells, No. | ||||||
| Median | 8 | 8 | 4 | |||
| Range | 0-28 | 0-26 | 0-29 | |||
| < 5 | 28 | 31 | 17 | 23 | 78 | 50 |
| 5-10 | 23 | 26 | 30 | 40 | 29 | 19 |
| 11-20 | 23 | 26 | 25 | 33 | 28 | 19 |
| 21-30 | 12 | 14 | 3 | 4 | 18 | 12 |
| Missing data | 3 | 3 | 0 | 0 | 0 | 0 |
| IPSS risk category | ||||||
| Low | 0 | 0 | 2 | 3 | 34 | 22 |
| Intermediate-1 | 30 | 34 | 30 | 40 | 56 | 37 |
| Intermediate-2 | 41 | 46 | 31 | 41 | 38 | 25 |
| High | 18 | 20 | 11 | 15 | 22 | 14 |
| Unclassified | 0 | 0 | 1 | 1 | 3 | 2 |
| FAB type | ||||||
| Refractory anemia | 13 | 15 | 5 | 7 | 53 | 34 |
| Refractory anemia with ringed sideroblasts | 5 | 6 | 3 | 4 | 17 | 11 |
| Refractory anemia with excess blasts | 58 | 65 | 51 | 68 | 70 | 46 |
| Chronic myelomonocytic leukemia | 13 | 15 | 13 | 17 | 13 | 9 |
| Unclassified | 0 | 0 | 3 | 4 | 0 | 0 |
| Karyotype | ||||||
| Good | 32 | 36 | 40 | 53 | 87 | 57 |
| Intermediate | 18 | 20 | 20 | 27 | 34 | 22 |
| Poor | 22 | 25 | 15 | 20 | 30 | 20 |
| Unclassified | 17 | 19 | 0 | 0 | 2 | 1 |
| MDS type | ||||||
| De novo | 77 | 86 | 51 | 68 | 108 | 71 |
| Secondary | 12 | 14 | 24 | 32 | 45 | 29 |
Abbreviations: IPSS, International Prognostic Scoring System; FAB, French-American-British; MDS, myelodysplastic syndrome.
The median follow-up time was 13.7 months in the training cohort, 14.4 months in the first validation cohort, and 7.1 months in the second validation cohort. In the training cohort, we found no significant association between specimen sources and either overall survival (P = .80) or progression-free survival (P = .56).
Selection of a Panel of Genes for Methylation Analysis
We screened promoter CpG island methylation of 24 genes in a group of 24 patients with MDS; these genes were selected based on previous reports18–20 and our ongoing effort to identify hypermethylated genes in cancer by a genome-wide methylated CpG island amplification/representational difference analysis technique21,22 (details are summarized in the Data Supplement). After the initial screening, low levels of methylation were detected in 14 genes, and we excluded them from further study. Aberrant promoter CpG island methylation of 10 genes, including E-cadherin (CDH1), N-cadherin (CDH13), estrogen receptor-α (ERα), oxidored-nitro domain-containing protein isoform 1 (NOR1), nucleoplasmin 2 (NPM2), oligodendrocyte lineage transcription factor 2 (OLIG2), cyclin-dependent kinase 2B inhibitor (p15INK4B), progesterone receptor A (PGRA), progesterone receptor B (PGRB), and PDZ and LIM domain 4 (RIL), was found in more than 10% of the patients, and these genes were selected for further analysis (Data Supplement).
To confirm that the observed methylation differences were not merely measurement variation or cellular heterogeneity (such as analyses performed on unfractionated cells), we repeated bisulfite treatment, polymerase chain reaction, and pyrosequencing for six genes in 20 MDS patient samples, compared methylation for all genes between bone marrow and blood samples obtained at the same time from 25 patients, and compared methylation level between sorted CD34+ and CD3–/19– cells from the same patients (Data Supplement). The results from two replicate experiments were almost identical, with the Spearman correlation coefficient (r value) of 0.92 (n = 93). A significant correlation was found between methylation levels measured in bone marrow and blood (n = 117, r = 0.93), as well as between methylation levels measured in CD34+ and CD3–/19– cells (n = 13, r = 0.91).
