Abstract
Aberrant signaling causes many diseases, and manipulating signaling pathways with kinase inhibitors has emerged as a promising area of drug research. Most kinase inhibitors target the conserved ATP-binding pocket; therefore specificity is a major concern. Proteomics has previously been used to identify the direct targets of kinase inhibitors upon affinity purification from cellular extracts. Here we introduce a complementary approach to evaluate the effects of kinase inhibitors on the entire cell signaling network. We used triple labeling SILAC (stable isotope labeling by amino acids in cell culture) to compare cellular phosphorylation levels for control, epidermal growth factor stimulus, and growth factor combined with kinase inhibitors. Of thousands of phosphopeptides, less than 10% had a response pattern indicative of targets of U0126 and SB202190, two widely used MAPK inhibitors. Interestingly, 83% of the growth factor-induced phosphorylation events were affected by either or both inhibitors, showing quantitatively that early signaling processes are predominantly transmitted through the MAPK cascades. In contrast to MAPK inhibitors, dasatinib, a clinical drug directed against BCR-ABL, which is the cause of chronic myelogenous leukemia, affected nearly 1,000 phosphopeptides. In addition to the proximal effects on ABL and its immediate targets, dasatinib broadly affected the downstream MAPK pathways. Pathway mapping of regulated sites implicated a variety of cellular functions, such as chromosome remodeling, RNA splicing, and cytoskeletal organization, some of which have been described in the literature before. Our assay is streamlined and generic and could become a useful tool in kinase drug development.
The advent of Gleevec® (imatinib) less than 10 years ago was a landmark for utilizing small molecule compounds as kinase inhibitor drugs (1–3). This type of drug is usually directed against one specific kinase whose malfunctioning plays a key role in the given disease. Generally these drugs are thought to be selective, easy to modify, and effective. As the molecular principles of various diseases are better understood, kinase inhibitors are being developed in various fields with cancer remaining the predominant one (4). Kinase inhibitor compounds constitute about 30% of all drug development programs in the pharmaceutical industry (5).
Kinase inhibitor drugs are typically developed with a targeted and “rational” strategy, often focusing on a kinase known to be involved in the etiology of a disease. Large libraries of chemical compounds, for example ATP analogs, are screened in vitro against the activity of this kinase, and their effects on a panel of manually selected kinases with similar sequences or structures are evaluated to assess specificity (6, 7). A few promising leads are then selected for further improvement. In recent years, high throughput technologies have been introduced to speed up these enzyme assays. Innovations include the phage display assay (8, 9), yeast three-hybrid assay (10), and chemical proteomics assay (11, 12). These methods achieve better coverage of the kinome and thus provide less biased results.
Although these in vitro assays are very informative, they have several limitations. First, chemical or genetic modifications are often required, such as generating fusion proteins or adding chemical linkers to the inhibitor, which may change the binding properties of the kinases and the inhibitor compounds. Second, these methods investigate the direct binding targets of the inhibitor compounds but do not determine their influence on the entire cellular signaling network. As more and more kinases are proven to function in multiple signaling pathways, inhibitor compounds may influence cellular functions that are not easily predicted. Third, cancer cells are notoriously known to evolve point mutations or to activate alternative signaling proteins to escape drug inhibition (13, 14). Therefore, the concept of utilizing multiple kinase inhibitors is increasingly established in the clinic (15, 16). This has complicated drug evaluation as different inhibitor compounds can generate synergistic or counteracting effects. Certainly a whole cell-based approach, which allows a systems-wide elucidation of inhibitor function, should improve the target evaluation process and help to monitor drug effects in vivo.
Increasingly powerful imaging methods can, in principle, provide a comprehensive assessment of signaling pathways. Multiplexed fluorescence provides direct visualization of localization and activities of the selected pathway molecules in vivo after kinase inhibition (17–20). However, imaging methods require hundreds or thousands of experiments to cover all molecules of interest. In contrast, quantitative mass spectrometry is able to measure protein expression and modification events in single experiments at a global level and in a simultaneous manner. Stable isotope labeling by amino acids in cell culture (SILAC)1 generates completely labeled cell populations that are otherwise equal to non-labeled cells (21, 22). This system enables a direct and large-scale comparison of several cell populations with different biological or chemical treatments (23–25). When SILAC was used to study the effect of the HER2 kinase inhibitor PD168393, changes of the tyrosine phosphorylated proteins could be quantified (26). In recent years, studies of phosphorylation at a site-specific level have been greatly enhanced by progress in MS instrumentation and algorithms. Combined with key advances in phosphopeptide enrichment methods, such as immobilized metal ion affinity chromatography (IMAC) and titanium dioxide (TiO2) chromatography, this has enabled detection and quantitation of thousands of phosphorylation sites, completely changing the capabilities of the phosphoproteomics field (27–34).
Chronic myelogenous leukemia (CML) is one of the diseases caused by constitutively active signaling and is characterized by overproliferation of myeloid cells. The fundamental principle of its etiology is the fusion of chromosomes 9 and 22 to produce the so-called Philadelphia chromosome and the constitutively activated tyrosine kinase BCR-ABL fusion protein (35). Treatment of CML has been greatly advanced by small inhibitor compounds that selectively inhibit the kinase activity of BCR-ABL. The striking success of the first BCR-ABL inhibitor drug Gleevec, or imatinib, proved the concept of using small kinase inhibitor compounds as drugs (36). Later, a second generation of drugs was developed to inhibit Gleevec-resistant, point-mutated versions of BCR-ABL. Among these is dasatinib, a highly potent, orally active inhibitor for both inactive and active BCR-ABL that inhibits most BCR-ABL variants found in CML patients (37, 38).
Dasatinib binds to the kinase domain of ABL kinase. It has similar potency toward SRC family kinases and the platelet-derived growth factor receptor family (39). Research into the mechanism of action of dasatinib focuses on two major themes: the direct binding targets and more downstream signaling molecules. Mass spectrometry has played an important role in these investigations. Recently Goss et al. (40) analyzed immunoprecipitated proteins by tandem MS and derived a common phosphotyrosine signature for BCR-ABL in six different CML cell lines. Hantschel et al. (41) and Bantscheff et al. (12) combined affinity purification techniques with quantitative MS to screen for binding targets of dasatinib. In the study of Bantscheff et al. (12) several broad band kinase inhibitors were combined and immobilized on one affinity resin (kinobeads). Different kinase inhibitor compounds were then used to compete with the unspecific interaction used in immobilization. The kinase beads covered 65% of the phylogenetic human kinome tree.
We reasoned that the combination of SILAC and state of the art phosphoproteomics techniques should provide an excellent tool to explore the effects of kinase inhibitors on individual phosphorylation sites and on the entire cellular network of signal transduction. We first examined two inhibitor compounds widely used in signal transduction laboratories: U0126 inhibits MEK1/2, and SB202190 inhibits p38α/β MAPK. We then applied the same technique to determine the effects of dasatinib, a clinical drug for inhibition of mutated BCR-ABL in CML, on the phosphoproteome.
EXPERIMENTAL PROCEDURES
SILAC Labeling and Cell Culture
HeLa cells (from the American Type Culture Collection (ATCC)) were cultured in DMEM (4.5 g/liter glucose)-based medium. K562 cells (kindly provided by Axel Ullrich) were cultured in suspension in RPMI-based medium. To generate triple encoding SILAC conditions, normal medium deficient in arginine and lysine (from Invitrogen) was supplemented with stable isotope-encoded arginine and lysine (Sigma-Aldrich). For “heavy” labeling we used l-[13C6,15N4]arginine (Arg10) and l-[13C6,15N2]lysine (Lys8), for “medium” labeling, l-[13C6]arginine (Arg6) and l-[2H4]lysine (Lys4) were used, and for the “light” condition l-[12C6,14N4]arginine (Arg0) and l-[12C6,14N2]lysine (Lys0) were used. Final concentrations of arginine were 28 mg/liter in DMEM and 84 mg/liter in RPMI. Final concentrations of lysine were 73 mg/liter in DMEM and 49 mg/liter in RPMI. In each SILAC condition, medium was supplemented with 10% dialyzed fetal bovine serum with 10-kDa cutoff, 1% (10 mg/ml) streptomycin/(10,000 units/ml) penicillin, and 1% l-glutamine (200 mm in 0.85% NaCl; all from Invitrogen). General cell culture conditions were 37 °C, 5% CO2, and humidified atmosphere.
