Abstract
Claims about the diagnostic or prognostic accuracy of markers often prove disappointing when “discrimination” found between cancers versus normals is due to bias, a systematic difference between compared groups. This article describes a framework to help simplify and organize current problems in marker research by focusing on the role of specimens as a source of bias in observational research and using that focus to address problems and improve reliability. The central idea is that the “fundamental comparison” in research about markers (ie, the comparison done to assess whether a marker discriminates) involves two distinct processes that are “connected” by specimens. If subject selection (first process) creates baseline inequality between groups being compared, then laboratory analysis of specimens (second process) may erroneously find positive results. Although both processes are important, subject selection more fundamentally influences the quality of marker research, because it can hardwire bias into all comparisons in a way that cannot be corrected by any refinement in laboratory analysis. An appreciation of the separateness of these two processes—and placing investigators with appropriate expertise in charge of each—may increase the reliability of research about cancer biomarkers.
INTRODUCTION
Molecular markers for cancer diagnosis and prognosis have been studied for more than 10 years in discovery research, an approach in which there is no need to identify targets a priori.1 Despite sizable investments of time and funding, and despite strong claims in research reports, few new markers have been proven to have clinical value. The slow progress is not simply a result of the normal ebb and flow of science, but rather there seem to be system-wide problems in the process by which we discover and develop markers for cancer.2,3
A key problem of current marker research is that reports of discovery of a high degree of diagnostic or prognostic discrimination often turn out to be wrong because of bias, or systematic inequality of the groups compared. Bias is unintentional, but it can commonly occur because the observational design used in marker research is much more subject to bias than the experimental design (also known as an interventional study or randomized clinical trial [RCT]) used in therapeutic research. After a brief review of bias, we discuss the “fundamental comparison” in a research study and how that comparison is arranged in different research designs. Unlike in an RCT, in observational research an investigator makes critical decisions about subject (human or animal) selection and specimen handling that determine whether the fundamental comparison of specimens is reliable. Sometimes, early events unknown to the laboratory scientist create biased specimens and, inevitably, unreliable findings.
BIAS THREATENS VALIDITY OF STUDY RESULTS
A study is valid if results represent an unbiased estimate of the underlying truth.4 Validity of a clinical research study may be affected by threats of three types: chance, generalizability, and bias.5,6 Bias is the most important3 and can occur at multiple locations in a research study, depending on details of research design, on biology, and on technology.3,5,7–9 Table 1 lists several sources of bias that are particularly important in marker research about diagnosis, prognosis, and response to therapy.
Table 1.
Sources and “Locations” of Bias in Marker Research
| Source of Bias | Location of Bias: Before or After Specimens Are Received in the Laboratory |
Example | |
|---|---|---|---|
| Before | After | ||
| Features of subjects, determined in selection: | X | Cancer subjects are male, whereas control subjects are mainly female. Bias: Assay results may depend on sex. | |
| Age | |||
| Sex | |||
| Comorbid conditions | |||
| Medications | |||
| Specimen collection | X | Cancer specimens come from one clinic, whereas controls come from a different clinic. Bias: Assay results may depend on conditions that differ between clinics. |
|
| Specimen storage and handling | X | X | Cancer specimens are stored for 10 years because it takes longer to collect them, whereas control specimens are collected and stored over 1 year. Bias: Assay results may vary with duration of storage, or with different numbers of thaw-freeze cycles. |
| Specimen analysis | X | Cancer specimens are run on one day, whereas control specimens are run on a different day. Bias: Assay results may depend on day of analysis in a machine that “wanders” over time. |
|
NOTE. The table shows examples of different sources of bias and the location of the bias before or after specimens are received in the laboratory. The list is not exhaustive; other biases may be important, and the biases listed may or may not be important in any given research study, depending on details of biology and technology (ie, what is being measured and how it might be influenced).