Model of DNA Methylation Profiling for Predicting Survival in the MDS Training Cohort
Among the 89 patients in the training cohort, promoter methylation (> 15%) was found in 7% at ERα, 15% at CDH1, 15% at NOR1, 20% at NPM2, 21% at CDH13, 23% at p15INK4B, 41% at OLIG2, 45% at PGRB, 45% at PGRA, and 70% at RIL. Using methylation level as a continuous variable and analyzing the correlation between each gene by Spearman correlation analysis, we found significant positive associations among methylation of different genes within the same patients (Data Supplement). These results indicate concordant methylation in a subset of patients (Data Supplement) and hence that combining multiple gene methylation profiles could provide greater accuracy than individual markers in predicting clinical outcomes. Therefore, we used average z scores based on methylation of all genes to compare clinical characteristics and build predictive models of survival for individual patients. There were no significant associations between average methylation z score and age, sex, hemoglobin level, absolute neutrophil count, platelet count, bone marrow blast percentage, IPSS status, French-American-British type, or cytogenetics (Table 2).
Table 2.
Correlation Between Average Methylation of 10 Genes (by z score) and Clinical Features
| Characteristic | Training Cohort |
First Validation Cohort |
Second Validation |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean z Score | SD | P | Mean z Score | SD | P | Mean z Score | SD | P | |
| Age, years | |||||||||
| < 45 | −0.16 | 0.12 | .11 | −0.38 | 0 | .09 | 0.49 | 0.63 | .11 |
| 45-65 | 0.08 | 0.83 | 0.12 | 0.37 | 0.32 | 0.61 | |||
| > 65 | −0.04 | 0.49 | 0.23 | 0.39 | 0.14 | 0.63 | |||
| Sex | |||||||||
| Male | −0.07 | 0.48 | .17 | 0.18 | 0.41 | .52 | 0.24 | 0.64 | .87 |
| Female | 0.16 | 0.84 | 0.13 | 0.30 | 0.22 | 0.60 | |||
| Hemoglobin level, g/dL | |||||||||
| < 10 | 0.05 | 0.65 | .41 | 0.20 | 0.37 | .39 | 0.25 | 0.67 | .73 |
| ≥ 10 | −0.07 | 0.56 | 0.11 | 0.42 | 0.21 | 0.58 | |||
| Absolute neutrophil count, × 109/L | |||||||||
| < 1.8 | −0.05 | 0.48 | .19 | 0.21 | 0.36 | .27 | 0.33 | 0.66 | .03 |
| ≥ 1.8 | 0.21 | 0.88 | 0.10 | 0.45 | 0.12 | 0.57 | |||
| Platelet count, × 109/L | |||||||||
| < 100 | 0.03 | 0.64 | .69 | 0.15 | 0.41 | .66 | 0.30 | 0.63 | .10 |
| ≥ 100 | −0.03 | 0.61 | 0.25 | 0.33 | 0.13 | 0.61 | |||
| Marrow blast percentage | |||||||||
| < 5 | −0.08 | 0.38 | .80 | 0.13 | 0.42 | .66 | 0.10 | 0.59 | .01 |
| 5-10 | 0.00 | 0.56 | 0.13 | 0.40 | 0.29 | 0.58 | |||
| 11-20 | 0.11 | 0.92 | 0.24 | 0.38 | 0.31 | 0.66 | |||
| 21-30 | 0.08 | 0.54 | 0.18 | 0.11 | 0.60 | 0.67 | |||
| IPSS risk category | |||||||||
| Low | NA | .18 | 0.83 | 0.89 | .30 | −0.02 | 0.53 | .01 | |
| Intermediate-1 | −0.29 | 0.41 | 0.03 | 0.43 | 0.19 | 0.61 | |||
| Intermediate-2 | −0.02 | 0.77 | 0.24 | 0.32 | 0.29 | 0.59 | |||
| High | 0.02 | 0.46 | 0.34 | 0.50 | 0.53 | 0.72 | |||
| FAB type | |||||||||
| Refractory anemia | −0.12 | 0.41 | .44 | 0.29 | 0.71 | .67 | 0.09 | 0.51 | .002 |
| Refractory anemia with ringed sideroblasts | −0.25 | 0.43 | −0.07 | 0.27 | −0.10 | 0.54 | |||
| Refractory anemia with excess blasts | 0.07 | 0.70 | 0.17 | 0.31 | 0.43 | 0.68 | |||
| Chronic myelomonocytic leukemia | −0.08 | 0.44 | 0.17 | 0.52 | 0.18 | 0.52 | |||
| Karyotype | |||||||||
| Good | −0.08 | 0.53 | .07 | 0.14 | 0.42 | .14 | 0.17 | 0.62 | .42 |
| Intermediate | 0.18 | 0.50 | 0.14 | 0.42 | 0.32 | 0.72 | |||
| Poor | −0.07 | 0.37 | 0.28 | 0.24 | 0.28 | 0.55 | |||
| MDS type | |||||||||
| De novo | −0.02 | 0.65 | .12 | 0.16 | 0.44 | .80 | 0.25 | 0.63 | .51 |
| Secondary | 0.11 | 0.38 | 0.19 | 0.24 | 0.18 | 0.62 | |||
Abbreviations: SD, standard deviation; ANC, absolute neutrophil count; IPSS, International Prognostic Scoring System; FAB, French-American-British; MDS, myelodysplastic syndrome.
