Abstract
We developed a sample preparation protocol for rapid and unbiased analysis of the membrane proteome using an alimentary canal-mimicking system in which proteases are activated in the presence of bile salts. In this rapid and unbiased protocol, immobilized trypsin is used in the presence of deoxycholate and lauroylsarcosine to increase digestion efficiency as well as to increase the solubility of the membrane proteins. Using 22.5 μg of Escherichia coli whole cell lysate, we quantitatively demonstrated that membrane proteins were extracted and digested at the same level as soluble proteins without any solubility-related bias. The recovery of membrane proteins was independent of the number of transmembrane domains per protein. In the analysis of the membrane-enriched fraction from 22.5 μg of E. coli cell lysate, the abundance distribution of the membrane proteins was in agreement with that of the membrane protein-coding genes when this protocol, coupled with strong cation exchange prefractionation prior to nano-LC-MS/MS analysis, was used. Because this protocol allows unbiased sample preparation, protein abundance estimation based on the number of observed peptides per protein was applied to both soluble and membrane proteins simultaneously, and the copy numbers per cell for 1,453 E. coli proteins, including 545 membrane proteins, were successfully obtained. Finally, this protocol was applied to quantitative analysis of guanosine tetra- and pentaphosphate-dependent signaling in E. coli wild-type and relA knock-out strains.
Despite the importance of cell surface biology, the conventional shotgun proteomics strategy generally underrepresents the membrane proteome because of inadequate solubilization and protease digestion (1, 2). The ageless gel strategy, consisting of SDS-PAGE followed by in-gel digestion, can partially solve this problem (3–5), but the recovery from in-gel digestion is generally lower than that from in-solution digestion, and this approach is far from suitable for a rapid, simple, and high throughput automated system. Numerous approaches have been reported to overcome the difficulties in membrane proteome analysis, such as the use of surfactants (2, 6–11), organic solvents (6, 7, 12–15), or chaotropic reagents (2, 6, 16). Acid-labile surfactants, such as RapiGest SF, are among the most promising additives to enhance protein solubilization without interfering with LC-MS performance (6, 10, 17–19). However, the cleavage step at acidic pH causes loss of hydrophobic peptides because of coprecipitation with the hydrophobic part of RapiGest SF (20). Recently, we developed a new protocol to dissolve and digest membrane proteins with the aid of a removable phase transfer surfactant (PTS),1 such as sodium deoxycholate (SDC) (20). The solubility of membrane proteins with SDC was comparable to that with sodium dodecyl sulfate. In addition, the activity of trypsin was enhanced ∼5-fold in the presence of 1% SDC because this rapid PTS method mimics conditions in the alimentary canal in which bile salts such as cholate and deoxycholate are secreted together with trypsin. After tryptic digestion, SDC is removed prior to LC-MS/MS analysis by adding an organic solvent followed by pH-induced transfer of the surfactant to the organic phase, whereas tryptic peptides remain in the aqueous phase. This protocol offers a significant improvement in identifying membrane proteins by increasing the recovery of hydrophobic tryptic peptides compared with the protocols using urea and RapiGest SF.
The goal of this study is to establish a membrane proteomics method that is unbiased with respect to protein solubility, hydrophobicity, and protein abundance; i.e. membrane proteins can be as efficiently extracted and digested as soluble proteins. So far, to our knowledge, little information about the recovery of the membrane proteome has been reported. Instead, the number of identified membrane proteins or the content of membrane proteins identified in the membrane-enriched fraction has been used as an indicator of the efficiency of procedures for membrane proteome analysis (4, 5, 21–23). However, these parameters usually depend on the experimental conditions, including the sample preparation procedure and LC-MS instrument used. Therefore, it is difficult to compare data obtained with these protocols except in the case of direct comparison. Furthermore, there has been no report quantitatively comparing the recovery of membrane proteome with that of soluble proteins.
In this study, we used a modified version of our PTS protocol with immobilized trypsin columns to reduce the digestion time and evaluated its suitability for unbiased quantitation of the membrane proteome. In addition, we applied this protocol to estimate the copy numbers per cell of 1,453 proteins, including 545 membrane proteins, using the exponentially modified protein abundance index (emPAI). Finally, this rapid and unbiased PTS protocol was applied to the quantitative analysis of Escherichia coli BW25113 wild-type and relA knock-out (KO) strains.
EXPERIMENTAL PROCEDURES
Materials
Sodium carbonate, SDC, sodium N-lauroylsarcosinate (SLS), ammonium bicarbonate, dithiothreitol, iodoacetamide, mass spectrometry grade lysyl endoprotease, ethyl acetate, acetonitrile, acetic acid, methanol, and trifluoroacetic acid were purchased from Wako (Osaka, Japan). Modified trypsin was from Promega (Madison, MA). 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride was from Nacalai (Kyoto, Japan). Poroszyme-immobilized trypsin enzyme and iTRAQ reagents were purchased from Applied Biosystems (Foster City, CA). C18 and strong cation exchange (SCX) Empore products were from 3M (St. Paul, MN). Water was purified by a Millipore Milli-Q system (Bedford, MA).