Bias may occur before an investigator receives specimens in the laboratory for analysis. In one report in which peptide patterns were said to have nearly 100% sensitivity and specificity for prostate cancer,10 cancer specimens came from a group of men with a mean age of 67 years, whereas control specimens came from a group composed of 58% women, with a mean age of 35 years.11 Although sex and age may not necessarily explain all the differences between the compared groups, they must be prominently considered in claims about discrimination. In another report, a serum test was said to discriminate with nearly 100% accuracy between women with and without ovarian cancer12; however, differences between the kinds of patients studied and the settings where blood from cancers and controls was drawn may have caused differences in levels of the particular analytes measured.13,14 In a report of plasma proteins to identify early Alzheimer's disease, cases came from Europe, whereas controls came from the United States; these differences were not discussed as potential explanations of the observed results.15 In another example, the ability of a blood test discovered to discriminate prostate cancer from noncancer was later reported by the investigators themselves probably to have been biased by sample-related issues, including the longer storage duration of specimens in the cancer group compared with the noncancer group, introducing spurious signal into specimens.16 The investigators concluded that “the results from our previous studies—in which differentiation between prostate cancer and noncancer was demonstrated…. likely had biases in sample selection…. ”16 In an accompanying article, the investigators discussed two kinds of problems that happen before investigators receive specimens. They wrote, “Our analysis uncovered possible sources of storage time variability that arose from different collection protocols,” and they concluded, “These are critical issues often overlooked in the biomarker discovery process that are likely to be the single greatest reason most biomarker discoveries fail to be validated.”17 This kind of attention to detail and candid reporting is to be encouraged. Although these types of problems are common in observational research, investigators may not routinely search for or report them.
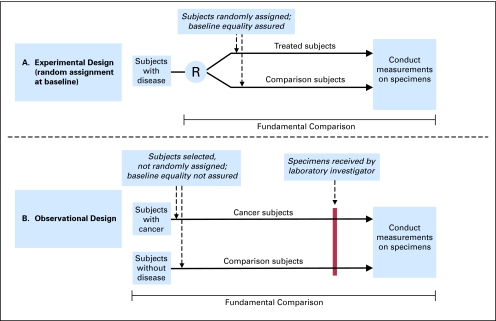
Bias occurring before specimens ever reach an investigator's laboratory (ie, to the left of the red line in Fig 1B) may be especially problematic for two reasons. First, it may simply be invisible to or unappreciated by a laboratory investigator. Second, even if recognized, bias already hardwired in at that point may be impossible to adjust for in subsequent laboratory or statistical analysis.5
Fig 1.
The fundamental comparison in experimental and observational study design. (A) Randomized controlled trial (RCT) of therapy (experimental design). The comparison in an RCT of treatment begins with random allocation of subjects (eg, humans or animals) to the treatment or comparison group, so that baseline equality is assured in the compared groups. The entire fundamental comparison is done under the observation or supervision of the laboratory investigator, making potential sources of bias easier to anticipate and control. Although selection of subjects affects generalizability—“to whom” results may apply—the selection of subjects is not involved in the comparison itself. Specimens (eg, tissue or blood) may be taken from subjects at different times (eg, at the end of the study to assess an outcome of treatment [as shown in the figure]), or just after random allocation to assess prognosis or prediction. (B) Observational study of diagnosis. The comparison begins with subject selection that is not randomized; the process of subject selection may, itself, introduce bias, or systematic differences between the compared groups. This kind of bias, occurring before specimens ever reach the laboratory, may be totally unknown to the laboratory investigator and may fatally flaw any comparison done in the investigator's laboratory. R, random allocation.
Bias may also occur after specimens are received in the laboratory (ie, to the right of the red line in Fig 1B). A study reported that a serum peptide pattern derived in a training set of specimens could identify ovarian cancer with nearly 100% sensitivity and specificity in an independent validation set.18 After two related data sets produced by the same investigators at later times were made available to the public through unrestricted Web access, all three data sets were reanalyzed by Baggerly et al,19 who concluded that baseline correction prevented reproduction of the original results.19 Their troubleshooting approaches suggested that discrimination could have been due to bias related to instrument calibration or artifact. They concluded, “ Taken together, these and other concerns suggest that much of the structure uncovered in these experiments could be due to artifacts of sample processing, not to the underlying biology of cancer.”19
The examples above come mainly from the field of proteomics for cancer diagnosis because problems related to bias are well documented in the literature11,14,16,17,19,20; however, similar problems may occur in discovery in other “-omics” fields and in any marker research,5 and in studies of diagnostic tests in general,21 because such studies must use observational (nonexperimental) designs that are inherently more challenging and more subject to bias than the experimental design. In contrast, the field of research methods used to discover and develop drugs is better developed3 than for the field of markers, in large part because the experimental design provides such strength in avoiding bias. The larger topic of observational versus experimental research is extensively covered in journal articles and textbooks about epidemiology, biostatistics, and research design.22–24
Methodologic issues in marker research have been discussed in general reviews,3,5,6,25–29 rules of thumb,30 guidelines for reporting,31–35 guidelines for quality requirements in use of markers,36 and use of phases to organize biomarker development.25,37 Although these discussions provide important perspectives and details, the field is short on clear, practical organizing themes. Ideas and principles from that larger field, when focused on potential sources of bias in specimens, may explain how one seemingly small detail of a research study may lead to fatal bias in results, and on how those ideas and principles may point to a solution.