HeLa cells were seeded with 20–30% density and split or harvested when cell density reached 90%. K562 cells were seeded at 0.2 × 106 cells/ml and were split or harvested at 1–1.2 × 106 cells/ml.
Cell Stimulation, Harvest, and Protein Recovery
The 10 mm stock solution of U0126 (from Promega), SB202190 (from Sigma-Aldrich), or dasatinib was prepared by dissolving each compound in DMSO. The U0126 and SB202190 experiments were performed identically but separately on a split cell population. To obtain 20 mg of starting material, four 15-cm cell culture dishes of 90% confluent cells per condition were prepared for each triple SILAC experiment. HeLa cells were serum-starved for 16 h. Heavy (Arg10, Lys8) HeLa cells were stimulated with 10 μm kinase inhibitor (U0126 or SB202190) for 20 min. Then, in the presence of the inhibitor compound, 150 ng/ml EGF (from Millipore) was added to the medium for another 15 min. Medium (Arg6, Lys4) HeLa cells were stimulated with 150 ng/ml EGF for 15 min. Light (Arg0, Lys0) labeled HeLa cells were left untreated.
In the dasatinib experiment, medium (Arg6, Lys4) and heavy (Arg10, Lys8) labeled K562 cells were treated with 5 and 50 nm dasatinib for 1 h, respectively. Light (Arg0, Lys0) K562 control cells were treated with DMSO for 1 h. To obtain 20 mg of starting material, ∼5 dishes (inner diameter, 15 cm) of cells per labeling condition were prepared for the triple SILAC experiment.
Cells were lysed in modified radioimmune precipitation assay buffer and harvested essentially as described (42). After centrifugation, to recover proteins from the supernatant, we added 4 volumes of ice-cold acetone. These precipitated proteins were collected by centrifugation and dissolved in an 8 m urea buffer. To recover proteins from the pellet, cellular debris were digested with Benzonase (from Merck) in urea buffer containing 6 m urea, 2 m thiourea, and 10 mm Hepes, pH 7.5. The concentration of the dissolved proteins was measured by the Bradford method (A590 nm). Equal amounts of protein from each SILAC condition were mixed.
Peptide Fractionation by Strong Cation Exchange (SCX) and Phosphopeptide Enrichment by TiO2
In each SILAC experiment, around 20 mg of protein were obtained and in-solution digested by trypsin into peptides as described (42). Crude separation of phosphopeptides from non-phosphorylated peptides was performed using SCX chromatography as described in detail elsewhere (42, 43). In each SCX experiment, 15 fractions were collected via an automated fraction collector. Based on UV absorbance, some fractions containing a low amount of peptides were pooled, resulting in around 10 fractions. The flow-through liquid during sample loading was also collected and treated as one fraction. Phosphopeptides were enriched by 2,5-dihydroxybenzoic acid-precoated TiO2 beads (31, 44).
C18 Reverse-phase HPLC Separation and Mass Spectrometric Methods
The peptide mixture was separated by nanoscale C18 reverse-phase liquid chromatography (Agilent 1200, Agilent Technologies, Waldbronn, Germany) coupled online to an LTQ-Orbitrap mass spectrometer (Thermo Electron, Bremen, Germany) as described (42, 45). Briefly, each sample was eluted with 0–40% organic buffer solvent (80% MeCN in water and 0.5% acetic acid) over 90 min and electrosprayed into the mass spectrometer via a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark).
The mass spectrometers were operated in positive ion mode and used a data-dependent automatic switch between MS and MS/MS acquisition modes. A full scan was acquired at a target value of 1,000,000 ions with resolution R = 60,000 at m/z 400. A total scan cycle comprising three mass ranges was applied: m/z 350–1,050, 850–1,850, and 350–1,850. The top five most intense ions from the first two ranges were selected for fragmentation in the LTQ, whereas in the last range the top seven most intense ions were selected. Fragmentation in the LTQ was induced by collision-induced dissociation with a target value of 5,000 ions.
For accurate mass measurement, the “lock mass” function was enabled for both MS and MS/MS scan modes (46). To improve the fragmentation of phosphopeptides, the multistage activation algorithm in the Xcalibur software was enabled for each MS/MS spectrum using the neutral loss values of 97.97, 48.99, and 32.66 m/z units (47). Former target ions selected for MS/MS were dynamically excluded for 300 s.
General mass spectrometric conditions were as follows: spray voltage, 2.1–2.4 kV; no sheath and auxiliary gas flow; ion transfer tube temperature, 150–180 °C; normalized collision energy (40%) using wide band activation mode for MS2. Ion selection thresholds were 3,000 counts for MS2. An activation of q = 0.25 and activation time of 30 ms were applied in MS2 acquisitions.
Mass Spectrometric Data Analysis by MaxQuant
Raw MS spectra were processed in Quant.exe, the first unit of our in-house built software MaxQuant (48). The derived peak list was searched with the Mascot search engine (Matrix Science, London, UK) against the International Protein Index (IPI) human protein database version 3.37 containing 69,289 proteins to which 175 commonly observed contaminants and all the reversed sequences had been added. Carbamidomethylation of cysteine residues was set as a fixed modification. Variable modifications for the phosphoproteome mapping data sets included oxidation (Met), N-acetylation (protein), and phosphorylation (Ser, Thr, and Tyr). Full tryptic specificity was required, and up to two missed cleavages were allowed. The initial mass tolerances of the precursor ion and fragment ions were set to 7 ppm and 0.5 Da, respectively.
The derived peptides and their assigned proteins were further processed in Identify.exe, the second module of MaxQuant. The posterior error probability and false discovery rate (FDR) were used for statistical evaluation. All phosphopeptide identifications suggested by Mascot were filtered in MaxQuant by applying thresholds on peptide length, mass error, SILAC state, and Mascot score. We accepted peptides based on the criteria that the number of forward hits in the database was at least 100-fold higher than the number of reversed database hits (incorrect peptide sequences); this gives an estimated FDR of less than 1%. To achieve highly reliable identifications, the following criteria were used: maximal peptide posterior error probability of 0.1, maximal peptide FDR of 0.01, and minimal peptide length of 6.
For quantitation in MaxQuant, peptide ratios were calculated according to the intensities of all centroids of each of the SILAC forms. Systematic deviations, such as mixing errors, are corrected by the quantitation algorithm in the MaxQuant software by normalizing all peptide ratios such that the mean of all log-transformed ratios are zero. To robustly represent the ratio of a peptide being quantified multiple times, the median value was chosen. To derive biologically robust regulation, a 2-fold change (ratio ≥ 2 or ratio ≤ 0.5) was required.
The assignment of the phosphosite within identified phosphopeptides was based on the localization post-translational modification scoring algorithm as described before (31, 44). Phosphosites fulfilling the following two criteria are defined as class I sites: 1) their localization probability for the assignment is at least 0.75, and 2) the score difference from the second possible localization assignment is 5 or higher. Note that we use multistage activation for phosphopeptide sequencing, which results in composite tandem mass spectra that consist of both MS/MS and MS/MS/MS ions. These spectra are therefore more complex (and information-rich) compared with standard CID spectra. MaxQuant matches and labels primary fragment ions (b- and y-type ions) in the displayed figures, and therefore all of the abundant neutral loss-directed fragment peaks (−H2O, −NH3, −HPO3, and −H3PO4) are not labeled.
For the proteome comparison with and without dasatinib treatment, standard conditions employed in our laboratory were used (49). Specifically, we used OFFGEL isoelectric focusing of tryptic peptides in 24 fractions and analyzed each of them in 140-min gradients. Results of this experiment are reported in supplemental Table 5.