Sample Preparation
Whole cell lysate and the membrane-enriched fraction of E. coli K12 strain BW25113 were prepared as described previously (20, 24). Proteins were extracted with 12 mm SDC, 12 mm SLS, and 50 mm ammonium bicarbonate containing 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride; reduced with 10 mm dithiothreitol at room temperature for 30 min; and alkylated with 55 mm iodoacetamide in the dark at room temperature for 30 min. The protein mixture was 5-fold diluted with 50 mm ammonium bicarbonate and digested by loading it onto pipette tips packed with the immobilized trypsin beads at 100 × g for 15 min. An equal volume of ethyl acetate was added to the eluent solution, and the mixture was acidified with 0.5% trifluoroacetic acid (final concentration) according to the PTS protocol reported previously (20). The mixture was shaken for 1 min and centrifuged at 15,700 × g for 2 min, and then the aqueous phase was collected. Tryptic peptides were fractionated with SCX-StageTips and desalted with C18-StageTips (25–27).
Nano-LC-MS/MS Analysis
An LTQ-Orbitrap XL (Thermo Fisher Scientific, Bremen, Germany) or a QSTAR-XL (AB/MDS-Sciex, Toronto, Canada) with a nano-LC interface (Nikkyo Technos, Tokyo, Japan), Dionex Ultimate3000 pump with FLM-3000 flow manager (Germering, Germany), and HTC-PAL autosampler (CTC Analytics, Zwingen, Switzerland) was used for nano-LC-MS/MS measurement with self-pulled needle columns (150-mm length, 100-μm inner diameter, 6-μm opening) (28). The injection volume was 5 μl, and the flow rate was 500 nl/min. Other LC conditions as well as MS acquisition conditions were described in the previous study (24).
Data Analysis and Bioinformatics
The raw data files were analyzed by Mass Navigator v1.2 (Mitsui Knowledge Industry, Tokyo, Japan) to create peak lists on the basis of the recorded fragmentation spectra for Mascot v2.2 (Matrix Science, London, UK) to identify E. coli K12 proteins registered in GenoBase (January 31, 2006; 4,316 entries). All parameters used in this process are described in supplemental Table S1. A precursor mass tolerance of 3 ppm (LTQ-Orbitrap) or 0.25 Da (QSTAR) and a fragment ion mass tolerance of 0.8 Da (LTQ-Orbitrap) or 0.25 Da (QSTAR) were used with strict trypsin specificity, allowing for up to two missed cleavages. Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation was allowed as a variable modification. Peptides were rejected if the Mascot score was below the 95% confidence limit based on the “identity” score of each peptide, and a minimum of two peptides with at least 7 amino acid residues was required for protein identification. In cases where the identified protein was a member of a multiprotein family featuring similar sequences, the protein was identified according to the highest number of matched peptides and Mascot score. False-positive rates (FPRs) were estimated by searching against a randomized decoy database created by the Mascot Perl program supplied by Matrix Science. The GRAVY values for identified proteins were calculated according to Kyte and Doolittle (29). Proteins exhibiting positive GRAVY values were recognized as hydrophobic. Mapping of transmembrane (TM) domains for the identified proteins was conducted using the TM hidden Markov model (TMHMM) algorithm (30). Information on the subcellular location of identified proteins was obtained from gene ontology component terms. Proteins with TM domains according to the TMHMM algorithm or positive GRAVY scores were classified as membrane proteins. Protein abundance was estimated according to emPAI using the number of observed precursor ions per protein (31).
Measurement of Protein Copy Numbers per Cell by Isotope Dilution
The copy numbers of proteins expressed in E. coli BW25113 were measured by the isotope dilution method as described (32). In this case, we used a stable isotope-labeled E. coli cell lysate, including 59 enzymes with known amounts ranging from nine to 70,000 copies per cell (33). In total, 36 proteins with at least two quantified peptides per protein were directly quantified using Mass Navigator.
E. coli mRNA Microarrays
E. coli BW25113 was grown as described previously (20, 24). For the microarray experiment, we followed the labeling and hybridization methods of Oshima et al. (34). DNA from E. coli BW25113 was used for the control channel. Duplicate two-color experiments were performed using an E. coli gene array named nara_operonEcoK12 registered on the ArrayExpress database. Raw data files were analyzed by the statistical algorithm in ImaGene version 4.0 (BioDiscovery, Los Angeles, CA) using the default parameters. In the absolute analysis of the mRNA present, we set the threshold of 0.1 for signal intensity, and the mRNA signal intensity for each gene was calculated as the mean of values obtained.