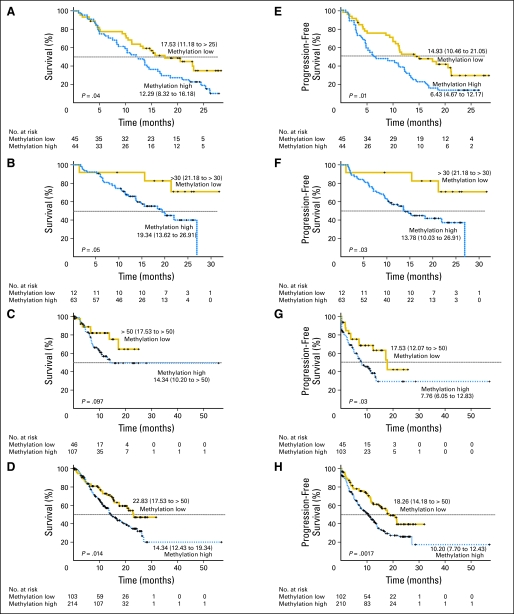
To evaluate the prognostic significance of the methylation profile, we performed both univariate and multivariate analyses. In the univariate analysis, only methylation was significantly associated with shorter overall survival (hazard ratio [HR] = 1.68; 95% CI, 1.01 to 2.80; P = .05). Methylation was most significantly associated with progression-free survival (HR = 1.03; 95% CI, 1.00 to 1.06; P = .06 for age; HR = 1.78; 95% CI, 1.01 to 3.13; P = .05 for IPSS status; and HR = 1.88; 95% CI, 1.15 to 3.07; P = .01 for methylation). Figure 1 shows Kaplan-Meier curves of overall survival and progression-free survival according to the baseline levels of methylation. The median overall survival time was 12.3 months in patients with high methylation compared with 17.5 months in patients with low methylation (P = .04; Fig 1A), and the median progression-free survival time was 6.4 months in patients with high methylation compared with 14.9 months in patients with low methylation (P = .01; Fig 1E). In multivariate analysis, methylation remained the only independent predictor of overall survival (HR = 1.68; 95% CI, 1.0 to 2.81; P = .05) and progression-free survival (HR = 1.95; 95% CI, 1.18 to 3.21; P = .009; Table 3). The effects of methylation on overall and progression-free survival were similar in patients who received decitabine and those who were on supportive care (Data Supplement).
Fig 1.
Kaplan-Meier survival estimates of overall and progression-free survival in patients with myelodysplastic syndromes. Overall survival in (A) training cohort, (B) first validation cohort, (C) second validation cohort, and (D) all patients. Progression-free survival in (E) training cohort, (F) first validation cohort, (G) second validation cohort, and (H) all patients. In each panel, patients are grouped into methylation low (gold) or methylation high (blue) groups according to their combined methylation z scores. Median survival (95% CI) of each group in each panel is shown. P values are based on the log-rank test.
Table 3.
