Abstract
Detection of lung cancer at an early stage is necessary for successful therapy and improved survival rates. We performed a bottom-up proteomics analysis using a two-dimensional LC-MS/MS strategy on the conditioned media of four lung cancer cell lines of different histological backgrounds (non-small cell lung cancer: H23 (adenocarcinoma), H520 (squamous cell carcinoma), and H460 (large cell carcinoma); small cell lung cancer: H1688) to identify secreted or membrane-bound proteins that could be useful as novel lung cancer biomarkers. Proteomics analysis of the four conditioned media allowed identification of 1,830 different proteins (965, 871, 726, and 847 from H1688, H23, H460, and H520, respectively). All proteins were assigned a subcellular localization, and 38% were classified as extracellular or membrane-bound. We successfully identified the internal control proteins (also detected by ELISA), kallikrein-related peptidases 14 and 11, and IGFBP2. We also identified known or putative lung cancer tumor markers such as squamous cell carcinoma antigen, carcinoembryonic antigen, chromogranin A, creatine kinase BB, progastrin-releasing peptide, neural cell adhesion molecule, and tumor M2-PK. To select the most promising candidates for validation, we performed tissue specificity assays, functional classifications, literature searches for association to cancer, and a comparison of our proteome with the proteome of lung-related diseases and serum. Five novel lung cancer candidates, ADAM-17, osteoprotegerin, pentraxin 3, follistatin, and tumor necrosis factor receptor superfamily member 1A were preliminarily validated in the serum of patients with lung cancer and healthy controls. Our results demonstrate the utility of this cell culture proteomics approach to identify secreted and shed proteins that are potentially useful as serological markers for lung cancer.
Lung cancer is the leading cause of cancer-related mortality worldwide in both men and women. An estimated 213,000 news cases and 160,000 deaths from lung cancer occur in the United States every year (National Cancer Institute). According to the World Health Organization, lung cancers are largely classified into two histologically distinct types, based on the size and appearance of the malignant cells: small cell (SCLC)1 and non-small cell lung cancer (NSCLC). NSCLC, which comprises more than 80% of lung cancers, can be further divided into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma.
Despite advances in treatments such as surgery, chemotherapy, and radiotherapy, the clinical outcome for patients with lung cancer still remains poor. The overall 5-year survival rate is only 10–15% (1) mainly because, at the time of diagnosis, most lung cancer patients are at advanced stages. In this context, there is a critical need to detect lung cancer earlier by improving the current diagnostic methods such as computed tomography and chest x-ray and by discovering useful diagnostic and prognostic biomarkers. To date, a number of serum biomarkers for lung cancer have been studied, including CEA, squamous cell carcinoma (SCC)-Ag, neuron-specific enolase, tissue polypeptide antigen, CYFRA21-1 (cytokeratin 19 fragment), and pro-GRP. They are elevated in serum of patients with lung cancer, but they are not sensitive or specific enough, alone or in combination, to reliably diagnose asymptomatic patients with lung cancer.
Recently, new approaches in clinical proteomics have been developed to identify novel biomarkers of lung pathology (COPD, asthma, pleural effusion, and cancer) and to gain insights into disease mechanisms in which proteins play a major role. Some proteomics analyses of various biological fluids associated with the human airway have been reported, including nasal lavage fluid (2–4), bronchoalveolar lavage fluid (5, 6), and saliva (7, 8). By using a combination of 2DE analysis and Gel electrophoresis coupled with LC-MS/MS, Nicholas et al. (9) identified 258 proteins in human sputum, and among them, 191 were of human origin. Proteins included lower and upper airway secretory products, cellular products, and inflammatory cell-derived products. In addition, Casado et al. (10) used capillary column LC-ESI-Q/TOF-MS to investigate the proteome profiles of hypertonic saline-induced sputum samples from healthy smokers and patients with COPD of different severity. A total of 203 unique proteins were identified of which some may be markers of COPD severity. The proteomics profile of human pleural effusion from 43 lung adenocarcinoma was also studied using a 2D nano-HPLC-ESI-MS/MS system (11). The results revealed 1,415 unique proteins of which 124 were identified with higher confidence (at least two unique peptide sequences matched). However, there are inherent limitations of using MS for biomarker discovery in complex biological mixtures such as fluids or serum (12, 13), requiring methodologies for depletion of high abundance proteins such as albumin and immunoglobulins. These limitations illustrate the need to find other sources to mine for biomarker discovery.
One approach to overcome this limitation posed by complex mixtures is by using a cell culture model, in which cells are grown in serum-free medium, to perform proteomics analysis. This model offers various advantages over the traditional cultures in serum-supplemented medium: it reduces complexity by avoiding interferences from nutritional proteins present in the medium, increases the reproducibility, and allows detection of low abundance proteins. This strategy has been successfully used in our laboratory for the discovery of novel breast and prostate biomarkers (14, 15). This technique was also reported in lung-related proteomics approaches. Tachibana et al. (16) reported the regulatory roles of β1 integrin in morphological differentiation in CADO LC6 cells, an SCLC cell line cultured in serum-free medium. To explore serum biomarkers of lung cancer at early stage, M-BE, an SV40T-transformed human bronchial epithelial cell line with the phenotypic features of early tumorigenesis at high passage, was cultured, and the conditioned medium was used to collect its secretory proteins (17). Proteins secreted from different passages of M-BE cells were extracted and then separated by 2DE followed by MALDI-TOF/TOF mass spectrometry. The authors identified 47 proteins, including cathepsin D, that exhibited increased abundance in culture media or cells during passaging. Moreover, Xiao et al. (18) analyzed the proteins released into the serum-free medium from the tumor microenvironment with short time-cultured lung cancer and adjacent normal bronchial epithelial cells, thus demonstrating the versatility of this approach.
In this study, we performed a shotgun proteomics analysis of the conditioned media of four lung cancer cell lines of differing histotypes. Our aim was to identify secreted or membrane-bound proteins that could be useful as novel lung cancer biomarkers. Five proteins were elevated in serum of lung cancer patients, suggesting that they may represent lung cancer biomarkers that are worth validating in the future.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The four lung cancer cell lines, H23 (CRL-5800), H520 (HTB-182), H460 (HTB-177), and H1688 (CCL-257), were purchased from the American Type Culture Collection (ATCC, Manassas, VA). These cell lines represent the four major histological lung cancer subtypes: (i) NSCLC adenocarcinoma (H23), (ii) NSCLC squamous cell carcinoma (H520), (iii) NSCLC large cell carcinoma (H460), and (iv) SCLC (H1688). All cell lines were maintained in 75-cm2 culture flasks in RPMI 1640 culture medium (BD Biosciences) supplemented with 8% fetal bovine serum (FBS) (Hyclone). All cells were cultured in a humidified incubator at 37 °C and 5% CO2.
Cells were seeded at different seeding densities (4 × 106 cells for H460, 8 × 106 cells for H23, 10 × 106 cells for H1688, and 12 × 106 cells for H520) into six 175-cm2 culture flasks per cell line (with the exception of three flasks for H460) and grown for 2 days in 30 ml of RPMI 1640 medium supplemented with 8% FBS. After 2 days, the culture medium was removed, and the cells were rinsed three times with 30 ml of 1× PBS (Invitrogen). Then, 30 ml of chemically defined Chinese hamster ovary (CDCHO) serum-free medium (Invitrogen) supplemented with glutamine (8 mm) (Invitrogen) were added to the flasks, and the flasks were incubated for 48 h. The H520 cell line was grown as described above except that the cells were incubated for 3 days in RPMI 1640 medium supplemented with 8% FBS before the medium was changed to CDCHO serum-free medium. All cell lines were grown in triplicate and independently processed and analyzed. The same conditions and procedures were applied to set up a negative control. In this case, 30 ml of RPMI 1640 medium supplemented with 8% FBS were prepared as mentioned above with no cells added to the 175-cm2 culture flask.