RESULTS AND DISCUSSION
Effects of U0126 and SB202190 on the EGF-induced Signaling Pathways
U0126 and SB202190 are cell-permeable, potent inhibitors for MEK1/2 and p38α/β MAPK, respectively, with IC50 values in the submicromolar range. SB202190 is a pyridinyl imidazole, which competes with ATP to bind to the p38α/β MAPK (3, 50). In contrast, U0126 exerts its inhibitory effect mainly by binding to inactive MEK1/2 and thus precludes allosteric activation of the kinase domain (51). Previous enzyme assays have suggested that U0126 is a highly selective chemical, whereas SB202190 is fairly selective and inhibits only a few other kinases in addition to p38 (7, 9).
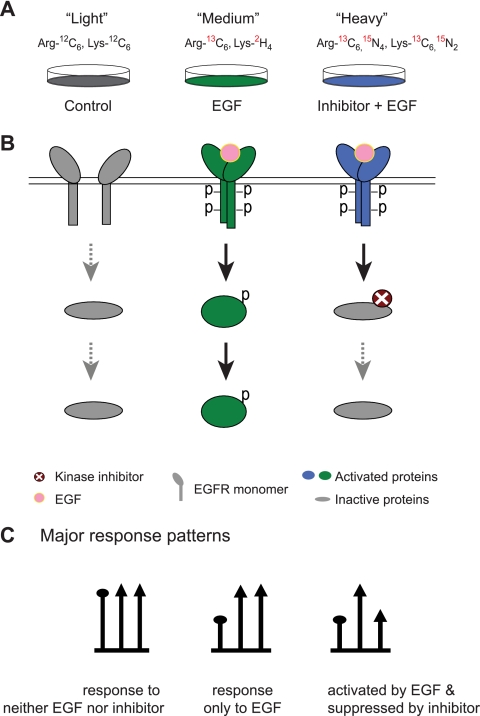
MEK1/2 and p38α/β MAPK belong to two different MAPK signaling cascades (52, 53). The Raf-MEK1/2-ERK1/2 cascade is predominantly involved in proliferation and growth, whereas the MEK3/6-p38 MAPK cascade is usually linked to stress or immune responses. To evaluate inhibitor effects on the signaling network, we used EGF stimulation, which activates both MEK1/2 and p38α/β MAPK in HeLa cells (31). We established three SILAC conditions in which HeLa cells were labeled with distinct forms of arginine and lysine. Light labeled cells (Arg0, Lys0) remained untreated and served as control. Medium labeled cells (Arg6, Lys4) were stimulated with EGF for a short time (150 ng/ml, 15 min), whereas heavy labeled cells (Arg10, Lys8) were pretreated with one of the kinase inhibitors (10 μm, 20 min) and then stimulated with EGF as for the medium cells (Fig. 1A). In this assay, the suppression of phosphorylation induced by the kinase inhibitor should become apparent by a “down-up-down” triangular response pattern (Fig. 1, B and C).
Fig. 1.
Strategy to detect the effects of kinase inhibitors on the cellular signaling network. A, three SILAC-labeled cell populations were subjected to no treatment, treatment with growth factor, and treatment with growth factor in the presence of kinase inhibitor. B, principal response of the targeted kinase and its downstream substrates in the three SILAC conditions. C, three major response patterns reflected in SILAC mass spectra. EGFR, EGF receptor.
Effects of U0126 and SB202190 on the Phosphoproteome
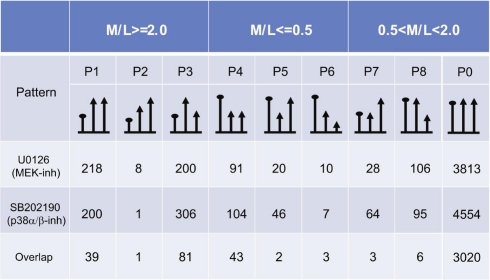
Mass spectrometric data from both inhibitor experiments were processed together in MaxQuant using stringent criteria. Respectively, 4,625 and 5,641 phosphopeptides were quantified after U0126 and SB202190 treatments. Between the two data sets, 4,009 phosphopeptides from 1,474 proteins were shared. In all, 6,451 phosphosites could be localized with high accuracy to particular serine, threonine, or tyrosine sites (class I sites). To define robust biological response, we set a conservative cutoff of a minimal 2-fold change. Nine response patterns can be defined, and quantitation results in relation to these patterns are listed in supplemental Tables 1 and 2 and summarized in Table I. Phosphopeptides with class I sites were first grouped into three major categories based on their response to EGF, namely up-regulated (minimal ratio(M/L) of 2), down-regulated (maximal ratio(M/L) of 0.5), and non-changed (0.5 < ratio(M/L) < 2). Within each category, these phosphopeptides were further sorted into three categories according to their response to the kinase inhibitor (ratio(H/M) ≥ 2, 0.5 < ratio(H/M) < 2 and ratio(H/M) ≤ 0.5). The majority of phosphopeptides (85%) were not influenced by the growth factor or the kinase inhibitors. In this case, we observed phosphopeptide triplets with very similar abundance (see Fig. 2A for an example). Although the overall numbers of quantified phosphopeptides were different in the two experiments, the percentages of EGF up- and down-regulated phosphopeptides were similar, with 9% up-regulation and 2% down-regulation in each case. Different phosphopeptides of the same protein can have different response patterns. A striking example is the neuroblast differentiation-associated protein AHNAK. Among the 44 quantified phosphopeptides of AHNAK, six of the nine possible response patterns were found (supplemental Table 3).
Table I. Quantified class I phosphosites of the U0126 and SB202190 experiments.
M/L represents the ratio of “medium labeled cells” (EGF stimulation) versus “light labeled cells” (untreated). Within each category, ratios are further classified according to their responses to the inhibitor (inh). Up-regulation, ratio ≥2.0; down-regulation, ratio ≤0.5.
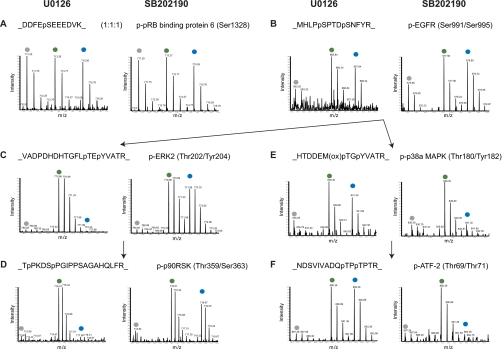
Fig. 2.
Effects of the inhibitors U0126 and SB202190 on known substrates of signaling branches of ERK1/2 and p38α/β. MS survey scans present phosphopeptide intensities in each of the SILAC triplets. Peaks marked on top by gray, green, and blue dots represent phosphopeptides from light, medium, and heavy labeled cells, respectively. For labeling details see “Experimental Procedures.” In each panel, the figure on the left is from the “U0126 experiment,” whereas the figure on the right is from the “SB202190 experiment.” A, a phosphopeptide was affected by neither the EGF stimulus nor the two inhibitors, demonstrating a 1:1:1 ratio. B, a phosphoserine peptide of EGF receptor (EGFR) was highly activated by EGF but not responsive to the two inhibitors. C, the autophosphorylation sites of ERK2 were highly activated by EGF and suppressed by U0126 but not responsive to SB202190. D, a known substrate of ERK1/2 demonstrated the same response pattern as the ERK1/2 autophosphorylation. E, the autophosphorylation sites of p38α were highly activated by EGF and suppressed by SB202190 but not responsive to U0126. F, a known substrate of p38α demonstrated the same response pattern as the p38α autophosphorylation. ox, oxidation; p-, phospho-; pRB, phospho-retinoblastoma; p90RSK, 90 kDa ribosomal protein S6 kinase.
Phosphorylation sites in proteins upstream of the MAPKs would not be expected to change upon kinase inhibitor treatment. Indeed, phosphorylation sites on the EGF receptor itself were up-regulated upon EGF stimulation but were not repressed upon inhibitor treatment (Fig. 2B).