THE FUNDAMENTAL COMPARISON
The fundamental comparison refers to the process of arranging and analyzing groups of subjects and specimens to learn whether a possible cause is responsible for a difference observed in the compared groups. Depending on how the process of the fundamental comparison is arranged, results of a comparison will be valid or strong (represents fairly the underlying reality), or they may be unreliable because of bias. Bias—a systematic difference between the compared groups—tends to produce results that are positive but do not reflect an underlying reality and are not reproducible.
For a study to be valid or strong, having an unbiased fundamental comparison is obviously necessary. A study's overall strength depends on other things as well, such as an investigator's insight and creativity about features such as what intervention or cause will be assessed (eg, a newly developed therapy or genetic mutation), what subjects and specimens will be used (eg, a new animal model), and what outcome will be assessed (survival or response to therapy). However, insight and creativity cannot overcome or avoid the need for a fair comparison. Regardless of the degree of an investigator's creativity and insight, a biased comparison may produce misleading results.
SUBJECTS, SPECIMENS, AND STUDY DESIGN
Subject selection ultimately determines whether specimens are “strongly unbiased”3 or have high enough quality to be used for a comparison that is reliable. Because laboratory investigators may have little knowledge about selection methods, they may be unaware of fatal flaws producing important biases in specimens they receive for analysis. The role that subject selection plays depends on the location where the fundamental comparison begins in each design—in other words, before or after specimens are received in the laboratory (Fig 1).
In experimental research, the fundamental comparison begins when subjects are randomly assigned to the compared groups (Fig 1A). The purpose of random assignment is to assure there are no systematic differences in the compared groups at baseline. Random assignment addresses many problems in subject selection and specimen processing that can lead to bias. Random assignment ensures baseline equality in the fundamental comparison, and specimens collected at the time of randomization can be processed before the outcome is known, further helping to avoid bias.
In contrast, in observational research, subjects are selected and specimens are collected before reaching the investigator's laboratory, so that systematic differences may already exist at baseline between the compared groups, as illustrated in Figure 1B. In observational research, the processes of subject selection and specimen collection have become a critical part of the fundamental comparison itself. The next sections discuss details of these differences in design and how to improve this aspect of marker research.
Randomized Study of Treatment (experimental design)
The goal of an experiment is to assess whether an agent (like a drug or induced genetic mutation) is the cause of some effect. Randomization organizes the fundamental comparison in a way that keeps all other factors equal except the cause, so that measured effects will be unbiased and not explained by incidental factors. In an experiment, the choice of subjects has no direct effect on the baseline comparison. The choice of course affects the generalizability of results, or to whom results may apply.5 If the comparison of treatment versus control is conducted using one strain of mice, results might not apply to another strain or to human beings. But the baseline comparison is at least fair and reliable regardless of what subjects are chosen, except in rare instances when randomization does not work or is actually subverted.38
A strength of the experimental design—and a reason it is so reliable—is that the entire comparison can be arranged and supervised by the investigator, allowing powerful preemptive measures to be taken to avoid bias. As illustrated in Figure 1 and as stated by Potter, “The distinction between observation and experiment rests on whether the researcher is in charge of the differences in the initial conditions between the two compared entities.”39 Design might further include blinding subjects and study personnel to treatment status (double-blind design) to avoid biases at later steps. An investigator may decide not to implement some design features, or some features may not be possible (eg, in oncology studies, one cannot blind to radiation therapy versus surgery). In animal studies, conducting randomization and blinding is easier than in human studies, but investigators still may not use those techniques. The entire field of mouse model experiments to study amyotrophic lateral sclerosis has been said to be compromised by unreliable comparisons in nonrandomized studies without blinded evaluation of outcomes.40 Ultimately any study's reliability is determined by investigators' choices about critical details of research design and conduct.