After incubation in CDCHO, the conditioned media (CM) were collected and spun down to remove cellular debris. The CM were then frozen at −80 °C until further processing. Aliquots (1 ml) were taken from the CM at the time of harvest for measurement of total protein and lactate dehydrogenase as well as kallikrein-related peptidases 11 and 14 and insulin-like growth factor-binding protein 2 (internal control proteins) by using specific ELISAs.
Measurement of Total Protein, Lactate Dehydrogenase, Kallikreins 11 and 14, and IGFBP2
Total protein was quantified in the CM using a Coomassie (Bradford) assay (Pierce) according to the manufacturer's instructions. Lactate dehydrogenase (indicator of cell death) was measured in the CM using an enzymatic assay based on lactate to pyruvate conversion and parallel production of NADH from NAD+. The production of NADH was monitored at 340 nm using an automated method (Roche Applied Science Modular Systems).
Kallikrein-related peptidases 11 and 14 were measured with in-house ELISAs as described previously (19–21). An IGFBP2 sandwich ELISA kit, purchased from R&D Systems (Minneapolis, MN), was used to measure levels of IGFBP2 in the CM of lung cancer cell lines.
Conditioned Media Sample Preparation
One CM aliquot (30 ml) was collected for the cell line H460, whereas two 30-ml CM aliquots were combined (60 ml) for the three cell lines H23, H1688, and H520. Three biological replicates per cell line were performed. Each replicate contained ∼800 μg to 1 mg of total protein.
These replicates were dialyzed using a 3.5-kDa-molecular mass cutoff membrane (Spectrum Laboratories, Inc., Compton, CA). The CM were dialyzed overnight at 4 °C in 5 liters of 1 mm ammonium bicarbonate solution with two buffer changes. The dialyzed CM were frozen and lyophilized to dryness. Following lyophilization, samples were denatured using 8 m urea and reduced with DTT (final concentration of 13 mm; Sigma-Aldrich) at 50 °C for 30 min. Then, samples were alkylated with 500 mm iodoacetamide (Sigma-Aldrich) in the dark at room temperature for 1 h and desalted using a NAP5 column (GE Healthcare). The 1-ml final samples were lyophilized and trypsin (Promega)-digested at a molar ratio of 1:50 (trypsin:protein concentration) overnight at 37 °C. Finally, the peptides were lyophilized to dryness.
Strong Cation Exchange Liquid Chromatography
The trypsin-digested lyophilized samples were resuspended in 120 μl of 0.26 m formic acid in 10% acetonitrile (mobile phase A). The samples were fractionated using an Agilent 1100 HPLC system connected to a PolySULFOETHYL A™ column with a 200-Å pore size and a diameter of 5 μm (The Nest Group Inc.). A 1-h linear gradient was used with 1 m ammonium formate and 0.26 m formic acid in 10% acetonitrile (mobile phase B) at a flow rate of 200 μl/min. Fractions were collected via a fraction collector every 5 min (12 fractions per run) and frozen at −80 °C for further use. A peptide cation exchange standard, consisting of three peptides, was run at the beginning of each day to assess column performance (Bio-Rad).
Mass Spectrometry (LC-MS/MS)
Of the 12 fractions collected per HPLC run, seven fractions (fractions 5–11, containing the bulk of peptides) were analyzed by mass spectrometry. The seven fractions per replicate per cell line were C18-extracted using a ZipTipC18 pipette tip (Millipore) and eluted in 4 μl of 90% ACN, 0.1% formic acid, 10% water, and 0.02% TFA (buffer B). Eighty microliters of 95% water, 0.1% formic acid, 5% ACN, and 0.02% TFA (buffer A) were added to this mixture, and 40 μl were injected via an autosampler on an Agilent 1100 HPLC system. The peptides were first collected onto a 2-cm C18 trap column (inner diameter, 200 μm) and then eluted onto a resolving 5-cm analytical C18 column (inner diameter, 75 μm) with an 8-μm tip (New Objective). The HPLC system was coupled on line to a 2D linear ion trap (LTQ, Thermo Inc.) mass spectrometer using a nano-ESI source in data-dependent mode. Each fraction was run with a 120-min gradient. The eluted peptides were subjected to MS/MS. DTAs were created using the Mascot Daemon v2.16 and extract_msn (Matrix Science, London, UK). The parameters for DTA creation were as follows: minimum mass, 300 Da; maximum mass, 4,000 Da; automatic precursor charge selection; minimum peaks, 10 per MS/MS scan for acquisition; and minimum scans per group, 1.
Data Analysis
Mascot (Matrix Science; version 2.1.03) and X!Tandem (Global Proteome Machine Manager, Beavis Informatics Ltd.; version 2.0.0.4) search engines were used to analyze the resulting raw mass spectra from each fraction. Each fraction was analyzed by both search engines on the International Protein Index human database (version 3.16; >62,000 entries) (22). One missed cleavage was allowed, and searches were performed with fixed carbamidomethylation of cysteines and variable oxidation of methionine residues. A fragment tolerance of 0.4 Da and a parent tolerance of 3.0 Da were used for both search engines with trypsin as the specified digestion enzyme. This operation resulted in seven DAT files (Mascot) and seven XML files (X!Tandem) for each replicate sample per cell line. Scaffold (version Scaffold-01_06_19, Proteome Software Inc., Portland, OR) was utilized to validate MS/MS-based peptide and protein identifications. The cutoffs in Scaffold were set for 95% peptide identification probability as specified by the PeptideProphet algorithm (23) and 80% protein identification probability as assigned by ProteinProphet algorithm (24). Identifications not meeting these criteria were not included in the displayed results. The DAT and XML files for each cell line plus their respective negative control files (RPMI 1640 culture medium only) were inputted into Scaffold to cross-validate Mascot and X!Tandem data files. Each replicate sample was designated as one biological sample containing both DAT and XML files in Scaffold and searched with the multidimensional protein identification technology option selected. Using a similar approach of analysis of conditioned media from breast and prostate cancer cell lines, we observed a false positive error rate of 1–2% using the sequence-reversed International Protein Index human database.
The sample reports were exported to Excel, and an in-house developed program was used to extract Genome Ontology (GO) terms for cellular component for each protein and the proportion of each GO term in the data set. Proteins that were not able to be classified by GO terms were checked with Swiss-Prot entries and against the Human Protein Reference Database and Bioinformatic Harvester to search for cellular component annotations. The overlap between proteins identified from each cell line and between the three replicates of each cell line was assessed using an in-house developed program. All extracellular and membrane-bound proteins were also searched against the Plasma Proteome Database. The list of displayed proteins was also compared with those found in other lung-related proteomics studies (9–11, 18, 25, 26). Finally, we classified the extracellular and membrane proteins identified by cellular function and disease using Ingenuity Pathway Analysis software (Ingenuity Systems). In addition, the molecular functions associated with each of the biomarker candidates were analyzed with the Ingenuity Pathway Analysis software.