Measurement of Protein Abundance of ATP-binding Cassette (ABC) Transporters Using Synthetic Peptides
Seven ABC transporter complexes consisting of the TM subunits and the soluble subunits were selected for quantitation. Two tryptic peptides for each subunit protein were extracted for peptide synthesis using the criteria as described previously (35). The synthetic peptides were purchased from Sigma-Genesis with certificate of analysis. d7-Leucine-labeled E. coli BW25113 cell lysate spiked with known amounts of these synthetic peptides was used to quantify the TM subunit proteins and the soluble subunits of these ABC transporters by the isotope dilution method, and the SILAC (stable isotope labeling by amino acids in cell culture) quantitation was performed between the labeled and unlabeled lysates as described (35). In total, seven TM subunit proteins and six soluble subunit proteins of ABC transporters were quantified.
Quantitative Membrane Proteome Analysis for relA KO Strains
E. coli K12 BW25113 wild-type and relA KO strains from the Keio collection (36) were used in this study. Cells were grown in LB medium at 37 °C with shaking and were harvested at the stationary phase. The preparation of membrane-enriched fraction and the tryptic digestion was performed as described above. Peptides were fractionated into five vials using SCX-StageTips and were analyzed by means of nano-LC-MS/MS with the LTQ-Orbitrap instrument. Labeling of the peptides with iTRAQ reagents was performed according to the manufacturer's protocol. The iTRAQ-labeled peptides were desalted using C18-StageTips and were analyzed using the nano-LC-MS/MS system with QSTAR. The database searches for protein identification were performed as described above except that N-terminal and lysine modifications with the iTRAQ reagent were set as fixed modifications. Protein quantitation by iTRAQ was performed only for proteins with three or more identified peptides.
RESULTS AND DISCUSSION
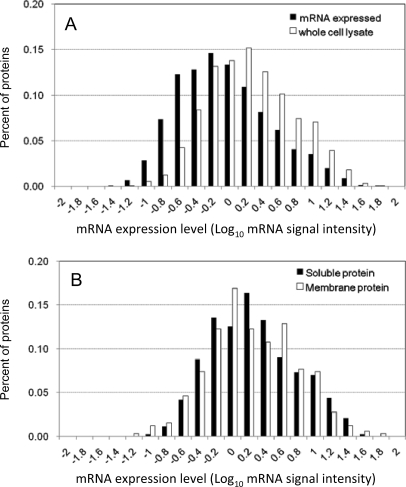
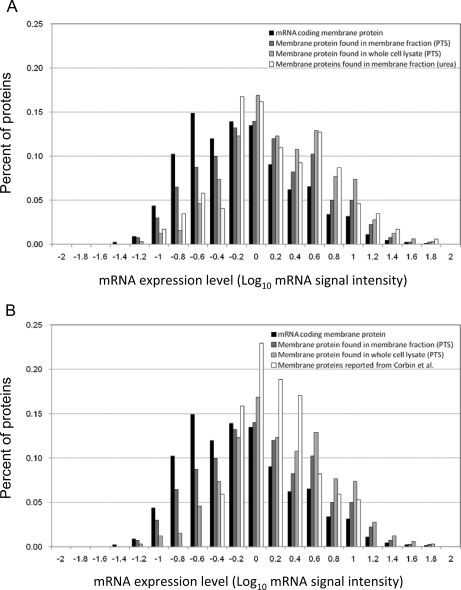
In this study, we used the PTS mixture of 12 mm SDC and 12 mm SLS for the extraction of the E. coli BW25113 proteome instead of 120 mm SDC because this mixture gave slightly superior results (supplemental Table S2). To accelerate the trypsin digestion, we used a spin-type minicolumn packed with trypsin-immobilized beads at 100 × g for 15 min. Note that we did not encounter clogging of the spin column during sample loading because the PTS mixture solubilized the membrane pellet completely as judged by visual inspection. Using this rapid PTS protocol, we digested 22.5 μg of the whole cell lysate of E. coli BW25113 and fractionated it into five vials using SCX-StageTips followed by nano-LC-MS/MS in triplicate. As a result, 1,270 proteins were identified for the whole cell lysate with 0.54% FPR (supplemental Table S3). To categorize these proteins roughly based on their abundance, we used the signal intensities of the corresponding genes measured in the DNA microarray experiment, and the distribution of the protein abundance for identified proteins was plotted against the gene expression. As shown in Fig. 1A, a bias to identify more abundant proteins was observed for the whole cell lysate proteome data. The reason for this bias is that the StageTip-based off-line SCX fractionation was insufficient to reduce the high complexity and the wide dynamic range of the sample (3). We also compared the membrane proteins identified from the whole cell lysate with the soluble proteins. As shown in Fig. 1B, the distribution patterns of both proteomes were in agreement with each other; i.e. there was no difference in the ability to identify membrane proteins and soluble proteins despite the difference in solubility. To reduce the bias due to protein abundance, we used the membrane-enriched fraction from the 22.5 μg of lysate of E. coli BW25113. As expected, the number of identified membrane proteins increased (485 membrane proteins with 1.2% FPR; supplemental Table S4), and the distribution pattern became closer to that of mRNA as compared with that obtained by using the urea protocol for the membrane-enriched lysate (173 membrane proteins; Fig. 2A) and that in the study by Corbin et al. (1) (170 membrane proteins; Fig. 2B). These results indicate that the recovery of low abundance proteins was significantly improved by applying the rapid PTS protocol to less complex samples, such as the membrane-enriched fraction.