The importance of the baseline equality achieved by randomization cannot be overstated. Randomization appears in the name of the research design that is strongest to study treatment or etiology: RCT. Journals routinely require investigators to report results of randomization in a table, so readers can see that baseline inequality was not the cause of the difference between groups in results. Even if every difference at baseline could be annotated accurately and in detail, there is no convincing method of statistical analysis to solve the problem of baseline inequality. As noted by Norman Breslow, a biostatistician, the problem is “… the fundamental quality of the data, and to what extent are there biases in the data that cannot be controlled by statistical analysis[.] One of the dangers of having all these fancy mathematical techniques is people will think they have been able to control for things that are inherently not controllable.”41
Nonrandomized Study of Diagnostic Test (observational design)
Understanding and addressing the problem of baseline equality is perhaps the most important challenge in observational research about markers for diagnosis and prognosis. Unlike in the experimental approach, the comparison in an observational study of diagnosis (or prognosis) begins during the process of subject selection—a process that may be totally outside the observation or supervision of the laboratory investigator.
Although arranging a meticulous comparison is obviously critical when evaluating a drug therapy, a laboratory investigator might fairly ask whether such a careful comparison is important in basic or biologic research that has no immediate clinical consequence. A strong case can be made that reliable results are important in any research, whether basic or clinical, if that result provides the basis for investing in some kind of additional work. A weak foundation may lead to wasted effort. Before approval of a $104 million proteomics initiative to develop improved technology, computational methods, and standardized reagents for proteomics studies,42 concern was raised about whether the preliminary results showing that the technology can discriminate diagnostically might be unreliable, in which case investment might not be warranted.42,43
Understanding the nonexperimental (observational) designs used in the field of marker studies is particularly difficult because many different designs are used, they may not be as easy to diagram as experimental or RCT research, and because sources of bias are more difficult to identify and manage. Designs for studies of diagnostic accuracy can involve, as explained by Knottnerus and Muris, “1 survey of the total study population, 2 case-referent approach, or 3 test-based enrollment.”21 Even the basic approaches to data collection may differ dramatically: “Data collection should generally be prospective, but ambispective [retrospective reference group is used as a control group, but remainder of study is prospective] and retrospective approaches are sometimes appropriate.”21 This translates into practical challenges for research methodologists and clinical researchers.
A special case of observational design is the nested case-control design (recently termed the PRoBE approach44) in which specimens are collected prospectively (specimens are collected before the diagnosis [or prognosis] is known) and later undergo retrospective blinded evaluation.44 Sometimes specimens may have been prospectively collected and already exist in a specimen bank that can be a source for a nested case-control analysis.3 The approach can help minimize the problem of baseline inequality because, as specimens are collected before diagnosis (or prognosis) is known, the presence or absence of disease cannot affect selection of subjects or handling of specimens. The unique recommendation of the PRoBE approach is that minimally acceptable performance standards for the true-positive rate and false-positive rate are defined before the study is conducted, taking into account the clinical application of the marker (eg, diagnostic v prognostic use).
The nested case-control approach is not new, and its strengths have been discussed elsewhere.3,45 In a study of breast cancer prognosis, tissue specimens collected before prognosis was known were retrospectively analyzed using already-collected tissue from the National Cancer Institute's (NCI's) National Surgical Adjuvant Breast and Bowel Project clinical trial B-1446; the positive result from that study provided the basis for introduction of the OncotypeDx test into clinical practice. In a study of stool DNA markers to screen for colon cancer, specimens were collected prospectively before colonoscopy and were analyzed retrospectively, blinded to diagnostic status.47 The stool test was substantially better than fecal occult blood testing,47 but the degree of discrimination was considered too modest, considering cost, to warrant implementation at that time.48 In a study of serum proteomics to diagnose colon adenomas, serum specimens were collected before the colonoscopy that established the diagnosis, and specimens were analyzed retrospectively and blinded.49 In this study, no discrimination was found, but the result seemed to be reliable because of the strength of the research design. Last, in a major ongoing study of serum proteomics to diagnose ovarian cancer, serum specimens collected in NCI's screening trial of prostate, lung, colon, and ovarian cancer are being analyzed retrospectively and in a blinded manner by multiple laboratories.50 This study's results, when available, will arguably provide the strongest evidence to date about a how well serum proteomics technology can diagnose ovarian cancer—a critically important issue for the entire field of serum proteomics, given the magnitude of investment and claims, particularly for ovarian cancer.