Many kinases are regulated by phosphorylation of the activation loop, which then directly reflects cellular kinase activity. We captured the relevant phosphopeptides from the activation loops of ERK1/2 and p38α/β in both inhibitor experiments in which they were highly activated. In contrast to the upstream sites, they were drastically down-regulated by inhibitor treatment, forming the triangular pattern in the triple SILAC experiment. There was no “cross-talk” among the inhibitors at this level because SB202190 did not down-regulate the ERK1/2 activation sites and U0126 did not affect the p38α activation sites (Fig. 2, C and E). Interestingly, the suppression was so strong that phosphorylation levels in heavy cells were even weaker than in the light labeled control cells, indicating that kinase activities were reduced below the basal level by the inhibitors. Specific downstream targets of these inhibited kinases, such as Thr359/Ser363 in the kinase p90RSK (54) and Thr69/Thr71 in the transcription factor ATF2 (55), displayed the same triangular response pattern (Fig. 2, D and F), characteristic of direct or indirect downstream targets of the inhibited kinase. In both inhibitor experiments, around 4–5% of the phosphopeptides matched this criterion (Fig. 3).
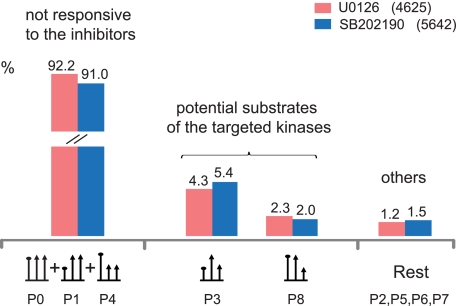
Fig. 3.
Quantitation patterns classified according to the responses to EGF and MAPK inhibitors. The corresponding SILAC patterns are depicted at the bottom.
MAPKs, such as ERK1/2, are known to have a large number of substrates (56). Furthermore, different MAPK family members have shared and specific targets, some of which do not participate in EGF signaling. In this context, we found phosphorylation sites that were unaffected by growth factor stimulation but still suppressed by kinase inhibitor (P8 pattern; Table I). This could be due to the suppression of kinase activities of MEK1/2 and p38α/β below the basal level or due to off-target effects on other kinases, which are not activated upon EGF treatment. Patterns P3 and P8 constitute the potential direct and indirect targets of the kinase inhibitors. The opposite pattern, unaffected by growth factor signal but up-regulated by kinase inhibitor (P7; Table I), was also observed. This pattern might be associated with downstream events that are triggered upon cellular suppression of an inhibitory phosphorylation. The precise chain of causation is difficult to disentangle by our or other methods; however, we still measured the end result of the kinase inhibitor treatment for each site. P8 could be indicative of potential substrates of either basal MAPK activity or other cellular kinase activities affected by drug treatment. The patterns P2, P5, P6, and P7 constitute a minor portion (1.5 or 2.2%) of all phosphopeptides and are labeled “others” in Fig. 3. They are not directly attributable to growth factor or inhibitor action but constitute a further category of potential off-targets induced by the inhibitor compounds. As the majority of drug-regulated phosphorylations can be explained by MAPK inhibition, we conclude that the two tested compounds, U0126 and SB202190, act as relatively specific kinase inhibitors in intact cells.
Effects of Inhibitors on MEK1/2 and p38α/β Signaling Pathways
Cross-talk between MEK1/2 and p38α/β Signaling Pathways
In the U0126 experiment, 199 phosphopeptides exhibited the same triangular response pattern as the activation loop phosphosites of ERK1/2 and should therefore be direct or indirect downstream substrates of MEK1/2 after EGF stimulation. Likewise, in the SB202190 experiment, 306 phosphopeptides displayed the same quantitation pattern as the activation loop of p38α/β. Therefore, these 306 phosphopeptides potentially participate in transducing the EGF signal through p38α/β.
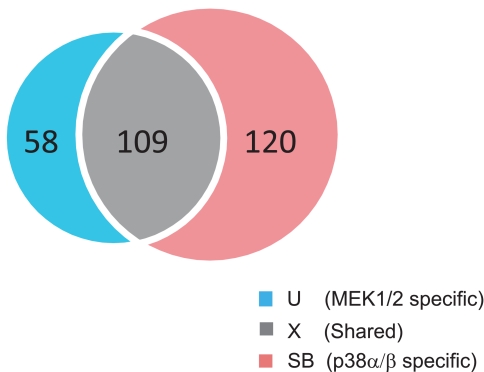
To better define shared and specific substrates, we selected those peptides that were quantified in both inhibitor experiments. Their corresponding phosphosites with accurate localization, i.e. class I sites, were compared in the following analyses (supplemental Table 4). We classified the phosphosites that were activated by EGF in both inhibitor experiments into three groups (Fig. 4): MEK1/2-specific (“U” group for U0126), p38α/β-specific (“SB” group for SB202190), and shared (“X” group) phosphosites.
Fig. 4.
Overlap between potential phosphosite substrates of MEK1/2 (left circle) and p38α/β (right circle).
Proline at the +1 position following Ser(P)/Thr(P) is a well accepted kinase motif for MAPK substrates. Within each group, we analyzed the adjacent amino acid residues surrounding each phosphosite. Interestingly, proline-directed phosphorylation (at (pS/pT)P motif where pS is phosphoserine and pT is phosphothreonine) accounts for only 11% in the shared group but 31 and 30%, respectively, in the MEK1/2-specific and p38α/β-specific groups. The fact that a large portion of the substrates undergo non-proline-directed phosphorylation indicates functions downstream of direct cellular substrates of p38α/β- or MEK1/2-regulated ERK MAPKs. Thus, the immediate downstream targets of the two kinases are quite distinct, whereas the targets further downstream (including other kinases and non-kinase proteins) overlap much more. These results indicate an unexpected divergence and convergence of MAPK-mediated signaling processes that, using conventional signaling technologies, would have been difficult to observe in a systems-wide manner.
Although ERK1/2 and p38α/β share 50% sequence similarity, they are used in different cellular responses (53). ERK1/2 are activated by various stimuli to regulate meiosis, mitosis, and postmitotic functions in differentiated cells. p38 MAPKs are strongly activated by cytokines and stress factors, including osmotic shock and heat shock, and in the regulation of immune and stress responses. The conserved motif in kinase activation loop is Thr-Glu-Tyr (TEY) for ERK1/2 and Thr-Gly-Tyr (TGY) for p38α/β. Previously, a comparison of the substrates for ERK1/2 and p38α/β was reported at the level of transcriptional regulation (57). Here we are able to compare their substrates at the phosphorylation level in the overall signaling network. Generally, phosphorylation involves rapid and short term regulation events, whereas transcription starts later, typically after 30 min, and integrates a large number of cellular signals, making it difficult to interpret. In contrast, our comparison is based on quantifying changes in the signaling network after 15 min of EGF stimulation when the signal is already propagated to various downstream effectors but the forward signal dominates over the feedback signal. It is within this network that we discovered many phosphosites regulated differentially by MEK1/2 and p38 α/β. These phosphosites were detected in important signaling proteins, such as phospholipase C, MSK2, HSP27, death-inducer obliterator 1, and DNA topoisomerase 2-α. These sites should be a valuable resource for the MAPK signaling community.
Having required EGF up-regulation as a prerequisite, we limited our analysis to a small, more easily analyzable data set. Still, caution should be exercised in data interpretation. U0126 was previously reported to inhibit ERK5, a fourth family member of MAPKs besides ERK1/2, c-Jun amino-terminal kinases (JNKs), and p38 MAPKs (58). ERK5 shares the same consensus motif, TEY, as ERK1/2. However, ERK5 is unique in its carboxyl-terminal half and possesses a bipartite nuclear translocation signal in that sequence. It is involved in cardiovascular development and neural differentiation (52). Because of their distinctive features, the downstream substrates of ERK5 and ERK1/2 may be different. Suppressing both at the same time can cause ambiguity in assigning the substrates of each MAPK because both would be captured in the triangular response pattern for U0126. However, if viewed from the perspective of drug screening, that it captures the overall effects of inhibitor treatment is exactly the strength of the unbiased, systematic approach of quantitative MS.