Fig. 1.
Comparison of E. coli mRNA expression profile with profile of the corresponding proteins identified by rapid PTS protocol from E. coli whole cell lysate. E. coli whole cell lysate (22.5 μg) was digested, and the resultant peptides were fractionated using SCX-StageTips (five fractions). These five samples were analyzed in triplicate by nano-LC-MS/MS using the LTQ-Orbitrap. A, the contents of proteins identified by the rapid PTS protocol (white bars) and mRNAs detected by DNA microarray (black bars) were plotted as a function of mRNA signal intensity. B, the protein abundance distribution for membrane proteins was compared with that for soluble proteins identified from the whole cell lysate.
Fig. 2.
Abundance distribution of membrane proteins identified under different conditions. E. coli membrane-enriched fraction (22.5 μg) was digested, and resultant peptides were fractionated using SCX-StageTips (five fractions) followed by nano-LC-MS/MS. Analyses were done in triplicate. The abundance distribution of identified proteins was plotted against the gene expression. The abundance distribution for membrane proteins identified from the membrane-enriched fraction and from the whole cell lysate by means of the rapid PTS protocol was compared with that for the membrane-enriched fraction examined by the conventional urea protocol (A) and with that found by Corbin et al. (1) (B).
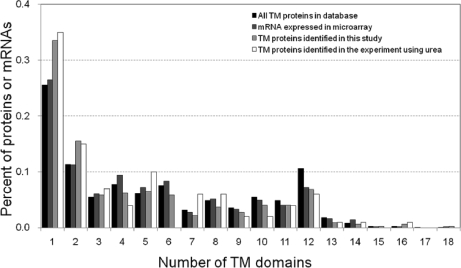
Because TM proteins generally have lower solubility as the number of TMDs per protein increases and multispanning TM proteins are generally difficult to digest with trypsin, we next examined the dependence of the identification efficiency in the PTS protocol on the number of TMDs per protein. The distribution of the number of TMDs per protein for 322 TM proteins identified in this study was compared with that of 541 TM proteins encoded by mRNAs detected in microarray analysis, 1,043 TM proteins registered in the GenoBase database, and 100 TM proteins identified by the urea protocol (Fig. 3). The distribution pattern of proteins identified by the rapid PTS protocol was consistent with both that of proteins encoded by the detected mRNAs and that of GenoBase-registered TM proteins, whereas the urea protocol provided less consistent distribution pattern. These results indicated that the efficiency of the rapid PTS protocol for identification of TM proteins is independent of the number of TMDs per protein. This is consistent with the fact that proteases such as trypsin and chymotrypsin can cleave a TMD if a cleavage site exists within it (supplemental Fig. S1).
Fig. 3.
Comparison of numbers of TM domains of membrane proteins identified according to rapid PTS protocol with number of mRNAs expressed and database values. All proteins with TM domains were extracted from the GenoBase database. In total, 1,043 TM proteins from the database, 542 TM proteins encoded by mRNA expressed in microarray analysis, and 322 TM proteins identified from E. coli cells according to the rapid PTS protocol were used to calculate the content (percentage) of proteins having various numbers of TM domains.
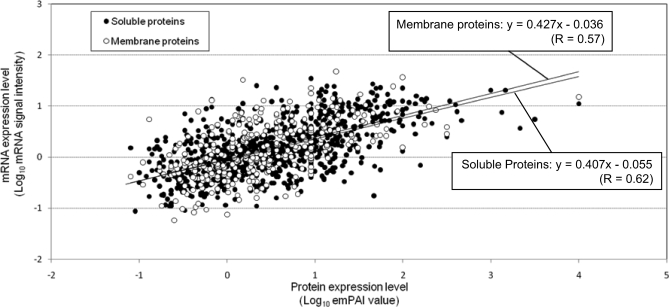
Protein abundance index (PAI), defined as the number of observed precursor ions per protein divided by the number of observable peptides per protein, is linearly related to the logarithm of protein amount. Based on this, emPAI, defined as 10PAI − 1, was introduced to estimate protein abundance (31). However, emPAI has been applied only to soluble proteins because the recovery of membrane proteins is generally lower than that of soluble proteins. This would also be true for spectral count-based protein abundance estimation (37). To show that there is no difference in the recovery of soluble and membrane proteins using the rapid PTS protocol, we compared the emPAI values of soluble proteins with those of membrane proteins. In this case, we used 1,146 mRNA expression data as a reference scale for emPAI data from 820 soluble and 326 membrane proteins. As shown in Fig. 4, a moderate correlation between genes and proteins was observed both for soluble and membrane proteins, and importantly, the relationship between soluble proteins and soluble protein-coding genes was highly consistent with that between membrane proteins and membrane protein-coding genes, indicating that membrane proteins were extracted and digested without any solubility-related bias. Thus, it was considered that the recoveries of soluble and membrane proteins from whole cell lysate were similar when the rapid PTS protocol was applied.