Validation of Lung Biomarker Candidates: Clinical Samples and ELISA Analysis
Fifty subjects, including 25 cases diagnosed with NSCLC and 25 normal healthy donors, were enrolled in this study. Samples were collected at the UCLA Medical Center between October 2004 and March 2006 in accordance with the UCLA Institutional Review Board approval and patient written informed consent. Peripheral blood was collected from patients at least 4 weeks prior to receiving therapy for patients with advanced disease. In patients who had previously undergone surgical resection, blood was collected after recurrence at least 1 year following surgery. Plasma was collected in EDTA-containing Vacutainer tubes. Samples were centrifuged at 3,000 rpm for 15 min within 1 h of collection, separated, and stored in aliquots at −80 °C. Staging was determined by the American Joint Committee on Cancer Guidelines. Distributions of patients by demographic and clinical characteristics are presented in supplemental Tables 1–5 for each of the candidates tested.
Serum levels of pentraxin-3 (tumor necrosis factor-stimulated gene 14), follistatin, and sTNFRI were measured by ELISA using a commercially available kit (R&D Systems). Serum levels of osteoprotegerin and ADAM-17 were measured using an in-house developed ELISA using commercial antibodies purchased from R&D Systems.
Statistical Analysis
The differences between groups were evaluated by the Mann-Whitney U test using GraphPad Prism version 4 for Windows (GraphPad Software, San Diego, CA). All comparisons were two-tailed, and p values of <0.05 were considered significant.
RESULTS
Optimization of Cell Culture Conditions
To delineate the secretome of the four lung cancer cell lines, cell culture conditions were first optimized to minimize cell death and maximize secreted protein concentration. For this purpose, cells were grown in serum-free medium for 48 h at different seeding densities. Total protein, lactate dehydrogenase levels, and the concentration of IGFBP2 in the CM of H1688, H520, H460, and H23 cells and kallikrein-related peptidase 11 (KLK11) and KLK14 in the CM of H1688 cells were measured. The ratio of IGFBP2 concentration to lactate dehydrogenase levels for each cell culture condition and the ratio of KLK11 and KLK14 concentrations to lactate dehydrogenase levels measured in the CM of H1688 cell line were compared (supplemental Figs. 1–4). The following optimal seeding densities were selected for proteomics analysis (4 × 106 cells for H460, 8 × 106 cells for H23, 10 × 106 cells for H1688, and 12 × 106 cells for H520) as those gave the highest ratio of IGFBP2 or KLK production (indicator of secreted proteins) to lactate dehydrogenase (indicator of cell death).
At the optimized seeding densities, total protein concentration was 38, 15, 14, and 15 μg/ml for H460, H23, H1688, and H520, respectively. Furthermore, proteomics analysis was accomplished with ∼800 μg to 1 mg of total protein.
Identification of Proteins by Mass Spectrometry
In this study, we used an experimental design for sample preparation, LC/MS/MS, and bioinformatic analysis similar to that described previously (14). Using Scaffold, which contains ProteinProphet and PeptideProphet software, a list of all proteins with an 80% probability and all peptides with a 95% probability was generated. In total, from the three replicates per cell line, 965, 871, 726, and 847 proteins were identified in the H1688, H23, H460, and H520 cell lines, respectively (supplemental Tables 6–9). The complete list of the 1,830 different proteins identified in this proteomics analysis is presented in supplemental Table 10. From the negative control flask that did not contain any cells but was treated in the same manner as the CM of the four cell lines, a total of 84 proteins were identified (supplemental Table 10). Many of these were proteins derived from FBS, which was used to initially culture the cells. These proteins were removed from the list of total proteins identified in the CM of each lung cancer cell line and were not considered further in data analysis.
Overlap of Proteins between Replicates and Reproducibility of Method
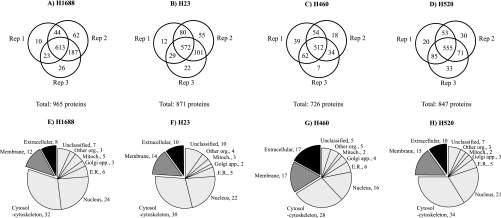
To investigate the reproducibility of our method, each cell line was cultured in triplicate, providing three independent biological replicates per cell line. Fig. 1 shows the overlap between the three replicates of each cell line. For H1688, we identified 965 proteins (Fig. 1A). Of these, 613 were identified in all three replicates, yielding a reproducibility of 63.5%. For the H23 cell line, a total of 871 proteins were identified (Fig. 1B) of which 572 were common to all three replicates (65.7% reproducibility). Furthermore, 726 proteins were identified in H460 (Fig. 1C) of which 512 were found in all three replicates (70.5% reproducibility). Finally, we identified 847 proteins in H520. Of these, 555 were common to all three replicates, yielding a reproducibility of 65.5% (Fig. 1D). Approximately 20–26% of proteins were found in two replicates, whereas ∼10% were exclusive to one replicate.
Fig. 1.
Number of proteins identified by LC-MS/MS in CM of four lung cancer cell lines (A–D) and their cellular localization (E–H). Shown is the overlap of three independent replicates (Rep 1–3) and total number of proteins identified. The lower panel depicts cellular localization. Mitoch., mitochondria; Golgi app., Golgi apparatus; E.R., endoplasmic reticulum; Other org., other organelles.
Identification of Internal Control Proteins by MS
To monitor the cell culture optimization process, the concentration of two kallikrein-related peptidases (KLK11 and KLK14) and IGFBP2, which are known to be secreted, was measured by ELISA in the CM of the four lung cancer cell lines. All cell lines expressed IGFBP2 (15–110 μg/liter), whereas H1688 was the only cell line expressing KLK11 (6.3 μg/liter) and KLK14 (1.9 μg/liter) at levels measurable by ELISA. Using our MS approach, we were able to confidently identify KLK11 and KLK14 in the CM of H1688 and IGFBP2 in the CM of all four cell lines. Supplemental Fig. 5 illustrates the sequences of KLK11, KLK14, and IGFBP2 and the peptides identified by MS. IGFBP2 displayed ∼10 unique peptides in all three replicates of each cell line, covering ∼40% of its sequence (supplemental Fig. 5A). Six (replicate 1) to seven unique peptides (replicates 2 and 3) were identified for KLK11, resulting in a 28–41% sequence coverage (supplemental Fig. 5B). Furthermore, KLK14 was identified in two replicates of H1688 by one and three unique peptides, respectively. This resulted in a 5–17% sequence coverage (supplemental Fig. 5C). H520, H23, and H460 cells did not secrete any detectable KLK14 by ELISA, and as expected, this kallikrein-related peptidase was not found in their CM by MS. Thus, successful identification of our endogenous internal control proteins by MS, especially those expressed at relatively low levels (KLK11 and KLK14), demonstrated that the detection limit of our MS method for marker identification was in the low μg/liter range.
Classification of Proteins Identified by MS by Cellular Localization
Each identified protein was classified by its cellular localization using the Genome Ontology, Swiss-Prot, Human Protein Reference, and Bioinformatic Harvester databases. These categories are non-exclusive because a protein can be classified in more than one cellular compartment. Fig. 1, E–H, shows the cellular localization of proteins identified in the CM of H1688 (E), H23 (F), H460 (G), and H520 (H). Twenty to 34% of the proteins identified were classified as extracellular or membrane-bound in each cell line. The remainder of the proteins identified in the CM were classified as intracellular (>50% (cytosol-cytoskeleton, nucleus, endoplasmic reticulum, Golgi apparatus, mitochondria, and other organelles such as endosomes and lysosomes)), whereas 5–10% were unclassified. Supplemental Tables 6–9 display the classification of proteins by cellular localization for each cell line.