Evaluation of EGF Signal Propagation
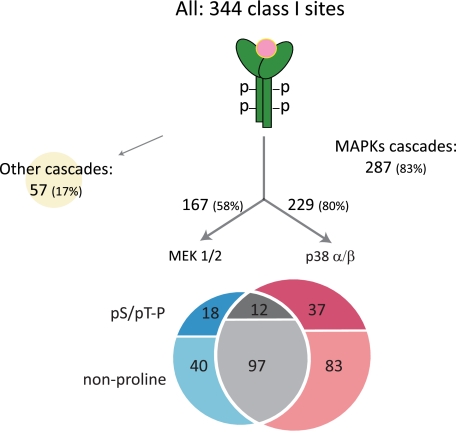
Based on the above analysis, we created a rough model of EGF signal propagation upon 15 min of 150 ng/ml EGF stimulation (Fig. 5). Of the 344 class I phosphorylation sites detected and up-regulated by EGF in both experiments, 83% participated in MAPK cascades as judged by their suppression after MAPK inhibitor treatments (triangular response pattern), leaving only a small role for other signaling cascades. Within the MAPK cascades, there were 67 potential direct MAPK substrates ((pS/pT)P) of which 27% were specific for MEK1/2 inhibition, 55% were specific for p38 inhibition, and 18% were shared. In comparison, for all targets downstream of MAPK inhibition, our inhibitor studies indicated that 20% were regulated by MEK1/2, 42% were regulated by p38, and the remaining 38% were regulated by both branches of MAPK signaling. This overall assessment again indicates higher specificity on the level of direct MAPK substrates compared with targets located further downstream.
Fig. 5.
A simple model of EGF signal propagation. Overall 298 class I phosphosites were up-regulated by EGF in both inhibitor experiments. The numbers of proline-directed phosphorylation events are listed. The three groups (U0126 (U), shared (X), and SB202190 (SB)) are the same as in Fig. 4.
Up-regulation of Phosphorylation Sites after Inhibitor Treatment
Apart from the expected triangular pattern P3, we also observed a number of phosphopeptide quantitation patterns that were unexpected and that can potentially perturb the signaling pathway in unforeseen ways. Specifically, we inspected phosphopeptides that were up-regulated by the inhibitor compounds compared with EGF-only treatment (ratio(H/M) ≥ 2). There are 81 phosphopeptides in the SB202190 data set and 51 phosphopeptides in the U0126 data set that fulfill this criterion. Among the list are regulatory molecules, such as cell cycle progression protein 1, BCL-9-like protein, and general transcription factor 3C. Activation of these proteins may have unexpected effects on the signaling network. Therefore, phosphopeptides with these response patterns (P2, P5, and P7) are also of potential interest for explaining unforeseen effects of kinase inhibitors.
Effect of Dasatinib on the Phosphoproteome of BCR-ABL-expressing Leukemia Cells
The human immortalized leukemia cell line K562 expresses the constitutively activated BCR-ABL fusion protein. Therefore, it serves as a suitable cell model for studying CML pathogenesis. In our experiments, three populations of K562 cells were cultured in light, medium, and heavy SILAC conditions. In contrast to the MAPK inhibitor studies described above, no stimulus was necessary because BCR-ABL itself provides a constitutive signal. Although light cells were treated with DMSO only, medium and heavy cells were treated with 5 and 50 nm dasatinib for 1 h, respectively. This time point was optimized by monitoring the decrease of ERK1/2 phosphorylation at different drug concentrations to establish efficient inhibition. To exclude that apparent phosphorylation changes were actually protein expression changes, we also quantified the cell line proteome after the same, short term dasatinib treatments. Less than 1% of the proteins changed by more than 2-fold, and they are indicated in supplemental Table 5.
We identified 5,689 phosphopeptides from 1,889 proteins from which 5,063 class I phosphosites were derived. About 12% of the phosphopeptides were suppressed at least 2-fold by 5 nm dasatinib treatment, whereas the 10 times higher dose of dasatinib down-regulated about 17% of the phosphopeptides (supplemental Fig. 1A). The class I sites from these down-regulated phosphopeptides of both treatment conditions are listed in supplemental Table 6. As expected, there were many more down-regulation events than up-regulation events (0.8%) (supplemental Fig. 1B).
Dasatinib is known to down-regulate several important signaling proteins. Table II lists some of the well known signaling proteins that were captured in our study. The activating autophosphorylation sites of ABL1, SRC, MAPK1, and MAPK3 were significantly suppressed, indicating that their cellular kinase activities were reduced. Additionally, tyrosine phosphorylation of BCR and LYN was drastically down-regulated. The Ser(P)62 site of c-MYC is known to be regulated by mitogen stimulation, which is crucial for various functions of c-MYC, such as transcriptional regulation (59). Following upstream kinase inhibitions, Ser62 was largely dephosphorylated after dasatinib treatment in our experiments. Together, these data demonstrate that large scale quantitative phosphoproteomics can pinpoint crucial sites involved in cell proliferation.
Table II. Marker phosphopeptides down-regulated by dasatinib.
Underscores denote the beginning and the end of the peptide sequence in the protein sequence. pY, phosphotyrosine; pS, phosphoserine; pT, phosphothreonine.
| Name | Phospho position | No. phosphosites | Sequence | M/L | H/L |
|---|---|---|---|---|---|
| BCR | 122 | 1: Ser | _ASASRPQPAPADGADPPPAEEPEARPDGEGpSPGK_ | 0.3 | 0.2 |
| 644 | 1: Tyr | _NSLETLLpYKPVDR_ | 0.3 | 0.1 | |
| ABL1 | 805, 808 | 2: Ser, Ser | _DIMEpSpSPGSSPPNLTPK_ | 0.4 | 0.4 |
| 248 | 1: Ser | _NKPTVYGVpSPNYDK_ | 0.6 | 0.4 | |
| 245 | 1: Tyra | _NKPTVpYGVSPNYDK_ | 0.4 | 0.4 | |
| SRC | 426 | 1: Tyra | _LIEDNEpYTAR_ | 0.4 | 0.1 |
| MAPK1 | 185, 187 | 2: Thr, Tyra | _VADPDHDHTGFLpTEpYVATR_ | 0.1 | 0.1 |
| MAPK3 | 202, 204 | 2: Thr, Tyra | _IADPEHDHTGFLpTEpYVATR_ | 0.1 | 0.1 |
| MYC | 62 | 1: Ser | _KFELLPTPPLpSPSR_ | 0.4 | 0.2 |
| LYN | 578 | 1: Tyr | _AEERPTFDYLQSVLDDFYTATEGQpYQQQP_ | 0.3 | 0.3 |
a Autophosphorylation sites.
Effect of Dasatinib on the Entire Signaling Network
Accumulating knowledge of BCR-ABL signaling has revealed several critical pathways that contribute to chronic myeloid leukemia transformation (60). In association with SHC and Grb2, BCR-ABL activates ERK1/2 and JAK-STAT pathways and therefore causes growth factor-independent proliferation and cell growth. BCR-ABL activates the phosphatidylinositol 3-kinase-AKT and JAK-STAT pathways to enhance cell survival, whereas its activation of focal adhesion components (actin, focal adhesion kinase, Crk-associated substrate, etc.) leads to a decrease in cell adhesion and abnormal interaction with extracellular matrix and stroma.
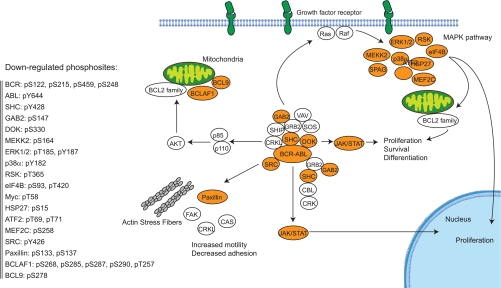
According to the comprehensive review from Weisberg et al. (60), we recapitulated the BCR-ABL signaling network on the level of down-regulated phosphosites (Fig. 6). Key signaling events in the three major MAPK cascades were suppressed, including the activation loop phosphorylation sites of p38α/β and ERK1/2. Inhibition was also evident at the level of putative ERK substrates as 97 of the suppressed phosphosites display the classical substrate motif for ERK1/2 (supplemental Table 7).
Fig. 6.