Multivariate Cox Proportional Hazards Analysis of Overall and Progression-Free Survival
| Variable | Training Cohort (n = 89) |
All Patients (N = 317) |
Patients Without Treatment of Hypomethylating Agents (n = 162) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall Survival |
Progression-Free Survival |
Overall Survival |
Progression-Free Survival |
Overall Survival |
Progression-Free Survival |
|||||||||||||
| Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | Hazard Ratio | 95% CI | P | |
| Age | 1.02 | 0.99 to 1.05 | .27 | 1.02 | 0.99 to 1.05 | .11 | 1.05 | 1.03 to 1.07 | < .0001 | 1.03 | 1.01 to 1.04 | .0023 | 1.05 | 1.03 to 1.08 | .0001 | 1.02 | 1.00 to 1.04 | .0395 |
| Sex | 1.08 | 0.63 to 1.85 | .78 | 0.96 | 0.57 to 1.62 | .87 | 0.88 | 0.59 to 1.32 | .54 | 1.09 | 0.77 to 1.56 | .62 | 0.68 | 0.38 to 1.22 | .19 | 0.89 | 0.54 to 1.47 | .64 |
| IPSS status | 1.31 | 0.71 to 2.42 | .38 | 1.67 | 0.93 to 2.97 | .08 | 1.40 | 0.91 to 2.14 | .12 | 2.60 | 1.78 to 3.78 | < .0001 | 1.56 | 0.82 to 2.97 | .18 | 4.73 | 2.78 to 8.05 | < .0001 |
| Methylation | 1.68 | 1.00 to 2.81 | .05 | 1.95 | 1.18 to 3.21 | .009 | 1.83 | 1.23 to 2.71 | .0027 | 1.87 | 1.31 to 2.67 | .0006 | 2.07 | 1.18 to 3.65 | .011 | 2.31 | 1.38 to 3.86 | .0015 |
Abbreviation: IPSS, International Prognostic Scoring System.
Validation of the DNA Methylation Prognostic Model in Two Independent Cohorts
As shown in the Data Supplement, concordant methylation of the 10 genes was also observed in the two validation cohorts. No significant association was found between average methylation z score at baseline and clinical variables in the first validation cohort (Table 2). In the second validation cohort (consecutive series that included patients with low-risk MDS), significantly lower levels of methylation were found in patients with lower bone marrow blast percentage, lower risk IPSS status, and refractory anemia with ringed sideroblasts.
Figure 1 shows overall survival and progression-free survival according to baseline levels of methylation in the validation cohorts. Methylation was significantly associated with progression-free survival in both cohorts (P = .03 and P = .03; Figs 1F and 1G). We found significant association between methylation and overall survival in the first validation cohort (P = .05; Fig 1B); however, the difference in overall survival was not statistically significant in the second validation cohort (P = .097; Fig 1C). Because the second validation cohort comprised more patients with low-risk MDS, one possible explanation is that the relatively short follow-up in these patients may not be sufficient to detect significant differences. We then performed separate analyses in patients with or without low-risk MDS and found that methylation was not significantly associated with overall or progression-free survival in patients with low-risk MDS during the follow-up period (Data Supplement). For patients with other than low-risk MDS, methylation was confirmed to predict shorter overall and progression-free survival (P = .035 and P = .012, respectively; Data Supplement). In the original and validation cohorts combined, patients with high methylation had a worse survival than patients with less methylation (14.3 v 22.8 months, respectively; P = .014 for overall survival and 10.2 v 18.3 months, respectively; P = .0017 for progression-free survival; Figs 1D and 1H).
Multivariate analyses were performed using data from the two validation cohorts or from all patients (Table 3). As in the training cohort, after adjusting for age, sex, and IPSS status, methylation was independently associated with overall survival (HR = 1.83; 95% CI, 1.23 to 2.71; P = .0027) and progression-free survival (HR = 1.87; 95% CI, 1.31 to 2.67; P = .0006).
Because hypomethylating agents may alter the natural history of disease, we then analyzed the correlation between baseline methylation score, clinical characteristics, and standard prognosis separately according to whether or not the patients received treatment with either decitabine or azacitidine. Including only patients who did not receive hypomethylating agents yielded the same results as including all patients (Data Supplement lists clinical characteristics and shows Kaplan-Meier analysis; Table 3 lists results of multivariate survival analysis).