Overlap of Proteins between Four Lung Cancer Cell Lines
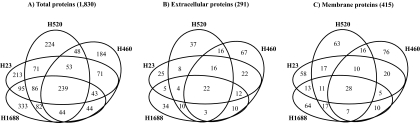
The proteins identified among the four lung cancer cell lines were analyzed for overlap using an in-house developed program (Fig. 2). Of the 1,830 unique proteins identified in this study, 239 (13%) were common to all four cell lines (Fig. 2A). Moreover, 226 (12.4%) and 411 (22.5%) proteins were found in three and two of the cell lines, respectively. Interestingly, about 52% of the proteins identified were unique to one of the cell lines.
Fig. 2.
Overlap of proteins identified in CM by LC-MS/MS between each of four lung cancer cell lines. The overlap of total proteins (A; total number in parentheses), extracellular proteins (B), and membrane proteins (C) is shown.
Fig. 2, B and C, display the overlap among the 291 extracellular proteins and the 415 membrane-bound proteins, respectively. We found that 22 (about 8%) of the extracellular proteins and 28 (about 7%) of the membrane-bound proteins were common to all four cell lines. Consistent with our previous findings on the overlap of proteins between the cell lines, a large portion of extracellular proteins (56%) and membrane-associated proteins (63%) was identified in only one cell line. These results illustrate the heterogeneity of lung cancer cell lines and the requirement of analyzing multiple cell lines to better depict the secretome of lung cancer. Supplemental Tables 10–12 present the overlap of total, extracellular, and membrane-bound proteins between cell lines.
Extracellular and Membrane-bound Proteins Identified by MS
According to GO annotation, 291 proteins (15.9%) were classified as extracellular, and 415 proteins (22.7%) were classified as membranous. From the list of extracellular and membrane-bound proteins, some known or putative lung cancer biomarkers were identified. These included CEA (27, 28), chromogranin A (29), chromogranin B (30), gastrin-releasing peptide (29, 31), kallikrein-related peptidases 11 and 14 (32–34), matrix metallopeptidase 1 collagenase (18), and neural cell adhesion molecule (35–37) (Table I). Moreover, all of the extracellular and membrane-bound proteins were compared with the human Plasma Proteome Database to determine whether they have been found previously in plasma. Of 291 secreted proteins, 129 (44.3%) were identified in human plasma. One hundred and sixty-eight of 415 membranous proteins (40.5%) were also found in human plasma. These are highlighted in supplemental Tables 11 and 12. Finally, supplemental Table 13 contains detailed information on all of the proteins identified for each of the cell lines, including the number of unique peptides, peptide sequences, precursor ion mass, and charge states.
Table I. Examples of known and putative lung cancer biomarkers identified in the conditioned media of H1688, H23, H460, and H520 cell lines by LC-MS/MS.
LDH, lactate dehydrogenase; EGFR, epidermal growth factor receptor; CGA, chromogranin A; CK-BB, creatine kinase BB.
| Protein | H1688a | H23a | H460a | H520a | Relationship | Refs. |
|---|---|---|---|---|---|---|
| CEA | 7, 8, 8 | Useful indicator of disease extent and may potentially have important prognostic value for NSCLC patients. Serum CEA level appears to be closely associated with the presence of EGFR gene mutations in patients with pulmonary adenocarcinomas. | 27, 28 | |||
| Chromogranin A | 17, 15, 14 | 11, 10, 12 | High serum levels of CGA before chemotherapy were found as an unfavorable prognostic determinant for NSCLC patients. | 29 | ||
| Chromogranin B | 30, 26, 23 | 6, 7, 7 | 29, 22, 31 | Marker for the immunohistochemical demonstration of carcinoids and well differentiated neuroendocrine carcinomas. | 30 | |
| Creatine kinase BB | 10, 13, 12 | 1, 3, 1 | 1, 1, 2 | 9, 12, 12 | Elevated levels of CK-BB found in the serum of SCLC patients. Relationship found between enhanced levels of CK-BB and the degree of lung carcinoma advance. | 65, 66 |
| Progastrin-releasing peptide | 4, 3, 4b | High serum levels of pro-GRP before treatment conferred a survival advantage for NSCLC patients. Higher serum levels of pro-GRP-(31–98) in patients with extensive SCLC than in patients with limited disease. Higher serum levels of pro-GRP-(31–98) in patients with pure small cell carcinoma than in patients with mixed small cell/large cell carcinoma. | 29, 31 | |||
| Kallikrein 11 | 6, 7, 7 | Higher serum levels in NSCLC than in healthy volunteers; associated with higher risk of NSCLC. KLK11 mRNA overexpression in a subgroup of neuroendocrine tumors with unfavorable outcome. | 32, 33 | |||
| Kallikrein 14 | 1, 0, 3 | KLK14 mRNA overexpression in lung tumors associated with a positive nodal status of the tumor. Higher serum levels in NSCLC than in healthy volunteers. Associated with higher risk of NSCLC. | 33, 34 | |||
| l-Lactate dehydrogenase B chain | 4, 6, 6 | 8, 8, 10 | 8, 6, 7 | 6, 5, 7 | Elevated serum LDHB correlated with the clinical stage of lung cancer. | 67 |
| MMP1 collagenase | 0, 4, 3 | 15, 15, 14 | Elevated serum levels in late stage lung cancer patients. Overexpression in tumor tissues and association with tumor invasion and metastasis. | 18, 68 | ||
| Neural cell adhesion molecule | 0, 1, 2 | 1, 0, 0 | Elevated serum levels in patients with SCLC. Patients with pathologic NCAM levels have significantly shorter survival times. Potential tumor marker for SCLC. Raised serum NCAM in active SCLC and in patients in relapse suggests that NCAM can be used as a target for antibody-directed therapy of micrometastases. | 35–37 | ||
| Peroxiredoxin 1 | 12, 12, 9 | 12, 12, 11 | 10, 10, 13 | 16, 17, 18 | Up-regulated in lung cancer and may serve as a prognostic biomarker and therapeutic target in NSCLC. | 69 |
| SCC antigen | 3, 1, 0 | Both the preoperative SCC-Ag level and its postoperative decrease have prognostic significance inferior to stage of disease. Elevated levels (>1.5 ng/ml) of SCC were observed in 52.7% of squamous cell lung cancer patients but in only 14.2% of nonsquamous cell lung cancer patients. | 47, 48 | |||
| Tumor M2-PK | 20, 21, 22 | 14, 21, 24 | 22, 22, 28 | 0, 2, 0 | Elevated serum M2-PK found with progressive lung tumor stages. | 49 |
a Each column contains the number of unique peptides per protein; three values are reported for each protein representing each of the three replicate samples per cell line.
b Combination of isoforms 1 and 2 of gastrin-releasing peptide identified in H1688 CM.
Comparison of Present Proteome with Other Lung Proteomics Publications
We compared proteins identified in the four lung cancer cell lines with the proteomes of lung-related diseases and lung-related biological fluids.