APPROACHES TO IMPROVE THE SITUATION
The following approaches may help simplify, organize, and improve research about markers for cancer diagnosis and prognosis.
Understand That Subject Selection Is Part of the Fundamental Comparison
Our first goal should be for every investigator to understand the critical role of subject selection in the fundamental comparison of every observational study. Even if subject selection does not seem like part of a study, the process will need to be reported in detail in a research report, so that potential biases in specimens can be identified. Indeed, current guidelines prescribing which details of study design should be reported in research about diagnosis (eg, Standards for Reporting of Diagnostic Accuracy [STARD]34) or prognosis (eg, Reporting Recommendations for Tumor Marker Prognostic Studies [REMARK]31) focus mainly on events that happen to the left of the red line (Fig 1) because of the fatal flaws in comparison that can occur. If investigators fail to assess details until the end—after the laboratory work is completed—they may find out too late that baseline inequality exists because the specimens were fatally biased to begin with. For this reason, investigators should learn, in advance of doing any laboratory analysis, enough about the features and history of specimens to decide whether specimens may be so flawed that the laboratory work should not even be conducted. Minor flaws must be appropriately understood, managed, and discussed in interpretation of results. Although an understanding of the effect of subject selection on baseline equality may be second nature to persons experienced in epidemiology, biostatistics, and observational research design, it may be totally outside the experience of laboratory investigators. In contrast, bias to right of the red line (Fig 1B) may be relatively easy to deal with because what happens can be directly observed and can often be corrected by refinements in laboratory technique.
Improve Late-Phase Research (validation)
The intended use of the PRoBE approach, as proposed by its authors, is in a pivotal or late-phase study done just before doing a (usually very expensive) RCT in which marker results will be used to direct a therapy or other intervention to improve the outcome.20,44 Such a study would determine whether the test discriminates among those subjects in whom an intervention would be relevant, for example, persons with asymptomatic screen-detected cancer. A nested case-control study may provide the least-biased late-phase observational assessment of a test's diagnostic discrimination before doing the RCT that assesses the combined effect of early detection and intervention. Although this study design may address the critical bias of baseline inequality, it does not necessarily address other problems; for example, the nested case-control study of stool DNA markers described above47 used specimen storage conditions suitable to preserve DNA mutations, but not adequate for a newly developed DNA integrity assay.47,51
Late-phase studies in general may be expensive and cumbersome, and the potential benefits must be weighed against costs. For example, a study of colorectal cancer screening involving stool collection and colonoscopy in asymptomatic, average-risk subjects required enrolling more than 5,000 persons to yield 31 cancers, at a cost of more than $10 million.47 However, that may still be money well invested if it avoids false leads that trigger other studies consuming millions more. In other examples, specimens may be collected as part of multimillion-dollar RCTs enrolling large numbers of subjects.46,50 The degree of effort required may be worth the cost, but investment of such magnitude requires serious deliberation.
Improve Early-Phase Research (discovery)
The reality in 2010 is that very few markers ever become candidates for a pivotal study and subsequent RCT. The main problem currently is not that promising candidates that fail in RCTs could have been weeded out by a well-done nested case-control study beforehand. The problem is that discovery is so weak, because of bias, that research results are not reliable and cannot be reproduced.