Effect of dasatinib on the BCR-ABL signaling pathway. Phosphorylation sites that were suppressed by both 5 and 50 nm dasatinib are displayed on the left. Signaling pathways are based on Weisberg et al. (60). FAK, focal adhesion kinase; DOK, docking protein 1; RSK, 90-kDa ribosomal protein S6 kinase; CRKL, Crk-like protein; CAS, Crk-associated substrate; SPAG, sperm-associated antigen; SOS, Son of sevenless; VAV, Vav proto-oncogene; SHIP, SH2 domain-containing inositol phosphatase; CRK, Crk proto-oncogene; CBL, Cbl protein, Cas-Br-M (murine) ecotropic retroviral transforming sequence; SHC, SHC-transforming protein.
There is increasing evidence that a major mechanism of BCR-ABL transformation is suppression of apoptosis (61). Primary CML progenitors require growth factors for proliferation (62). But in contrast to normal cells, they have increased cell viability in the absence of serum and growth factor. Antisense oligonucleotide-mediated down-regulation of BCR-ABL confers susceptibility of BCR-ABL-positive cells to apoptotic stimuli and does not affect cell cycle progression (63). In our analysis, we captured several down-regulated sites on BCL superfamily proteins as well as on p53-binding protein 2, an essential regulator of p53 in apoptosis and cell growth. Thus, our analysis recapitulates and extends the known connection between BCR-ABL signaling and apoptosis.
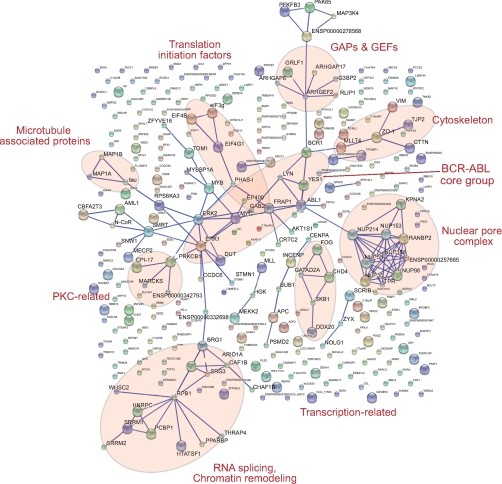
To better understand the overall influence on the cellular signaling network, we looked for protein interactions among the phosphoproteins commonly suppressed by both doses of dasatinib. To do this, we chose the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, which is constructed on the basis of both physical and functional interactions (64, 65). To avoid spurious interactions in our large data set, we required the highest stringency level (0.9). Several interaction groups are immediately apparent (see Fig. 7). The BCR-ABL core group includes BCR, ABL1, LYN, YES, ERK1, ERK2, MYC, and others, clustered in the center of the interaction map and providing a striking positive control for our analysis. From nearly each of the core group members, different interaction subgroups branch off. Interestingly, the phosphorylation of 10 members of the nuclear pore complex (NPC) was suppressed by dasatinib. It is known that NPC phosphorylation changes during the cell cycle (66), and we previously observed that NPC members were phosphorylated upon EGF stimulation (31), indicating that NPC function may be regulated by phosphorylation. The fact that a kinase inhibitor reversed that phosphorylation raises the possibility that this regulation is counteracted by the drug.
Fig. 7.
STRING database mapping reveals interaction groups within the down-regulated phosphoproteins by both 5 and 50 nm dasatinib. GAPs, GTPase-activating proteins; GEFs, guanine nucleotide exchange factors; PKC, protein kinase C.
Other interaction subgroups provide potential connections to diverse biological functions, such as chromatin remodeling, RNA splicing, transcription, translation initiation, and adhesion. Furthermore, protein clusters were also observed that directly relate to signaling and cancer-relevant changes such as protein kinase C and c-MYB, respectively, and GTPase-activating protein-guanine nucleotide exchange factor family proteins. The specific functions of these proteins in BCR-ABL signaling cannot be clearly assigned from our data as many of them are multifunctional signaling proteins. However, our data clearly indicate that the cellular effects of dasatinib are widespread and not limited to immediate targets of BCR-ABL.
Kinases and phosphatase are key molecules in regulating signaling pathways. To determine how broadly these regulators of signaling network are affected by dasatinib, we extracted the kinases and phosphatases with down-regulated phosphosites in our study. In total, phosphopeptides from 32 phosphatases and 118 kinases were quantified. Among these, nine phosphatases and 38 kinases had at least one phosphopeptide that was down-regulated by at least 2-fold by both doses of dasatinib.
In vitro dasatinib is known to bind to a large number of kinases. For example, Carter et al. (67) tested a panel of 148 kinases, and dasatinib influenced 76 of them with Kd values below 10 μm and 33 with Kd values of less than 50 nm, the maximal dasatinib concentration used in our cellular analysis. Because of the presence of millimolar ATP levels, cellular IC50 values for an ATP-competitive kinase inhibitor such as dasatinib are typically shifted to higher concentrations, depending on the individual Km values for ATP of the cellular target kinases. Therefore, the number of affected kinases in vivo can be expected to be smaller than that detected at similar concentrations in in vitro binding assays. Moreover, down-regulated phosphosites on kinases can be due to either a direct inhibitor binding and reduced autophosphorylation or the suppression of distinct kinases that act as upstream regulators. Thus, our observation that the number of kinases detected with down-regulated phosphosites is comparable with the number of interacting kinases at similar dasatinib concentrations in vitro is likely due to secondary effects on kinases downstream of direct cellular inhibitor targets.
In general, in vitro kinase assays and the quantitative phosphoproteome assay described here have complementary strengths. The phosphoproteome analysis is not exhaustive, and we may have missed particular phosphorylation sites on low abundance kinases and phosphatases. Additionally, different cell types retain their cell type-specific features to different degrees and therefore may not use all possible in vivo substrates to respond to a stimulus (42). More drastically, some of the in vivo targets may not even be expressed in the cell types under study. On the other hand, in vitro kinase assays operate at normalized concentrations of the enzymes, whereas the phosphoproteome analysis uses endogenous concentrations of the entire cellular kinome. For example, an off-target effect found at high kinase concentration may not have functional consequences in the “correct cellular milieu” where the kinase may be present at a very low concentration. These observations indicate that the study of inhibitor effect on the entire signaling network can serve as a useful complement to the study of the direct binding targets of the inhibitor.
Conclusions and Perspectives
Phosphorylation is a critical modification for various signal transduction events in normal and transformed cells, and selective inhibition of the crucial kinases or phosphatases has been proven to be a useful strategy for correcting aberrant enzyme activities. The majority of proteomics studies in kinase drug development have focused on direct binding targets of inhibitor compounds. The quantitative phosphoproteomics approach demonstrated here is able to examine the cellular effect at the phosphorylation level and, in principle, at the protein expression level.
In this study, we quantified the effects of three kinase inhibitors, including a clinical drug, on the phosphorylation changes in the entire cellular signaling network. Two of the inhibitors, U0126 and SB202190, are very commonly used in MAPK research. In these studies, researchers use a small set of phosphospecific antibodies and typically do not measure their overall effects. Our results suggest that the traditional approach misses many potentially relevant signaling molecules whose functions are regulated in response to the drug. We found that effects of the kinase inhibitors could be efficiently studied by classifying the response patterns in triple SILAC experiments in which we compared control, stimulus, and stimulus combined with kinase inhibitor. Most cellular phosphorylation events were not affected by either EGF stimulation (as expected from previous investigations (31)) or by the kinase inhibitors. In total, about 4% of all quantified sites were activated by growth factor stimulation and down-regulated by kinase inhibitor treatment. Based on these results, we derived a simple EGF signal propagation model, which indicates that MAPK signaling cascades are the dominant mediators in the early phase of EGF signal transduction. Overall, our results indicate that U0126 and SB202190 are indeed rather specific inhibitors at the phosphoproteome level.
Off-target effects are a major concern for inhibitor compound development. As most kinase inhibitors are ATP competitors and ATP-binding pockets are conserved among kinases, it is likely that inhibitor compounds suppress more kinases than the intended one. The fact that these suppressed kinases can have limited sequence homology further complicates analysis. Besides the off-target issue, there is an equally important question in the field, which is the secondary effect of the kinase inhibitor compound on the signaling pathway. This issue has been addressed to a lesser degree and in a less systematic manner because of technical limitations. The strength of our approach is to examine the global effects of kinase inhibitors over the entire network regardless of their mechanisms.