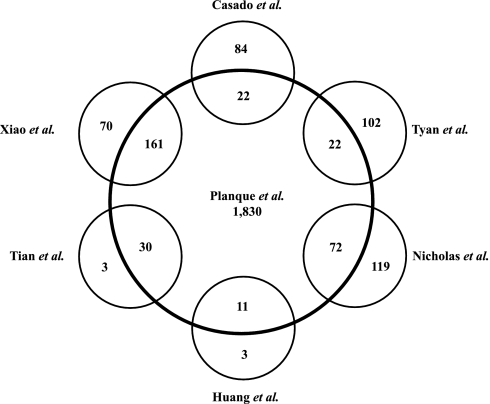
Xiao et al. (18) used proteomics techniques to analyze CM from primary cultures of lung cancer cells and adjacent normal bronchial epithelial cells of six lung cancer patients. Using one-dimensional PAGE and nano-ESI-MS/MS, they identified 231 proteins of which 161 (70%) were also found in our proteomics study. Huang et al. (25) analyzed secreted proteins in the CM of an NSCLC cell line (A549) by 2D PAGE and MALDI-TOF MS. Fourteen human proteins were identified of which 11 (79%) were also found by us, including α-enolase, peroxiredoxin 1, galectin 1, ubiquitin carboxyl-terminal hydrolase (PGP9.5), and dihydrodiol dehydrogenase. In addition, comparative proteomics analysis of the two NSCLC cell lines with different metastatic potential was carried out using 2DE followed by MALDI-TOF MS and MS/MS analysis. Thirty-three differentially expressed proteins were identified, including 16 proteins that were significantly up-regulated and 17 proteins that were down-regulated in highly metastatic cells compared with non-metastatic cells (26). Of these 33 proteins reported to be altered, 30 (91%) were also found among the 1,830 proteins in our CM. Importantly, all proteins identified as up-regulated in highly metastatic cells were identified in our study. Among these candidates, Tian et al. (26) observed by immunohistochemistry a correlation between up-regulation of S100A11 expression in NSCLC tissues and higher tumor-node-metastasis stage and positive lymph node status.
We also compared our data with proteomics analyses of human induced sputum (9) and human induced sputum of chronic bronchitis subjects (10). With the combination of 2D gel analysis and GeLC-MS/MS, a total of 191 human proteins were confidently assigned in induced sputum (9) of which 72 were also found by us. Interestingly, several extracellular and membranous proteins such as annexins A1 and A2, cathepsin D, clusterin, cystatins C and SN, IGFBP2, kallikrein-related peptidase 11, prominin 1, gelsolin, and lipocalin 1 were present in both studies. However, we found less overlap with the proteome of induced sputum of chronic bronchitis subjects (22 of 106 proteins; Ref. (10), likely due to the presence of abundant proteins (including immunoglobulins) in the sputome (38 of 106; Ref. 10) that were not present in our list of proteins.
Using 2D nano-HPLC-ESI-MS/MS, Tyan et al. (11) reported identification of 124 proteins from 43 pooled adenocarcinoma patient pleural effusions with high confidence (at least two or more unique peptides for each protein identified). From these, 22 were also identified by us, including extracellular lipocalin 1, gelsolin, lumican, pigment epithelium-derived factor, α1-antitrypsin, zinc α2-glycoprotein 1, and apolipoprotein E. Fig. 3 summarizes the overlap between other publications and our data. Supplemental Table 14 lists all proteins identified by us and the six previous studies.
Fig. 3.
Overlap of proteins identified in the current study (Planque et al.) and six other lung-related proteomics studies: Casado et al. (10), Tyan et al. (11), Nicholas et al. (9), Huang et al. (25), Tian et al. (26), and Xiao et al. (18).
Validation of Proteins Identified by MS as Potential Lung Cancer Biomarkers
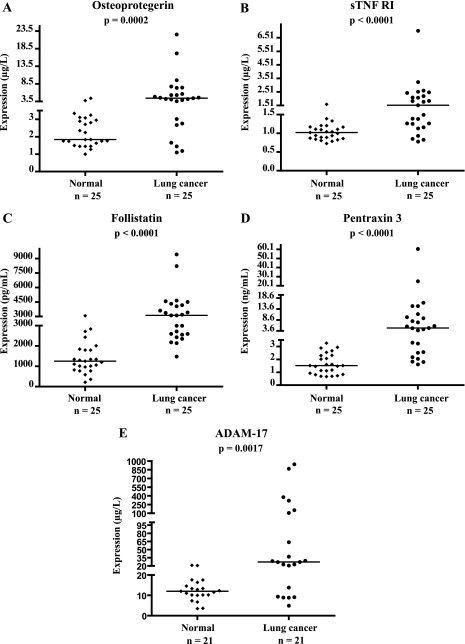
Using commercially available or in-house developed sandwich immunoassays, we performed preclinical validation of five candidates, sTNFRI, follistatin, ADAM-17, pentraxin 3, and osteoprotegerin, selected from our list of proteins identified by MS. Candidate biomarker concentration was examined in serum samples from patients with or without lung cancer (Fig. 4 and supplemental Tables 1–5). Serum levels of osteoprotegerin were significantly elevated in patients with lung cancer (median = 4.43 μg/liter) in comparison with healthy individuals (median = 1.84 μg/liter) (p = 0.0002).
Fig. 4.
Validation of osteoprotegerin (A), sTNFRI (B), follistatin (C), pentraxin 3 (D), and ADAM-17 (E) in serum. Levels of these candidate biomarkers were measured by ELISA in serum of patients with or without lung cancer. n, number of subjects. Median values are shown by a horizontal line. p values were calculated with the Mann-Whitney U test.
The sTNFRI serum levels in NSCLC were significantly higher (median = 1.53 μg/liter) than those in healthy controls (median = 1.02 μg/liter) (p < 0.0001). A significant elevation of follistatin was observed in serum of lung cancer patients (median = 3,116 pg/ml) as compared with healthy volunteers (median = 1,251 pg/ml) (p < 0.0001).
Pentraxin 3 (PTX3) was identified in all three NSCLC cell lines and especially with higher abundance in the squamous cell carcinoma cell line with 15 to 16 unique peptides. As demonstrated in Fig. 4D, the distribution of PTX3 between cases and controls was significantly different (p < 0.0001). Serum levels of PTX3 were much higher in lung cancer patients (median = 4.91 ng/ml) as compared with healthy individuals (median = 1.52 ng/ml).
By using an ELISA developed in house, we observed a significant increase of ADAM-17 in serum of patients with NSCLC (median = 27.3 μg/liter) in comparison with healthy volunteers (median = 12.0 μg/liter) (p = 0.002). In a very preliminary assessment of these five candidate markers for lung carcinoma, we calculated the diagnostic sensitivity (percentage of patients with elevated marker levels) at 100% specificity (using as cutoff the highest value in the normal group). These diagnostic sensitivities were as follows: osteoprotegerin, 52%; sTNFRI, 52%; follistatin, 56%; pentraxin 3, 68%; and ADAM-17, 67%.