It is not clear, however, that a nested case-control approach can be used in early-phase or discovery research that is often (but not always) done on subjects with advanced disease and who are symptomatic. The approach cannot be meaningfully applied if diagnosis is already known. In some circumstances, it may be possible to use the approach in early-phase or discovery research if discovery can be done with specimens collected from asymptomatic subjects (often with early-stage disease) before diagnosis is known. It may even be possible to use the same larger set of specimens for both discovery (to derive analytes or patterns that may discriminate) and validation3 (to show that discrimination occurs in samples totally independent of those used in discovery3,6,44). For example, in the study of stool DNA markers described above,47 the main goal was to use specimens in late-phase validation of markers discovered previously in research using more advanced-stage disease and from patient groups where the diagnosis was already known.52,53 A secondary goal of that study was to create a specimen bank of aliquots to be used later for discovery of markers developed in the future. Unfortunately, one of the assays required using all available aliquots, and the planned bank was depleted. In another study of serum markers for ovarian cancer, specimens banked in NCI's prostate, lung, colon, and ovary study50 are being used both in validation and discovery (C. Berg, personal communication, August 2009). These examples illustrate how specimens of appropriate quality might be used for both discovery and validation.3
National Efforts to Improve Cancer Marker Discovery Methods
Some national efforts are being undertaken to create or improve specimen banks that can be used for development of markers of diagnosis and prognosis, for example in NCI's Early Detection Research Network54 and Office of Biorepositories and Biospecimen Research.55,56 Although these efforts devote substantial attention to standard operating procedures54–56 for events that happen to the right of the red line after an investigator receives samples, we suggest that similar attention must be paid to events that occur to the left of the red line. One way to provide that attention is to add to standard procedures a process that involves suitable expertise, from the fields of epidemiology, biostatistics, and clinical research design, to review study design on the left side of the red line. One part of that process should be to consider whether the design of subject selection and specimen collection will allow a reliable answer to some specific proposed research question about diagnosis or prognosis. This kind of expertise and process was instrumental in the design and interpretation of the successful studies discussed above.46,47,49,50
Consider Separating Clinical and Laboratory Processes, With Specimens as the Handoff Point
Many investigators conducting early discovery research understand and enjoy the process of laboratory research more than the process of clinical research. Because this preference may sometimes be a source of problems in effective communication or collaboration, it may be advantageous to deliberately separate the two processes involved in the fundamental comparison instead of requiring laboratory researchers to understand details of specimen collection and requiring clinical researchers to understand details of laboratory methods. In this formulation, clinical researchers, epidemiologists, and biostatisticians would focus on the first process—research design including specimen collection to ensure high-quality or strongly unbiased specimens, then specimens would be “handed off ” by the clinical research group to the laboratory group for the second process—laboratory analysis. Of course, some degree of cross-talk among collaborators and across the divide would be essential, but the success and efficiency of marker research might be enhanced by emphasizing separation, except among highly trained and highly dedicated experts who can successfully complete both processes in their research group.
In conclusion, because studies of cancer markers for diagnosis and prognosis are observational, not experimental, the process of subject selection is a critical part of the fundamental comparison. Understanding how to best manage this process in the design and conduct of marker research, especially in early discovery, is still in its infancy relative to other areas of observational epidemiology research. A major problem in current biomarker discovery research is baseline inequality of the specimen groups that are compared in laboratory analysis, originating from flawed subject selection earlier in the study. Understanding the role of specimens—as a product of one process (subject selection) in the fundamental comparison and the substrate for the second process (laboratory analysis)—may help simplify and strengthen the process of discovery and validation of biomarker research. Sufficient attention to each process, and perhaps a division of labor between clinical and laboratory researchers, may help improve the reliability of biomarker research.
Acknowledgment
We thank the Early Detection Research Network, the Clinical Proteomic Technology Assessment for Cancer, the Early Detection Research Group, and the Biometry Research Group of the National Cancer Institute; many ideas were developed through participation in activities of these organizations.