Fig. 4.
Correlation between abundance of mRNAs and membrane proteins or soluble proteins identified from whole cell lysate. mRNA expression levels were estimated from the signal intensities measured in the DNA microarray analysis. In total, 1,146 mRNA expression data were used as a reference scale for emPAI values from 820 soluble and 326 membrane proteins identified from the whole cell lysate. Closed circles, soluble proteins. Open circles, membrane proteins.
Based on this quantitative evaluation, we calculated emPAI values of 1,453 proteins, including 545 membrane proteins, and estimated the absolute copy numbers per cell for each protein using the calibration line between emPAI values and absolute amounts for 36 soluble proteins, which were measured by the isotope dilution method with stable isotope-labeled E. coli BW25113 lysate (33). To validate the emPAI-based copy numbers of proteins estimated in this study, we compared the emPAI-based copy numbers of 26 proteins with the concentrations taken from the literature (38). As shown in supplemental Fig. S2, the emPAI-based copy numbers of these proteins correlate well with literature values over a range of more than 3 orders of magnitude (r = 0.87).
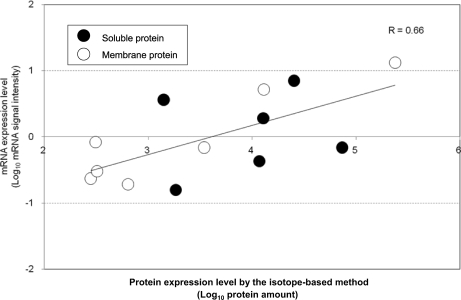
Because emPAI is a semiquantitative parameter (31), we used more accurate abundance values obtained by the stable isotope-based approach to validate that membrane proteins were extracted and digested without any solubility-related bias. ABC transporters were selected as examples because all ABC transporters have TM subunit proteins and soluble subunit proteins associated with the TM proteins on one side of the membrane. The obtained abundance values for seven membrane subunit proteins and six soluble subunit proteins were well correlated with their mRNA expression data, independently of their solubility (Fig. 5). We also confirmed the moderate correlation between the isotope-based protein abundance and emPAI, supporting the validity of emPAI values in this experiment (supplemental Fig. S3).
Fig. 5.
Comparison of protein expression with mRNA expression for 7 transmembrane and 6 soluble subunit proteins from 7 ABC transporter complexes. Protein expression levels were quantified by the isotope-based method, and the amounts injected (amol) for LC-MS/MS were used as protein amounts for this graph. The plotted proteins (with the number of TMDs calculated by TMHMM in parentheses) were ArtI (0), FtsE (0), HisP (0), MalE (0), LolD (0), OppF (0), ArtM (5), FtsX (4), HisJ (1), HisQ (5), MalF (8), LolC (4), and OppA (1). Closed circles, soluble proteins (TMD = 0). Open circles, TM proteins (TMD > 0).