Assignments of Biological Function and Network Construction for Biological Processes
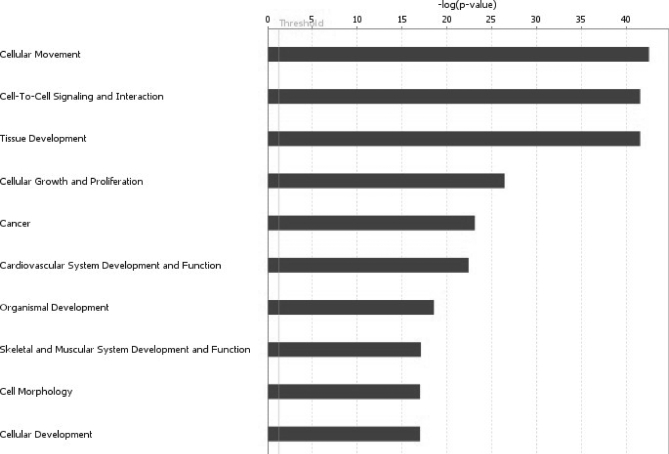
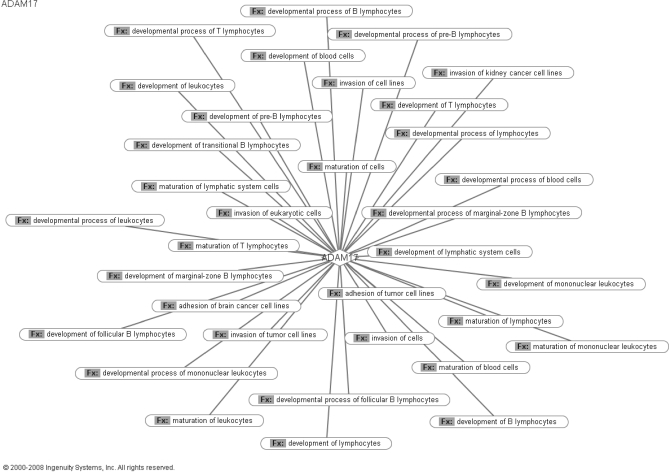
The potential biological functions of extracellular and membrane-bound proteins identified in CM of all cell lines were analyzed using Ingenuity Pathway Analysis. The top 10 functions are illustrated in Fig. 5. Major categories included cellular movement, cell-to-cell signaling and interaction, cellular growth and proliferation, and cancer. Ingenuity Pathway Analysis was also used to develop biological networks showing the functions and disease association of each of the five candidates selected for preliminary validation. ADAM-17 plays a significant role in recruitment of immune cells during the inflammatory response. ADAM-17 also plays a role in modulating cell adhesion and potentially contributing to the invasiveness of cancer cells. Both of these functions have been well recognized to play a significant role in tumor progression and invasion. The biological network constructed for ADAM-17 is presented in Fig. 6. Follistatin is associated with various processes involving malignant progression and invasion. As presented in supplemental Fig. 6, follistatin is involved with regulation of cell growth and proliferation of various cancer cell lines. The molecular functions associated with PTX3 are highlighted in supplemental Fig. 7. These include participation in mediation of inflammatory response. Interestingly, PTX3 was shown to be involved with respiratory disorders in mice. The protein sTNFRI displays connections with cancer progression. As shown in supplemental Fig. 8, networks involved with cancer include apoptosis, malignant progression, cell survival, and proliferation. Finally, osteoprotegerin (TNFRSF11B) has shown to be involved with several molecular networks, including cell adhesion, apoptosis, cell migration, and malignant transformation (supplemental Fig. 9).
Fig. 5.
Biological function analyses. The top 10 functions for the extracellular and membrane-bound proteins, as determined by Ingenuity Pathway Analysis, are shown. The y axis shows the negative log of p value.
Fig. 6.
Molecular functions related to diseases associated with ADAM-17. The web diagram generated through Ingenuity Pathway Analysis software depicts the biological functions (Fx) with which ADAM-17 is associated in the context of disease.
DISCUSSION
In proteome projects, the 2DE approach has been the primary technique of separation and comparison of complex protein mixtures. However, this approach suffers from large variations caused by sample preparation, protein loading, and gel staining (38). Another limit of 2DE for proteomics concerns the poor recovery of proteins from the gel for MS. Methods to supplement or replace 2DE, such as multidimensional LC have therefore been sought (39). Multidimensional LC-MS/MS analysis allows identification of proteins in a high throughput fashion unlike the rather slow and laborious 2DE-MS/MS methods. This technique has been used to discover cancer biomarkers by analyzing complex protein mixtures such as biological fluids, tissues, or cell cultures (14, 15, 40–44). However, this technology is still challenged in the case of complex mixtures such as serum that require well established methodologies for depletion of highly abundant proteins and efficient sample fractionation before proteomics analysis (12, 13).
In this study, a 2D LC-MS/MS strategy was utilized to identify the secretome of four lung cancer cell lines of differing histological subtypes grown in serum-free medium. Because lung cancer is a heterogenous disease, we analyzed the secretome of cell lines of differing origin to have a better depiction of the proteome of lung cancer and more chances to discover biomarkers of this pathology. By searching with both Mascot and X!Tandem, we identified over 1,800 proteins in the CM of all four cell lines combined that, to our knowledge, is one of the largest repositories of proteins identified for lung cancer. As reported by Kapp et al. (45), the use of multiple search engines increases confidence of protein identification. These search engines utilize different algorithms and scoring functions to determine whether a mass spectrum matches an entry in the database (46). Moreover, by combining the use of PeptideProphet and ProteinProphet algorithms embedded within Scaffold, the confidence of protein identification probabilities is increased (23, 24).
Particular attention was placed on extracellular and membrane-bound proteins from the four lung cancer cell lines because these proteins have the highest probability of being found in the circulation and functioning as potential biomarkers. In our study, 38% of identified proteins were classified as extracellular and membrane-bound. Among them, we identified various cytokines, proteases, protease inhibitors, growth factors, extracellular matrix proteins, and receptors. We also found a large number of intracellular proteins, including ones classified as nuclear and cytoplasmic by GO annotation. In general, our current proteomics data revealed a similar distribution of proteins by cellular component in each cell line. During the cell culture phase, a small portion of the cell population dies, resulting in the release of intracellular proteins into the conditioned media. Despite our efforts to optimize cell culture conditions to minimize cell death and maximize secreted protein concentration, the identification of intracellular proteins in the CM by MS is inevitable because of the high sensitivity of the technique. By using quantitative proteomics techniques (ICAT reagents and MS/MS) to identify secreted and cell surface proteins from a prostate cancer cell line (LNCaP), Martin et al. (42) found that more than 50% of proteins identified in LNCaP-conditioned media were classified as intracellular. However, previous studies from our laboratory using a similar cell culture-based approach showed that proteins identified in the cell lysate did not contain as many secreted proteins as the CM for that cell line (14). Furthermore, the extracellular proteins found in the cell lysate showed minimal overlap with the proteins identified in the CM (14). These data demonstrate that our strategy significantly enriches for secreted proteins.
In this study, each cell line was cultured in triplicate. Using an in-house developed program, we examined the overlap of identified proteins between the three replicates of each cell line. As shown in Fig. 1, a 63.5–70.5% overlap of proteins between the replicates of each cell line was observed, suggesting excellent reproducibility between runs. Because of the nature of mass spectrometric measurements, not all peptides are ionized in each run, and subsequently, different peptides are selected for ionization and finally detected (14). The diverse steps during sample preparation, including reduction-alkylation, lyophilization, sample fractionation, and using a ZipTip, can also be important contributing factors to the variations observed between the replicates.