Footnotes
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: David F. Ransohoff
Manuscript writing: David F. Ransohoff, Margaret L. Gourlay
Final approval of manuscript: David F. Ransohoff, Margaret L. Gourlay
REFERENCES
- 1.Ransohoff DF. Developing molecular biomarkers for cancer. Science. 2003;299:1679–1680. doi: 10.1126/science.1083158. [DOI] [PubMed] [Google Scholar]
- 2.Ransohoff DF. The process to discover and develop biomarkers for cancer: A work in progress. J Natl Cancer Inst. 2008;100:1419–1420. doi: 10.1093/jnci/djn339. [DOI] [PubMed] [Google Scholar]
- 3.Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: Roles of phases, guidelines, and study design. J Clin Epidemiol. 2007;60:1205–1219. doi: 10.1016/j.jclinepi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Guyatt G, Rennie D, editors. Users' Guides to the Medical Literature: Essentials of Evidence-Based Clinical Practice. Chicago, IL: AMA Press; 2002. [Google Scholar]
- 5.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 6.Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4:309–314. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 7.Feinstein AR. Philadelphia, PA: WB Saunders; 1985. Clinical Epidemiology: The Architecture of Clinical Research. [Google Scholar]
- 8.Fletcher RH, Fletcher SW, Wagner EH. Clinical Epidemiology: The Essentials. ed 3. Baltimore, MD: Williams & Wilkins; 1996. [Google Scholar]
- 9.Hulley SB, Cummings SR, Browner WS, et al. Designing Clinical Research: An Epidemiologic Approach. ed 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 10.Villanueva J, Shaffer DR, Philip J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamandis EP, Kulasingam V, Sardana G. Letter to the editor about Differential exoprotease activities confer tumor-specific serum peptidome. J Clin Invest. [Eletter published on February 21, 2006] http://www.jci.org/eletters/view/26022#sec2. [Google Scholar]
- 12.Visintin I, Feng Z, Longton G, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14:1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 13.Food and Drug Administration Office of In Vitro Diagnostic Device Evaluation and Safety: OvaSure Manufacturer Letter A, 2008. http://www.fda.gov/ICECI/EnforcementActions/WarningLetters/2008/ucm1048114.htm.
- 14.McIntosh M, Anderson G, Drescher C, et al. Ovarian cancer early detection claims are biased. Clin Cancer Res. 2008;14:7574. doi: 10.1158/1078-0432.CCR-08-0623. [DOI] [PubMed] [Google Scholar]
- 15.Ray S, Britschgi M, Herbert C, et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 16.McLerran D, Grizzle WE, Feng Z, et al. SELDI-TOF MS whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin Chem. 2008;54:53–60. doi: 10.1373/clinchem.2007.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLerran D, Grizzle WE, Feng Z, et al. Analytical validation of serum proteomic profiling for diagnosis of prostate cancer: Sources of sample bias. Clin Chem. 2008;54:44–52. doi: 10.1373/clinchem.2007.091470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 19.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: Comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 20.Feng Z, Kagan J, Srivastava S. Toward a robust system for biomarker triage and validation—EDRN experience. In: Ankerst DP, Tangen CM, Thompson IM Jr, editors. Prostate Cancer Screening. ed 2. Philadelphia, PA: Humana; 2009. pp. 297–306. [Google Scholar]
- 21.Knottnerus JA, Muris JW. Assessment of the accuracy of diagnostic tests: The cross-sectional study. J Clin Epidemiol. 2003;56:1118–1128. doi: 10.1016/s0895-4356(03)00206-3. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan Pepe M. Oxford, England: Oxford University Press; 2003. The Statistical Evaluation of Medical Tests for Classification and Prediction. [Google Scholar]
- 23.Sackett DL, Haynes RB, Tugwell P, et al. Clinical Epidemiology: A Basic Science for Clinical Medicine. ed 2. Boston, MA: Little, Brown and Company; 1991. [Google Scholar]
- 24.Hennekens CH, Buring JE. Epidemiology in Medicine. Boston, MA: Little, Brown and Company; 1987. [Google Scholar]
- 25.Baker SG, Kramer BS, McIntosh M, et al. Evaluating markers for the early detection of cancer: Overview of study designs and methods. Clin Trials (London) 2006;3:43–56. doi: 10.1191/1740774506cn130oa. [DOI] [PubMed] [Google Scholar]