Finally, we applied this rapid PTS protocol to quantitative proteome analysis for membrane and soluble proteins regulated by guanosine tetra- and pentaphosphate ((p)ppGpp) using E. coli relA KO and wild-type BW25113 cells. RelA is the major synthetase of (p)ppGpp, which is accumulated under conditions of nutrient starvation in bacteria (39). It is known that relA KO induces dynamic changes in the gene expression of several membrane protein-coding genes (40, 41). In particular, flagellum- and chemotaxis-related protein-coding genes were reported to be up-regulated in relA KO under nutrient-starved conditions (40). We first used an iTRAQ quantitation approach to quantify these proteins in the wild-type and relA KO cells. A limited number of quantified proteins were obtained (384 proteins). The use of pulsed Q dissociation with the LTQ-Orbitrap instrument (42) did not significantly improve the number of quantified proteins in our case. On the other hand, the emPAI values were obtained for more than 1,000 proteins, including 25 flagellum- and chemotaxis-related proteins from the identification results of the wild-type and the KO cells, and the emPAI ratios of the KO cells to the wild-type cells could be used for relative quantitation. To set the proper threshold for the emPAI ratio, iTRAQ ratios and emPAI ratios were compared with each other (supplemental Fig. S4). As a result, it was found that 91.1% of proteins with more than a 4-fold change in emPAI ratios had iTRAQ ratios with more than a 1.5-fold change (41 of 45 proteins; supplemental Table S5). Based on this, we defined proteins with more than a 4-fold change in emPAI ratios as “up- or down-regulated proteins.” Consequently, using the emPAI-based approach, we identified 1,302 proteins and quantified them by using emPAI ratios (supplemental Table S6). The FPRs calculated for E. coli wild-type and relA KO cells were 0.38 and 0.46%, respectively. The duplicate sample preparation coupled with duplicate LC-MS runs for each sample demonstrated that the reproducibility of the emPAI ratios was, on average, 45.8% relative S.D., which was satisfactory considering the accuracy of emPAI (74.0% on average). For membrane proteins, 567 proteins were quantified both from E. coli wild-type and relA KO cells, and 173 membrane proteins were regulated, including carbon starvation protein A, CstA, which has 18 transmembrane domains. In total, 114 up-regulated membrane proteins were found in the relA KO cells, including 25 flagellum- and chemotaxis-related proteins. Note that all of the quantified proteins related to flagellum and chemotaxis (Fig. 6) were up-regulated in relA mutant as expected (40). In E. coli wild-type, down-regulation of these proteins under conditions of nutrient starvation is advantageous for cell survival because flagellum production and high motility require considerable energy (43). On the other hand, we confirmed that three membrane proteins, acid-resistance membrane protein, cytochrome bd-II oxidase subunit I, and hydrogenase 1 small subunit, decreased in the relA KO strain as reported previously at the gene expression level (41). We also observed other membrane proteins potentially regulated by (p)ppGpp signaling, including 19 transporters and antiporters (supplemental Table S7).
Fig. 6.
Quantitation of flagellum and chemotaxis proteins in relA KO and wild-type cells. Membrane-enriched fractions from relA KO and wild-type cells were digested with the trypsin-immobilized column. The resultant peptides were fractionated using SCX-StageTips (five fractions). These five samples were analyzed by nano-LC-MS/MS using the LTQ-Orbitrap in triplicate analysis. The emPAI values of proteins for relA KO cells were compared with those for wild-type. More than a 4-fold change in emPAI ratios was defined as a significant change of protein expression level based on the iTRAQ ratios. White, up-regulated proteins in relA KO cells. Black, not identified in this study. Asterisk, membrane proteins.
Conclusions
We have demonstrated here for the first time that membrane proteins can be quantitatively extracted, digested, and identified with efficiency similar to that in the case of soluble proteins by means of the rapid PTS protocol. Using this high throughput and unbiased approach for quantitative membrane proteomics, the absolute amounts of 1,453 proteins, including 545 membrane proteins, were simultaneously estimated based on emPAI. This protocol, coupled with emPAI quantitation, was applied to quantitative membrane proteome analysis of E. coli BW25113 wild-type and relA KO cells in the stationary phase, and many membrane proteins were quantified as “regulated” proteins in terms of the response to stimulation. Because this rapid PTS protocol is applicable not only to bacterial cells but also to eukaryote cells and tissues, this approach may be universally suitable for sample preparation in quantitative proteomics.
Data Availability
All peptides identified in this study with annotated MS/MS spectra are available free in Keio University Pepbase peptide database.
Supplementary Material
Acknowledgments
We thank members of our research group for helpful discussions. We also thank Kenji Nakahigashi for helpful support of mRNA microarray analysis.
Footnotes
* This work was supported by research funds from Yamagata Prefecture and Tsuruoka City (to Keio University) and a research grant from the Naito Foundation (to Y. I.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S4 and Tables S1–S7.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S4 and Tables S1–S7.
1 The abbreviations used are:
- PTS
- phase transfer surfactant
- emPAI
- exponentially modified protein abundance index
- FPR
- false-positive rate
- KO
- knock-out
- SLS
- sodium N-lauroylsarcosinate
- (p)ppGpp
- guanosine tetra- and pentaphosphate
- SCX
- strong cation exchange
- SDC
- sodium deoxycholate
- TM
- transmembrane
- TMD
- transmembrane domain
- TMHMM
- TM hidden Markov model
- iTRAQ
- isobaric tag for relative and absolute quantitation
- ABC
- ATP-binding cassette.