Methylation-Based Prediction Within the Same Cytogenetic Risk Groups
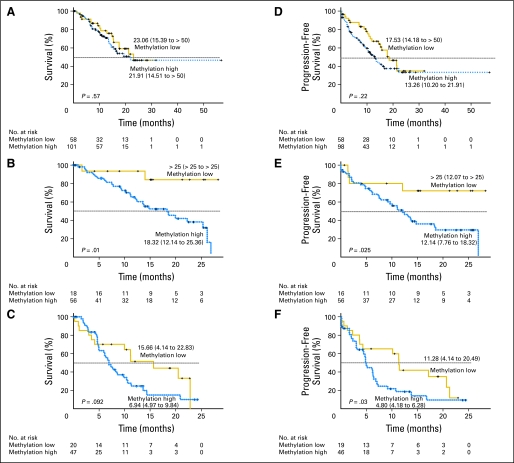
Taking advantage of the large data set, we were able to determine the impact of the DNA methylation prognostic model in patients within similar cytogenetic subgroups. Cytogenetic risk groups were defined by the IPSS as good, intermediate, and poor. Among patients who were in cytogenetic intermediate- and poor-risk groups, the methylation model was a significant predictor of survival (Figs 2B and 2C show overall survival, and Figs 2E and 2F show progression-free survival). Interestingly, survival curves showed no significant differences between high methylation and low methylation groups in patients with good cytogenetic risk (Figs 2A and 2D).
Fig 2.
The DNA methylation prognostic model and cytogenetic risk groups. The Kaplan-Meier estimates show survivals for groups of patients with cytogenetic good risk (A: overall survival; D: progression-free survivals), intermediate risk (B: overall survival; E: progression-free survival), and high risk (C: overall survival; F: progression-free survival).
Correlation Between DNA Methylation and Treatment Responses
To determine whether DNA methylation could predict responses to decitabine treatment, we compared DNA methylation at baseline with clinical responses in 163 patients enrolled onto the phase III and phase II trials. Nine percent of the 89 patients from the phase III trial achieved complete remission (CR) or partial remission (PR), and 47% of the 75 patients from the phase II trial achieved CR or PR. Although the patient groups are comparable between these two trials (Table 1), the difference in clinical responses may be related to the different dose-intensity of decitabine given and to the median number of decitabine courses, which was six in the phase II trial and three in the phase III trial.5,6 Treatment response was not correlated with either methylation of single genes or a combination of all genes (Data Supplement).
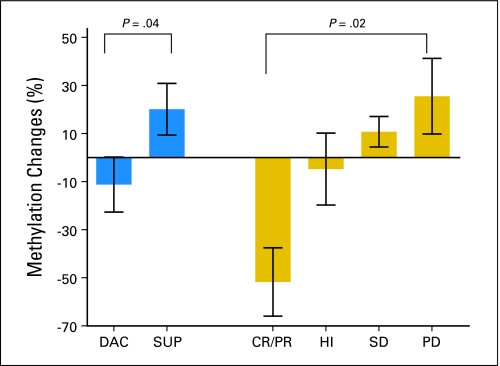
We then analyzed methylation changes at multiple time points for correlation with response in 34 patients from the phase III trial. These patients were selected solely based on tissue availability, and all available tissues were evaluated. Of 14 patients who received decitabine, two patients achieved CR, three achieved PR, four achieved hematologic improvement (HI), four had stable disease (SD), and one had progressive disease (PD). Of 20 patients on supportive care, two patients achieved HI, six had SD, and 12 had PD. Methylation levels at each time point were averaged across the 10 genes. At the latest available time point (> 4 months on therapy), we found that methylation decreased by 11.2% in patients on decitabine but increased by 20.1% in patients on supportive care (P = .04; Fig 3). A greater decrease was observed in patients with CR or PR (40.6% ± 15.7%) compared with HI (9.8% ± 13.2%). In contrast, methylation increased by 15.4% in patients with SD and by 27.2% in patients with PD (P = .02; Fig 3).
Fig 3.
Methylation changes at multiple time points after treatment. Average methylation changes (before and after 4 months on therapy) were compared between patients treated with decitabine (DAC) and supportive care (SUP). Methylation decreased by 11.2% in patients on DAC but increased by 20.1% in patients on SUP (P = .04 by Mann-Whitney U test). These methylation changes were then analyzed for correlation with response in 34 patients (DAC arm: two patients with complete remission [CR], three patients with partial remission [PR], four patients with hematologic improvement [HI], four patients with stable disease [SD], and one patient with progressive disease [PD]; supportive care arm: two patients with HI, six patients with SD, and 12 patients with PD). A greater decrease was observed in patients with CR or PR (40.6% ± 15.7%) compared with HI (9.8% ± 13.2%). Methylation increased by 15.4% in patients with SD and by 27.2% in patients with PD (P = .02 by Kruskal-Wallis test).