As determined by specific ELISA, the presence of three internal controls (IGFBP-2, KLK11, and KLK14) was confirmed by mass spectrometry in the CM of all lung cancer cell lines. Among them, KLK14 was the least abundant protein (1.9 μg/liter as determined by ELISA) and was detected in two of the three replicates of H1688 by one and three unique peptides, respectively. It is conceivable that the detection limit of our method is close to this value of 1.5–2 μg/liter as reported previously by our group (14, 15). Based on these observations, we conclude that our proteomics strategy can identify proteins in CM in the low μg/liter range or higher. With regard to current biomarkers used in the clinic, this is the expected concentration range, giving us hope that new lung cancer markers should be detectable in serum. Through our method, we successfully identified proteins that are putative or currently used as biomarkers of lung cancer, including CEA (27, 28), pro-GRP (29, 31), SCC antigen (47, 48), tumor M2-PK (49), NCAM (35–37), chromogranin A (29), and chromogranin B (30). In addition, we identified putative markers previously reported in lung-related proteomics studies such as member C1 of aldo-keto reductase family 1 identified by Huang et al. (25) as dihydrodiol dehydrogenase and matrix metallopeptidase 1 found to be overexpressed in lung cancer patients and especially in late stage (18). Furthermore, 129 of 291 extracellular and 168 of 415 membranous proteins identified in this study were found in the plasma proteome. Overall, these data further support our strategy of using the CM of lung cancer cell lines to discover candidate biomarkers.
From our list of proteins, we used some arbitrary criteria to select the most promising candidates for validation. Given that serological biomarkers identified so far are generally secreted or shed proteins, such as prostate-specific antigen, CA-125, and SCC-Ag in prostate, ovarian, and lung cancer, respectively, we hypothesized that new lung cancer markers might be secreted proteins (or their fragments) originating from cancer cells or their microenvironment and then entering the circulation (50). Consequently, we focused on proteins that were classified as extracellular or membrane-bound. As secondary criteria, we selected proteins that showed relatively lung-specific expression at the mRNA or protein level by examining the UniGene expressed tag database and the Human Protein Atlas database. Then, we performed literature searches to ensure that these proteins have not been examined as serological markers for lung cancer and showed biological connections with lung or other cancers. We also compared selected proteins with the proteomes of lung-related diseases (lung cancer (18, 25, 26) and pleural effusion (11)) or the proteome of a lung-related biological fluid (induced sputum (9, 10)) and serum (Plasma Proteome Database). Finally, potential candidates that had commercially available antibodies or immunoassays were selected.
From this selection, five candidates were retained for further investigation: ADAM-17, pentraxin 3, sTNFRI, osteoprotegerin, and follistatin. Serum levels of each candidate were higher in NSCLC patients in comparison with healthy controls. Follow-up large scale validation studies will be needed to determine the sensitivity, specificity, and prognostic value of these markers in lung cancer. To examine the putative connections with lung cancer, we constructed biological networks of each candidate in association to functions and diseases (Fig. 6 and supplemental Figs. 6–9). Each candidate is associated with various processes including tumor development or malignant progression. Previously, ADAM-17 was found to be overexpressed in breast cancer and associated with tumor progression and metastasis (51, 52). ADAM-17 was also shown to predict adverse outcome in breast cancer (53). More recently, ADAM-17, a major ErbB ligand sheddase, was found to be up-regulated in NSCLC tumor samples and was required not only for heregulin-dependent HER3 signaling but also for epidermal growth factor receptor ligand-dependent signaling in NSCLC cell lines (54). Pentraxin 3 was the first long pentraxin discovered; it was initially named tumor necrosis factor-stimulated gene 14 and later identified as an IL-1-inducible gene in human umbilical vein endothelial cells (55). Despite the fact that PTX3 was reported to prevent infection by certain fungi, bacteria, or viruses in the lung, increased expression was also associated with more severe lung injury such as high volume mechanical ventilation or severe bacterial infection (56). Follistatin showed several links to cancer, such as prostate (15, 57, 58), colon (59), and ovarian cancer (60). Concerning its link to lung cancer, follistatin has been suggested to suppress the production of multiple organ metastasis by small cell lung cancer cells in natural killer cell-depleted severe combined immunodeficiency mice predominantly by inhibiting angiogenesis (61). As shown in supplemental Fig. 8, sTNFRI is associated with cancer by participating in apoptosis, malignant progression, and proliferation. A spontaneous regression of lung metastasis was observed by Tomita et al. (62) in the absence of tumor necrosis factor receptor p55, suggesting that sTNFRI-mediated signals could maintain tumor neovascularization at least in part by inducing hepatocyte growth factor expression and eventually support lung metastasis. Osteoprotegerin, a secreted member of the tumor necrosis factor receptor superfamily, has not been well documented in lung disease. However, it displays several connections to cancer, such as pancreatic, colorectal (63), and bladder carcinoma (64). The involvement of each candidate in lung cancer pathobiology will need to be further studied to elucidate their role during cancer progression.
Because of the heterogeneity of lung cancer and the lack of sensitivity and specificity of individual markers, there is a growing consensus that panels of markers can improve screening, diagnosis, prognosis, or monitoring responses to therapy. Future studies will determine whether a combination of these putative lung cancer biomarkers can improve the sensitivity and specificity of current markers.
In summary, we present here one of the most comprehensive proteomics analyses of conditioned media from four lung cancer cell lines for new biomarker discovery. Five candidates were preliminarily validated in this study and are promising serum markers for lung cancer.
Supplementary Material
Acknowledgments
We thank Ihor Batruch for mass spectrometry analysis; Adrian Pasculescu, David Nguyen, and Peter Bowden for bioinformatics support; and Girish Sardana, Geeth Gunawardana, and Antoninus Soosaipillai for helpful discussions.
Footnotes
* This work was supported by a grant (to E. P. D.) from the Natural Sciences and Engineering Research Council of Canada and Proteomic Methods Inc.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1–9 and Tables 1–14.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1–9 and Tables 1–14.