REFERENCES
- 1.Corbin R. W., Paliy O., Yang F., Shabanowitz J., Platt M., Lyons C. E., Jr., Root K., McAuliffe J., Jordan M. I., Kustu S., Soupene E., Hunt D. F. ( 2003) Toward a protein profile of Escherichia coli: comparison to its transcription profile. Proc. Natl. Acad. Sci. U.S.A 100, 9232– 9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasonder E., Ishihama Y., Andersen J. S., Vermunt A. M., Pain A., Sauerwein R. W., Eling W. M., Hall N., Waters A. P., Stunnenberg H. G., Mann M. ( 2002) Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature 419, 537– 542 [DOI] [PubMed] [Google Scholar]
- 3.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. ( 2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251– 1254 [DOI] [PubMed] [Google Scholar]
- 4.Han C. L., Chien C. W., Chen W. C., Chen Y. R., Wu C. P., Li H., Chen Y. J. ( 2008) A multiplexed quantitative strategy for membrane proteomics: opportunities for mining therapeutic targets for autosomal dominant polycystic kidney disease. Mol. Cell. Proteomics 7, 1983– 1997 [DOI] [PubMed] [Google Scholar]
- 5.Lu X., Zhu H. ( 2005) Tube-gel digestion: a novel proteomic approach for high throughput analysis of membrane proteins. Mol. Cell. Proteomics 4, 1948– 1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen E. I., Cociorva D., Norris J. L., Yates J. R., 3rd ( 2007) Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J. Proteome Res 6, 2529– 2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang N., Chen R., Young N., Wishart D., Winter P., Weiner J. H., Li L. ( 2007) Comparison of SDS- and methanol-assisted protein solubilization and digestion methods for Escherichia coli membrane proteome analysis by 2-D LC-MS/MS. Proteomics 7, 484– 493 [DOI] [PubMed] [Google Scholar]
- 8.Katayama H., Tabata T., Ishihama Y., Sato T., Oda Y., Nagasu T. ( 2004) Efficient in-gel digestion procedure using 5-cyclohexyl-1-pentyl-beta-D-maltoside as an additive for gel-based membrane proteomics. Rapid Commun. Mass Spectrom 18, 2388– 2394 [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Zhou T., Cao R., Liu Z., Shen J., Chen P., Wang X., Liang S. ( 2006) Evaluation of the application of sodium deoxycholate to proteomic analysis of rat hippocampal plasma membrane. J. Proteome Res 5, 2547– 2553 [DOI] [PubMed] [Google Scholar]
- 10.Yu Y. Q., Gilar M., Lee P. J., Bouvier E. S., Gebler J. C. ( 2003) Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal. Chem 75, 6023– 6028 [DOI] [PubMed] [Google Scholar]
- 11.Norris J. L., Porter N. A., Caprioli R. M. ( 2003) Mass spectrometry of intracellular and membrane proteins using cleavable detergents. Anal. Chem 75, 6642– 6647 [DOI] [PubMed] [Google Scholar]
- 12.Blonder J., Conrads T. P., Yu L. R., Terunuma A., Janini G. M., Issaq H. J., Vogel J. C., Veenstra T. D. ( 2004) A detergent- and cyanogen bromide-free method for integral membrane proteomics: application to Halobacterium purple membranes and the human epidermal membrane proteome. Proteomics 4, 31– 45 [DOI] [PubMed] [Google Scholar]
- 13.Mitra S. K., Gantt J. A., Ruby J. F., Clouse S. D., Goshe M. B. ( 2007) Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J. Proteome Res 6, 1933– 1950 [DOI] [PubMed] [Google Scholar]
- 14.Russell W. K., Park Z. Y., Russell D. H. ( 2001) Proteolysis in mixed organic-aqueous solvent systems: applications for peptide mass mapping using mass spectrometry. Anal. Chem 73, 2682– 2685 [DOI] [PubMed] [Google Scholar]
- 15.Soskic V., Godovac-Zimmermann J. ( 2001) Improvement of an in-gel tryptic digestion method for matrix-assisted laser desorption/ionization-time of flight mass spectrometry peptide mapping by use of volatile solubilizing agents. Proteomics 1, 1364– 1367 [DOI] [PubMed] [Google Scholar]
- 16.Washburn M. P., Wolters D., Yates J. R., 3rd ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol 19, 242– 247 [DOI] [PubMed] [Google Scholar]
- 17.Nomura E., Katsuta K., Ueda T., Toriyama M., Mori T., Inagaki N. ( 2004) Acid-labile surfactant improves in-sodium dodecyl sulfate polyacrylamide gel protein digestion for matrix-assisted laser desorption/ionization mass spectrometric peptide mapping. J. Mass Spectrom 39, 202– 207 [DOI] [PubMed] [Google Scholar]
- 18.Speers A. E., Blackler A. R., Wu C. C. ( 2007) Shotgun analysis of integral membrane proteins facilitated by elevated temperature. Anal. Chem 79, 4613– 4620 [DOI] [PubMed] [Google Scholar]
- 19.Ruth M. C., Old W. M., Emrick M. A., Meyer-Arendt K., Aveline-Wolf L. D., Pierce K. G., Mendoza A. M., Sevinsky J. R., Hamady M., Knight R. D., Resing K. A., Ahn N. G. ( 2006) Analysis of membrane proteins from human chronic myelogenous leukemia cells: comparison of extraction methods for multidimensional LC-MS/MS. J. Proteome Res 5, 709– 719 [DOI] [PubMed] [Google Scholar]
- 20.Masuda T., Tomita M., Ishihama Y. ( 2008) Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res 7, 731– 740 [DOI] [PubMed] [Google Scholar]