DISCUSSION
In the present study, we conducted DNA methylation profiling of 10 genes in 317 patients with MDS and assessed associations between DNA methylation and clinical outcomes. Our data identify a subset of patients with concordant methylation at multiple genes, suggesting the presence of CpG island methylator phenotype (CIMP) in MDS, as described in colon cancer23 and other human neoplasms including acute myeloid leukemia.18,24–27 The presence of CIMP was significantly associated with poor prognosis and risk of leukemia transformation.
The mechanistic bases of CIMP in MDS remain unknown. One proposed mechanism involves aberrant recruitment of DNA methyltransferases to CpG islands28 and/or loss of methylation protection.29 Another possible explanation is that hypermethylation may not be directly linked to the methylation machinery, but rather reflects environmental exposures. Indeed, previous studies have suggested different methylation profiles in experimental lung tumors induced by various carcinogens,30 and studies have indicated correlations between CpG island methylation and specific carcinogenic exposures in hepatocellular carcinomas.31 Finally, a plausible hypothesis by which CIMP confers poor prognosis is that tumors with high degrees of methylation are more likely to inactivate genes critical for tumor progression and for response to chemotherapy.
Although we did not find any significant association between methylation at baseline and clinical response to decitabine treatment, reduced methylation over time was correlated with better clinical response. Further studies of methylation dynamics both before and after treatment will be useful to determine the ability of these markers to direct treatment. It is possible that future studies including unbiased methods for genome-scale DNA methylation may identify biomarkers that may predict for the success of epigenetic treatment.
In conclusion, we developed gene methylation signatures based on a combination of 10 genes that predict both overall and progression-free survival in patients with MDS. This methylation prognostic model was validated in independent samples of patients from two consecutive studies. Methylation at baseline does not predict response to decitabine, but decreased methylation after therapy is associated with clinical response. These findings may be useful to clinicians for predicting individual survival probabilities and for directing therapy and to researchers for designing and interpreting clinical trials.
Supplementary Material
Footnotes
Supported in part by National Institutes of Health Grants No. CA100632, CA108631, and CA121104. L.S. is a Kimmel Scholar supported by the Sidney Kimmel Foundation for Cancer Research. J.-P.J.I. is an American Cancer Society Clinical Research Professor supported by a generous gift from the F.M. Kirby Foundation.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Jean-Pierre J. Issa, MGI Pharma/Eisai (C) Stock Ownership: None Honoraria: None Research Funding: Hagop Kantarjian, Novartis, Bristol-Myers Squibb, Genzyme; Jean-Pierre J. Issa, Celgene, MGI Pharma/Eisai Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Lanlan Shen, Eli Estey, Jean-Pierre J. Issa
Financial support: Jean-Pierre J. Issa
Administrative support: Hagop Kantarjian, Jean-Pierre J. Issa
Provision of study materials or patients: Hagop Kantarjian, Sherry Pierce, Hussain Saba, Eli Estey, Jean-Pierre J. Issa
Collection and assembly of data: Lanlan Shen, Yi Guo, Donald Berry, Saira Ahmed, Wei Zhu, Sherry Pierce, Yutaka Kondo, Yasuhiro Oki, Jaroslav Jelinek
Data analysis and interpretation: Lanlan Shen, Yi Guo, E. Lin, Jianqin Shan, Xuelin Huang, Donald Berry, Yutaka Kondo, Yasuhiro Oki, Jean-Pierre J. Issa
Manuscript writing: Lanlan Shen, Hagop Kantarjian, Jean-Pierre J. Issa
Final approval of manuscript: Lanlan Shen, Hagop Kantarjian, Yi Guo, E. Lin, Jianqin Shan, Xuelin Huang, Donald Berry, Saira Ahmed, Wei Zhu, Sherry Pierce, Yutaka Kondo, Yasuhiro Oki, Jaroslav Jelinek, Hussain Saba, Eli Estey, Jean-Pierre J. Issa
REFERENCES
- 1.Gilliland DG. Hematologic malignancies. Curr Opin Hematol. 2001;8:189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Komrokji RS, Bennett JM. Evolving classifications of the myelodysplastic syndromes. Curr Opin Hematol. 2007;14:98–105. doi: 10.1097/MOH.0b013e328017f633. [DOI] [PubMed] [Google Scholar]