Here we used SILAC-based phosphoproteomics as a simple assay to elucidate the effect of dasatinib on BCR-ABL signaling. To our knowledge, this is the first time that the effect of a cancer drug on the phosphoproteome has been assessed in a systems-wide manner. Using the inhibition of the intrinsic BCR-ABL signal in a constitutively active human CML cell line at two different concentrations, we found that a relatively large proportion of the phosphoproteome was affected. Thus, dasatinib seems less specific than the MAPK inhibitors. Our screen was able to quantify key phosphorylation events known to be involved in ABL signaling. Furthermore, known downstream targets, including ERK1/2, STAT5, paxillin, p38α, and others, were repressed, providing strong positive controls for our screen. When analyzing the systems-wide effects of dasatinib, we found broad effects on the BCR-ABL core network but also downstream effects on the cytoskeleton and on chromatin remodeling as well as yet unreported cellular functions such as splicing.
We have demonstrated by the above studies that quantitative phosphoproteomics is a streamlined and generic tool that can be broadly applied to kinase drug studies. Our screen is based on creating cell populations distinguishable by MS using SILAC; incorporates the latest advances in phosphopeptide enrichment by SCX fractionation and TiO2 enrichment; uses the rapid, automated, and highly accurate mass spectrometry; and finally makes optimal use of the data through the MaxQuant analysis system. Routine quantitation of thousands of phosphosites with around 80% of sites assigned with single amino acid resolution can be achieved in a single experiment. With further technology development, we expect that this number will rise significantly. This screening method can easily be extended to study the effects of combination therapy, which are difficult to study by traditional screens. Signaling effects of genetic approaches, i.e. silencing or knock-out of particular kinases alone or in combination, can also be “read out.” In conclusion, our strategy seems well suited to support current clinical needs in cancer drug application.
Supplementary Material
Acknowledgments
We thank Juergen Cox and Nadine Neuhauser for informatics support and Chunaram Choudhary and other members of the Department of Proteomics and Signal Transduction for support and fruitful discussion. The Center for Protein Research is supported by a generous grant from the Novo Nordisk Foundation.
Footnotes
* This work was supported in part by the HepatoSys Network and European Union 6th Framework Program, Interaction Proteome Grant LSHG-CT-2003-505520.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Fig. 1 and Tables 1–7.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Fig. 1 and Tables 1–7.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- FDR
- false discovery rate
- NPC
- nuclear pore complex
- EGF
- epidermal growth factor
- MAPK
- mitogen-activated protein kinase
- CML
- chronic myelogenous leukemia
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- DMEM
- Dulbecco's modified Eagle's medium
- RPMI
- RPMI 1640 medium
- SCX
- strong cation exchange
- LTQ
- linear trap quadrupole
- ERK
- extracellular signal-regulated kinase
- H
- heavy
- M
- medium
- L
- light
- JAK
- Janus kinase
- STAT
- signal transducers and activators of transcription
- BCR
- breakpoint cluster region.
REFERENCES
- 1.Mauro M. J., Druker B. J. ( 2001) STI571: targeting BCR-ABL as therapy for CML. Oncologist 6, 233– 238 [DOI] [PubMed] [Google Scholar]
- 2.Baselga J., Averbuch S. D. ( 2000) ZD1839 (‘Iressa’) as an anticancer agent. Drugs 60, Suppl. 1, 33– 42 [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. ( 2002) Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov 1, 309– 315 [DOI] [PubMed] [Google Scholar]
- 4.Petrelli A., Giordano S. ( 2008) From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr. Med. Chem 15, 422– 432 [DOI] [PubMed] [Google Scholar]
- 5.Knight Z. A., Shokat K. M. ( 2005) Features of selective kinase inhibitors. Chem. Biol 12, 621– 637 [DOI] [PubMed] [Google Scholar]
- 6.Bain J., McLauchlan H., Elliott M., Cohen P. ( 2003) The specificities of protein kinase inhibitors: an update. Biochem. J 371, 199– 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. ( 2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J 408, 297– 315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian M. A., Biggs W. H., 3rd, Treiber D. K., Atteridge C. E., Azimioara M. D., Benedetti M. G., Carter T. A., Ciceri P., Edeen P. T., Floyd M., Ford J. M., Galvin M., Gerlach J. L., Grotzfeld R. M., Herrgard S., Insko D. E., Insko M. A., Lai A. G., Lélias J. M., Mehta S. A., Milanov Z. V., Velasco A. M., Wodicka L. M., Patel H. K., Zarrinkar P. P., Lockhart D. J. ( 2005) A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol 23, 329– 336 [DOI] [PubMed] [Google Scholar]
- 9.Karaman M. W., Herrgard S., Treiber D. K., Gallant P., Atteridge C. E., Campbell B. T., Chan K. W., Ciceri P., Davis M. I., Edeen P. T., Faraoni R., Floyd M., Hunt J. P., Lockhart D. J., Milanov Z. V., Morrison M. J., Pallares G., Patel H. K., Pritchard S., Wodicka L. M., Zarrinkar P. P. ( 2008) A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol 26, 127– 132 [DOI] [PubMed] [Google Scholar]
- 10.Becker F., Murthi K., Smith C., Come J., Costa-Roldán N., Kaufmann C., Hanke U., Degenhart C., Baumann S., Wallner W., Huber A., Dedier S., Dill S., Kinsman D., Hediger M., Bockovich N., Meier-Ewert S., Kluge A. F., Kley N. ( 2004) A three-hybrid approach to scanning the proteome for targets of small molecule kinase inhibitors. Chem. Biol 11, 211– 223 [DOI] [PubMed] [Google Scholar]
- 11.Daub H. ( 2005) Characterisation of kinase-selective inhibitors by chemical proteomics. Biochim. Biophys. Acta 1754, 183– 190 [DOI] [PubMed] [Google Scholar]
- 12.Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. ( 2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol 25, 1035– 1044 [DOI] [PubMed] [Google Scholar]
- 13.Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., Varmus H. ( 2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capdeville R., Buchdunger E., Zimmermann J., Matter A. ( 2002) Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov 1, 493– 502 [DOI] [PubMed] [Google Scholar]
- 15.Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., Jänne P. A. ( 2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039– 1043 [DOI] [PubMed] [Google Scholar]
- 16.Stommel J. M., Kimmelman A. C., Ying H., Nabioullin R., Ponugoti A. H., Wiedemeyer R., Stegh A. H., Bradner J. E., Ligon K. L., Brennan C., Chin L., DePinho R. A. ( 2007) Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318, 287– 290 [DOI] [PubMed] [Google Scholar]
- 17.Valet G. ( 2006) Cytomics as a new potential for drug discovery. Drug Discov. Today 11, 785– 791 [DOI] [PubMed] [Google Scholar]
- 18.Nicholson R. L., Welch M., Ladlow M., Spring D. R. ( 2007) Small-molecule screening: advances in microarraying and cell-imaging technologies. ACS Chem. Biol 2, 24– 30 [DOI] [PubMed] [Google Scholar]
- 19.Lang P., Yeow K., Nichols A., Scheer A. ( 2006) Cellular imaging in drug discovery. Nat. Rev. Drug Discov 5, 343– 356 [DOI] [PubMed] [Google Scholar]
- 20.Krutzik P. O., Crane J. M., Clutter M. R., Nolan G. P. ( 2008) High-content single-cell drug screening with phosphospecific flow cytometry. Nat. Chem. Biol 4, 132– 142 [DOI] [PubMed] [Google Scholar]
- 21.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376– 386 [DOI] [PubMed] [Google Scholar]
- 22.Mann M. ( 2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol 7, 952– 958 [DOI] [PubMed] [Google Scholar]
- 23.Cox J., Mann M. ( 2007) Is proteomics the new genomics? Cell 130, 395– 398 [DOI] [PubMed] [Google Scholar]
- 24.Bonaldi T., Straub T., Cox J., Kumar C., Becker P. B., Mann M. ( 2008) Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol. Cell 31, 762– 772 [DOI] [PubMed] [Google Scholar]