- 21.Wu C. C., MacCoss M. J., Howell K. E., Yates J. R., 3rd ( 2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol 21, 532– 538 [DOI] [PubMed] [Google Scholar]
- 22.Fischer F., Wolters D., Rögner M., Poetsch A. ( 2006) Toward the complete membrane proteome: high coverage of integral membrane proteins through transmembrane peptide detection. Mol. Cell. Proteomics 5, 444– 453 [DOI] [PubMed] [Google Scholar]
- 23.Nielsen P. A., Olsen J. V., Podtelejnikov A. V., Andersen J. R., Mann M., Wisniewski J. R. ( 2005) Proteomic mapping of brain plasma membrane proteins. Mol. Cell. Proteomics 4, 402– 408 [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki M., Masuda T., Tomita M., Ishihama Y. ( 2009) Chemical cleavage-assisted tryptic digestion for membrane proteome analysis. J. Proteome Res 8, 3169– 3175 [DOI] [PubMed] [Google Scholar]
- 25.Rappsilber J., Mann M., Ishihama Y. ( 2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc 2, 1896– 1906 [DOI] [PubMed] [Google Scholar]
- 26.Ishihama Y., Rappsilber J., Mann M. ( 2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res 5, 988– 994 [DOI] [PubMed] [Google Scholar]
- 27.Rappsilber J., Ishihama Y., Mann M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem 75, 663– 670 [DOI] [PubMed] [Google Scholar]
- 28.Ishihama Y., Rappsilber J., Andersen J. S., Mann M. ( 2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233– 239 [DOI] [PubMed] [Google Scholar]
- 29.Kyte J., Doolittle R. F. ( 1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 157, 105– 132 [DOI] [PubMed] [Google Scholar]
- 30.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. ( 2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol 305, 567– 580 [DOI] [PubMed] [Google Scholar]
- 31.Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. ( 2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265– 1272 [DOI] [PubMed] [Google Scholar]
- 32.Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F. U., Kerner M. J., Frishman D. ( 2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii N., Nakahigashi K., Baba T., Robert M., Soga T., Kanai A., Hirasawa T., Naba M., Hirai K., Hoque A., Ho P. Y., Kakazu Y., Sugawara K., Igarashi S., Harada S., Masuda T., Sugiyama N., Togashi T., Hasegawa M., Takai Y., Yugi K., Arakawa K., Iwata N., Toya Y., Nakayama Y., Nishioka T., Shimizu K., Mori H., Tomita M. ( 2007) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593– 597 [DOI] [PubMed] [Google Scholar]
- 34.Oshima T., Wada C., Kawagoe Y., Ara T., Maeda M., Masuda Y., Hiraga S., Mori H. ( 2002) Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol 45, 673– 695 [DOI] [PubMed] [Google Scholar]
- 35.Ishihama Y., Sato T., Tabata T., Miyamoto N., Sagane K., Nagasu T., Oda Y. ( 2005) Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat. Biotechnol 23, 617– 621 [DOI] [PubMed] [Google Scholar]
- 36.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. ( 2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H., Sadygov R. G., Yates J. R., 3rd ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem 76, 4193– 4201 [DOI] [PubMed] [Google Scholar]
- 38.Taoka M., Yamauchi Y., Shinkawa T., Kaji H., Motohashi W., Nakayama H., Takahashi N., Isobe T. ( 2004) Only a small subset of the horizontally transferred chromosomal genes in Escherichia coli are translated into proteins. Mol. Cell. Proteomics 3, 780– 787 [DOI] [PubMed] [Google Scholar]
- 39.Cashel M., Gentry D. R., Hernandez V. J., Vinella D. ( 1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology ( Neidhardt F. C., Curtiss R. I., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Rfznikopp W. S., Riley M., Schaechter M., Umbarger H. D. eds) 2nd Ed., pp. 1458– 1496, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 40.Durfee T., Hansen A. M., Zhi H., Blattner F. R., Jin D. J. ( 2008) Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol 190, 1084– 1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traxler M. F., Summers S. M., Nguyen H. T., Zacharia V. M., Hightower G. A., Smith J. T., Conway T. ( 2008) The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol 68, 1128– 1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bantscheff M., Boesche M., Eberhard D., Matthieson T., Sweetman G., Kuster B. ( 2008) Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteomics 7, 1702– 1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macnab R. M. ( 1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology ( Neidhardt F. C., Curtiss R. I., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Rfznikopp W. S., Riley M., Schaechter M., Umbarger H. D. eds) 2nd Ed., pp. 123– 145, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All peptides identified in this study with annotated MS/MS spectra are available free in Keio University Pepbase peptide database.