- 3.Valent P, Horny HP, Bennett JM, et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk Res. 2007;31:727–736. doi: 10.1016/j.leukres.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: Evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 5.Kantarjian H, Issa JP, Rosenfeld CS, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 6.Kantarjian H, Oki Y, Garcia-Manero G, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 7.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2003;17:1813–1819. doi: 10.1038/sj.leu.2403054. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Yao DM, Qian J, et al. Methylation status of fragile histidine triad (FHIT) gene and its clinical impact on prognosis of patients with myelodysplastic syndrome. Leuk Res. 2008;32:1541–1545. doi: 10.1016/j.leukres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Quesnel B, Guillerm G, Vereecque R, et al. Methylation of the p15(INK4b) gene in myelodysplastic syndromes is frequent and acquired during disease progression. Blood. 1998;91:2985–2990. [PubMed] [Google Scholar]
- 12.Tien HF, Tang JH, Tsay W, et al. Methylation of the p15(INK4B) gene in myelodysplastic syndrome: It can be detected early at diagnosis or during disease progression and is highly associated with leukaemic transformation. Br J Haematol. 2001;112:148–154. doi: 10.1046/j.1365-2141.2001.02496.x. [DOI] [PubMed] [Google Scholar]
- 13.Wu SJ, Yao M, Chou WC, et al. Clinical implications of SOCS1 methylation in myelodysplastic syndrome. Br J Haematol. 2006;135:317–323. doi: 10.1111/j.1365-2141.2006.06293.x. [DOI] [PubMed] [Google Scholar]
- 14.Aggerholm A, Holm MS, Guldberg P, et al. Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur J Haematol. 2006;76:23–32. doi: 10.1111/j.1600-0609.2005.00559.x. [DOI] [PubMed] [Google Scholar]
- 15.Grövdal M, Khan R, Aggerholm A, et al. Negative effect of DNA hypermethylation on the outcome of intensive chemotherapy in older patients with high-risk myelodysplastic syndromes and acute myeloid leukemia following myelodysplastic syndrome. Clin Cancer Res. 2007;13:7107–7112. doi: 10.1158/1078-0432.CCR-07-1193. [DOI] [PubMed] [Google Scholar]
- 16.Colella S, Shen L, Baggerly KA, et al. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 17.Cheadle C, Vawter MP, Freed WJ, et al. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyota M, Kopecky KJ, Toyota MO, et al. Methylation profiling in acute myeloid leukemia. Blood. 2001;97:2823–2829. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- 19.Shen L, Toyota M, Kondo Y, et al. Aberrant DNA methylation of p57KIP2 identifies a cell-cycle regulatory pathway with prognostic impact in adult acute lymphocytic leukemia. Blood. 2003;101:4131–4136. doi: 10.1182/blood-2002-08-2466. [DOI] [PubMed] [Google Scholar]
- 20.Boumber YA, Kondo Y, Chen X, et al. RIL, a LIM gene on 5q31, is silenced by methylation in cancer and sensitizes cancer cells to apoptosis. Cancer Res. 2007;67:1997–2005. doi: 10.1158/0008-5472.CAN-06-3093. [DOI] [PubMed] [Google Scholar]
- 21.Jelinek J, Mannari R, Issa JP. Identification of 41 novel promoter-associated CpG islands methylated in leukemias. Blood. 2004;2004:319a. abstr. [Google Scholar]
- 22.Toyota M, Issa JP. Methylated CpG island amplification for methylation analysis and cloning differentially methylated sequences. Methods Mol Biol. 2002;200:101–110. doi: 10.1385/1-59259-182-5:101. [DOI] [PubMed] [Google Scholar]
- 23.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 25.Liang G, Salem CE, Yu MC, et al. DNA methylation differences associated with tumor tissues identified by genome scanning analysis. Genomics. 1998;53:260–268. doi: 10.1006/geno.1998.5502. [DOI] [PubMed] [Google Scholar]
- 26.Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–5442. [PubMed] [Google Scholar]
- 27.Ueki T, Toyota M, Sohn T, et al. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 28.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: Evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turker MS. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin Cancer Biol. 1999;9:329–337. doi: 10.1006/scbi.1999.0133. [DOI] [PubMed] [Google Scholar]
- 30.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res. 1996;56:3655–3658. [PubMed] [Google Scholar]
- 31.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.