- 25.de Godoy L. M., Olsen J. V., de Souza G. A., Li G., Mortensen P., Mann M. ( 2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol 7, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bose R., Molina H., Patterson A. S., Bitok J. K., Periaswamy B., Bader J. S., Pandey A., Cole P. A. ( 2006) Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc. Natl. Acad. Sci. U.S.A 103, 9773– 9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. ( 2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol 20, 301– 305 [DOI] [PubMed] [Google Scholar]
- 28.Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. ( 2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem 76, 3935– 3943 [DOI] [PubMed] [Google Scholar]
- 29.Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. ( 2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310– 327 [DOI] [PubMed] [Google Scholar]
- 30.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. ( 2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873– 886 [DOI] [PubMed] [Google Scholar]
- 31.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. ( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635– 648 [DOI] [PubMed] [Google Scholar]
- 32.Zubarev R. A., Nielsen M. L., Fung E. M., Savitski M. M., Kel-Margoulis O., Wingender E., Kel A. ( 2008) Identification of dominant signaling pathways from proteomics expression data. J. Proteomics 71, 89– 96 [DOI] [PubMed] [Google Scholar]
- 33.Huang P. H., White F. M. ( 2008) Phosphoproteomics: unraveling the signaling web. Mol. Cell 31, 777– 781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macek B., Mann M., Olsen J. V. ( 2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol 49, 199– 221 [DOI] [PubMed] [Google Scholar]
- 35.Nowell P. C., Hungerford D. A. ( 1960) Chromosome studies on normal and leukemic human leukocytes. J. Natl. Cancer Inst 25, 85– 109 [PubMed] [Google Scholar]
- 36.Chabner B. A. ( 2001) The oncologic four-minute mile. Oncologist 6, 230– 232 [PubMed] [Google Scholar]
- 37.Steinberg M. ( 2007) Dasatinib: a tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. Clin. Ther 29, 2289– 2308 [DOI] [PubMed] [Google Scholar]
- 38.O'Hare T., Walters D. K., Stoffregen E. P., Jia T., Manley P. W., Mestan J., Cowan-Jacob S. W., Lee F. Y., Heinrich M. C., Deininger M. W., Druker B. J. ( 2005) In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res 65, 4500– 4505 [DOI] [PubMed] [Google Scholar]
- 39.Lombardo L. J., Lee F. Y., Chen P., Norris D., Barrish J. C., Behnia K., Castaneda S., Cornelius L. A., Das J., Doweyko A. M., Fairchild C., Hunt J. T., Inigo I., Johnston K., Kamath A., Kan D., Klei H., Marathe P., Pang S., Peterson R., Pitt S., Schieven G. L., Schmidt R. J., Tokarski J., Wen M. L., Wityak J., Borzilleri R. M. ( 2004) Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem 47, 6658– 6661 [DOI] [PubMed] [Google Scholar]
- 40.Goss V. L., Lee K. A., Moritz A., Nardone J., Spek E. J., MacNeill J., Rush J., Comb M. J., Polakiewicz R. D. ( 2006) A common phosphotyrosine signature for the Bcr-Abl kinase. Blood 107, 4888– 4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hantschel O., Rix U., Schmidt U., Bürckstümmer T., Kneidinger M., Schütze G., Colinge J., Bennett K. L., Ellmeier W., Valent P., Superti-Furga G. ( 2007) The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc. Natl. Acad. Sci. U.S.A 104, 13283– 13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. ( 2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type specific functions. Mol. Cell. Proteomics 8, 443– 450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. ( 2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697– 707 [DOI] [PubMed] [Google Scholar]
- 44.Pan C., Gnad F., Olsen J. V., Mann M. ( 2008) Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics 8, 4534– 4546 [DOI] [PubMed] [Google Scholar]
- 45.Olsen J. V., Ong S. E., Mann M. ( 2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608– 614 [DOI] [PubMed] [Google Scholar]
- 46.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010– 2021 [DOI] [PubMed] [Google Scholar]
- 47.Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. ( 2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem 76, 3590– 3598 [DOI] [PubMed] [Google Scholar]
- 48.Cox J., Mann M. ( 2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367– 1372 [DOI] [PubMed] [Google Scholar]
- 49.Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. ( 2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672– 683 [DOI] [PubMed] [Google Scholar]
- 50.Tong L., Pav S., White D. M., Rogers S., Crane K. M., Cywin C. L., Brown M. L., Pargellis C. A. ( 1997) A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket. Nat. Struct. Biol 4, 311– 316 [DOI] [PubMed] [Google Scholar]
- 51.Davies S. P., Reddy H., Caivano M., Cohen P. ( 2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J 351, 95– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimoto S., Nishida E. ( 2006) MAPK signalling: ERK5 versus ERK1/2. EMBO Rep 7, 782– 786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson G. L., Lapadat R. ( 2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298, 1911– 1912 [DOI] [PubMed] [Google Scholar]
- 54.Smith J. A., Poteet-Smith C. E., Malarkey K., Sturgill T. W. ( 1999) Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem 274, 2893– 2898 [DOI] [PubMed] [Google Scholar]
- 55.Morton S., Davis R. J., Cohen P. ( 2004) Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS Lett 572, 177– 183 [DOI] [PubMed] [Google Scholar]
- 56.Yoon S., Seger R. ( 2006) The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24, 21– 44 [DOI] [PubMed] [Google Scholar]
- 57.Gazel A., Nijhawan R. I., Walsh R., Blumenberg M. ( 2008) Transcriptional profiling defines the roles of ERK and p38 kinases in epidermal keratinocytes. J. Cell. Physiol 215, 292– 308 [DOI] [PubMed] [Google Scholar]
- 58.Wang X., Tournier C. ( 2006) Regulation of cellular functions by the ERK5 signalling pathway. Cell. Signal 18, 753– 760 [DOI] [PubMed] [Google Scholar]
- 59.Gupta S., Seth A., Davis R. J. ( 1993) Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc. Natl. Acad. Sci. U.S.A 90, 3216– 3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weisberg E., Manley P. W., Cowan-Jacob S. W., Hochhaus A., Griffin J. D. ( 2007) Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat. Rev. Cancer 7, 345– 356 [DOI] [PubMed] [Google Scholar]
- 61.Jagani Z., Singh A., Khosravi-Far R. ( 2008) FoxO tumor suppressors and BCR-ABL-induced leukemia: a matter of evasion of apoptosis. Biochim. Biophys. Acta 1785, 63– 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emanuel P. D., Bates L. J., Castleberry R. P., Gualtieri R. J., Zuckerman K. S. ( 1991) Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood 77, 925– 929 [PubMed] [Google Scholar]
- 63.McGahon A., Bissonnette R., Schmitt M., Cotter K. M., Green D. R., Cotter T. G. ( 1994) BCR-ABL maintains resistance of chronic myelogenous leukemia cells to apoptotic cell death. Blood 83, 1179– 1187 [PubMed] [Google Scholar]
- 64.Snel B., Lehmann G., Bork P., Huynen M. A. ( 2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 28, 3442– 3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Mering C., Jensen L. J., Kuhn M., Chaffron S., Doerks T., Krüger B., Snel B., Bork P. ( 2007) STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35, D358– D362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glavy J. S., Krutchinsky A. N., Cristea I. M., Berke I. C., Boehmer T., Blobel G., Chait B. T. ( 2007) Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc. Natl. Acad. Sci. U.S.A 104, 3811– 3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carter T. A., Wodicka L. M., Shah N. P., Velasco A. M., Fabian M. A., Treiber D. K., Milanov Z. V., Atteridge C. E., Biggs W. H., 3rd, Edeen P. T., Floyd M., Ford J. M., Grotzfeld R. M., Herrgard S., Insko D. E., Mehta S. A., Patel H. K., Pao W., Sawyers C. L., Varmus H., Zarrinkar P. P., Lockhart D. J. ( 2005) Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl. Acad. Sci. U.S.A 102, 11011– 11016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.