- 26.Simon R, Radmacher MD, Dobbin K, et al. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 27.Contopoulos-Ioannidis DG, Ntzani E, Ioannidis JP. Translation of highly promising basic science research into clinical applications. Am J Med. 2003;114:477–484. doi: 10.1016/s0002-9343(03)00013-5. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis JP. Microarrays and molecular research: Noise discovery? Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- 29.Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: An empirical assessment. Lancet. 2003;362:1439–1444. doi: 10.1016/S0140-6736(03)14686-7. [DOI] [PubMed] [Google Scholar]
- 30.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 31.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 32.Taylor CF, Paton NW, Lilley KS, et al. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 33.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 34.Bossuyt PM, Reitsma JB, Bruns DE, et al. The STARD statement for reporting studies of diagnostic accuracy: Explanation and elaboration. Ann Intern Med. 2003;138:W1–12. doi: 10.7326/0003-4819-138-1-200301070-00012-w1. [DOI] [PubMed] [Google Scholar]
- 35.Diamandis EP. Proteomic patterns to identify ovarian cancer: 3 years on. Expert Rev Mol Diagn. 2004;4:575–577. doi: 10.1586/14737159.4.5.575. [DOI] [PubMed] [Google Scholar]
- 36.Sturgeon CM, Hoffman BR, Chan DW, et al. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for use of tumor markers in clinical practice: Quality requirements. Clin Chem. 2008;54:e1–e10. doi: 10.1373/clinchem.2007.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe M, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 38.Berger V. Hoboken, NJ: Wiley; 2005. Selection Bias and Covariate Imbalances in Randomized Clinical Trials. [DOI] [PubMed] [Google Scholar]
- 39.Potter JD. Epidemiology informing clinical practice: From bills of mortality to population laboratories. Nat Clin Pract Oncol. 2005;2:625–634. doi: 10.1038/ncponc0359. [DOI] [PubMed] [Google Scholar]
- 40.Schnabel J. Neuroscience: Standard model. Nature. 2008;454:682–685. doi: 10.1038/454682a. [DOI] [PubMed] [Google Scholar]
- 41.Taubes G. Epidemiology faces its limits. Science. 1995;269:164–169. doi: 10.1126/science.7618077. [DOI] [PubMed] [Google Scholar]
- 42.Hede K. $104 million proteomics initiative gets green light. J Nat Can Inst. 2005;97:1324–1325. doi: 10.1093/jnci/dji325. [DOI] [PubMed] [Google Scholar]
- 43.Goldberg KB. Advisors reject NCI's $89 million plan for proteomics as too much, too soon. Cancer Lett. 2005;31:1–10. [Google Scholar]
- 44.Pepe MS, Feng Z, Janes H, et al. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: Standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker SG, Kramer BS, Srivastava S. Markers for early detection of cancer: Statistical guidelines for nested case-control studies. BMC Med Res Methodol. 2002;2:4. doi: 10.1186/1471-2288-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 47.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 48.Woolf SH. A smarter strategy? Reflections on fecal DNA screening for colorectal cancer. N Engl J Med. 2004;351:2755–2758. doi: 10.1056/NEJMe048259. [DOI] [PubMed] [Google Scholar]
- 49.Ransohoff DF, Martin C, Wiggins WS, et al. Assessment of serum proteomics to detect large colon adenomas. Cancer Epidemiol Biomarkers Prev. 2008;17:2188–2193. doi: 10.1158/1055-9965.EPI-07-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winstead ER. Ovarian cancer study could speed early detection. NCI Cancer Bull. 2008;5:5–7. [Google Scholar]
- 51.Itzkowitz SH, Jandorf L, Brand R, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Ahlquist DA, Skoletsky JE, Boynton KA, et al. Colorectal cancer screening by detection of altered human DNA in stool: Feasibility of a multitarget assay panel. Gastroenterology. 2000;119:1219–1227. doi: 10.1053/gast.2000.19580. [DOI] [PubMed] [Google Scholar]
- 53.Boynton KA, Summerhayes IC, Ahlquist DA, et al. DNA integrity as a potential marker for stool-based detection of colorectal cancer. Clin Chem. 2003;49:1058–1065. doi: 10.1373/49.7.1058. [DOI] [PubMed] [Google Scholar]
- 54.Tuck MK, Chan DW, Chia D, et al. Standard operating procedures for serum and plasma collection: Early Detection Research Network Consensus Statement Standard Operating Procedure Integration Working Group. J Proteome Res. 2009;8:113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Cancer Institute Office of Biorepositories and Biospecimen Research: National Cancer Institute best practices for biospecimen resources. US Department of Health and Human Services, June 2007. http://biospecimens.cancer.gov/practices/
- 56.Bethesda, MD: National Cancer Institute; 2008. National Cancer Institute, National Institutes of Health, US Department of Health and Human Services: An annual plan and budget proposal; fiscal year 2010. NIH Publication No. 08-6363. [Google Scholar]