1 The abbreviations used are:
- SCLC
- small cell lung cancer
- CDCHO
- chemically defined Chinese hamster ovary
- CEA
- carcinoembryonic antigen
- CM
- conditioned media
- COPD
- chronic obstructive pulmonary disease
- 2D
- two-dimensional
- FBS
- fetal bovine serum
- GO
- Genome Ontology
- KLK
- kallikrein-related peptidase
- NSCLC
- non-small cell lung cancer
- pro-GRP
- progastrin-releasing peptide
- PTX3
- pentraxin 3
- SCC
- squamous cell carcinoma
- Ag
- antigen
- sTNFRI
- tumor necrosis factor receptor superfamily member 1A
- 2DE
- two-dimensional gel electrophoresis
- NCAM
- neural cell adhesion molecule.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Murray T., Xu J., Thun M. J. ( 2007) Cancer statistics. CA Cancer J. Clin 57, 43– 66 [DOI] [PubMed] [Google Scholar]
- 2.Bryborn M., Adner M., Cardell L. O. ( 2005) Psoriasin, one of several new proteins identified in nasal lavage fluid from allergic and non-allergic individuals using 2-dimensional gel electrophoresis and mass spectrometry. Respir. Res 6, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casado B., Pannell L. K., Iadarola P., Baraniuk J. N. ( 2005) Identification of human nasal mucous proteins using proteomics. Proteomics 5, 2949– 2959 [DOI] [PubMed] [Google Scholar]
- 4.Lindahl M., Irander K., Tagesson C., Ståhlbom B. ( 2004) Nasal lavage fluid and proteomics as means to identify the effects of the irritating epoxy chemical dimethylbenzylamine. Biomarkers 9, 56– 70 [DOI] [PubMed] [Google Scholar]
- 5.Sabounchi-Schütt F., Aström J., Hellman U., Eklund A., Grunewald J. ( 2003) Changes in bronchoalveolar lavage fluid proteins in sarcoidosis: a proteomics approach. Eur. Respir. J 21, 414– 420 [DOI] [PubMed] [Google Scholar]
- 6.Wu J., Kobayashi M., Sousa E. A., Liu W., Cai J., Goldman S. J., Dorner A. J., Projan S. J., Kavuru M. S., Qiu Y., Thomassen M. J. ( 2005) Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol. Cell. Proteomics 4, 1251– 1264 [DOI] [PubMed] [Google Scholar]
- 7.Xie H., Rhodus N. L., Griffin R. J., Carlis J. V., Griffin T. J. ( 2005) A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol. Cell. Proteomics 4, 1826– 1830 [DOI] [PubMed] [Google Scholar]
- 8.Hu S., Xie Y., Ramachandran P., Ogorzalek Loo R. R., Li Y., Loo J. A., Wong D. T. ( 2005) Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics 5, 1714– 1728 [DOI] [PubMed] [Google Scholar]
- 9.Nicholas B., Skipp P., Mould R., Rennard S., Davies D. E., O'Connor C. D., Djukanoviæ R. ( 2006) Shotgun proteomic analysis of human-induced sputum. Proteomics 6, 4390– 4401 [DOI] [PubMed] [Google Scholar]
- 10.Casado B., Iadarola P., Pannell L. K., Luisetti M., Corsico A., Ansaldo E., Ferrarotti I., Boschetto P., Baraniuk J. N. ( 2007) Protein expression in sputum of smokers and chronic obstructive pulmonary disease patients: a pilot study by CapLC-ESI-Q-TOF. J. Proteome Res 6, 4615– 4623 [DOI] [PubMed] [Google Scholar]
- 11.Tyan Y. C., Wu H. Y., Lai W. W., Su W. C., Liao P. C. ( 2005) Proteomic profiling of human pleural effusion using two-dimensional nano liquid chromatography tandem mass spectrometry. J. Proteome Res 4, 1274– 1286 [DOI] [PubMed] [Google Scholar]
- 12.Jacobs J. M., Adkins J. N., Qian W. J., Liu T., Shen Y., Camp D. G., 2nd, Smith R. D. ( 2005) Utilizing human blood plasma for proteomic biomarker discovery. J. Proteome Res 4, 1073– 1085 [DOI] [PubMed] [Google Scholar]
- 13.Qian W. J., Jacobs J. M., Liu T., Camp D. G., 2nd, Smith R. D. ( 2006) Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics 5, 1727– 1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulasingam V., Diamandis E. P. ( 2007) Proteomics analysis of conditioned media from three breast cancer cell lines: a mine for biomarkers and therapeutic targets. Mol. Cell. Proteomics 6, 1997– 2011 [DOI] [PubMed] [Google Scholar]
- 15.Sardana G., Jung K., Stephan C., Diamandis E. P. ( 2008) Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J. Proteome Res 7, 3329– 3338 [DOI] [PubMed] [Google Scholar]
- 16.Tachibana I., Mori M., Tanio Y., Hosoe S., Sakuma T., Osaki T., Ueno K., Kumagai T., Kijima T., Kishimoto T. ( 1996) A 100-kDa protein tyrosine phosphorylation is concurrent with beta 1 integrin-mediated morphological differentiation in neuroblastoma and small cell lung cancer cells. Exp. Cell Res 227, 230– 239 [DOI] [PubMed] [Google Scholar]
- 17.Lou X., Xiao T., Zhao K., Wang H., Zheng H., Lin D., Lu Y., Gao Y., Cheng S., Liu S., Xu N. ( 2007) Cathepsin D is secreted from M-BE cells: its potential role as a biomarker of lung cancer. J. Proteome Res 6, 1083– 1092 [DOI] [PubMed] [Google Scholar]
- 18.Xiao T., Ying W., Li L., Hu Z., Ma Y., Jiao L., Ma J., Cai Y., Lin D., Guo S., Han N., Di X., Li M., Zhang D., Su K., Yuan J., Zheng H., Gao M., He J., Shi S., Li W., Xu N., Zhang H., Liu Y., Zhang K., Gao Y., Qian X., Cheng S. ( 2005) An approach to studying lung cancer-related proteins in human blood. Mol. Cell. Proteomics 4, 1480– 1486 [DOI] [PubMed] [Google Scholar]
- 19.Borgoño C. A., Michael I. P., Shaw J. L., Luo L. Y., Ghosh M. C., Soosaipillai A., Grass L., Katsaros D., Diamandis E. P. ( 2007) Expression and functional characterization of the cancer-related serine protease, human tissue kallikrein 14. J. Biol. Chem 282, 2405– 2422 [DOI] [PubMed] [Google Scholar]
- 20.Diamandis E. P., Borgoño C. A., Scorilas A., Harbeck N., Dorn J., Schmitt M. ( 2004) Human kallikrein 11: an indicator of favorable prognosis in ovarian cancer patients. Clin. Biochem 37, 823– 829 [DOI] [PubMed] [Google Scholar]
- 21.Shaw J. L., Diamandis E. P. ( 2007) Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem 53, 1423– 1432 [DOI] [PubMed] [Google Scholar]
- 22.Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. ( 2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985– 1988 [DOI] [PubMed] [Google Scholar]
- 23.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383– 5392 [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646– 4658 [DOI] [PubMed] [Google Scholar]
- 25.Huang L. J., Chen S. X., Huang Y., Luo W. J., Jiang H. H., Hu Q. H., Zhang P. F., Yi H. ( 2006) Proteomics-based identification of secreted protein dihydrodiol dehydrogenase as a novel serum markers of non-small cell lung cancer. Lung Cancer 54, 87– 94 [DOI] [PubMed] [Google Scholar]
- 26.Tian T., Hao J., Xu A., Hao J., Luo C., Liu C., Huang L., Xiao X., He D. ( 2007) Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci 98, 1265– 1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salgia R., Harpole D., Herndon J. E., 2nd, Pisick E., Elias A., Skarin A. T. ( 2001) Role of serum tumor markers CA 125 and CEA in non-small cell lung cancer. Anticancer Res 21, 1241– 1246 [PubMed] [Google Scholar]
- 28.Shoji F., Yoshino I., Yano T., Kometani T., Ohba T., Kouso H., Takenaka T., Miura N., Okazaki H., Maehara Y. ( 2007) Serum carcinoembryonic antigen level is associated with epidermal growth factor receptor mutations in recurrent lung adenocarcinomas. Cancer 110, 2793– 2798 [DOI] [PubMed] [Google Scholar]
- 29.Nisman B., Heching N., Biran H., Barak V., Peretz T. ( 2006) The prognostic significance of circulating neuroendocrine markers chromogranin a, pro-gastrin-releasing peptide and neuron-specific enolase in patients with advanced non-small-cell lung cancer. Tumour Biol 27, 8– 16 [DOI] [PubMed] [Google Scholar]
- 30.Tötsch M., Müller L. C., Hittmair A., Ofner D., Gibbs A. R., Schmid K. W. ( 1992) Immunohistochemical demonstration of chromogranins A and B in neuroendocrine tumors of the lung. Hum. Pathol 23, 312– 316 [DOI] [PubMed] [Google Scholar]
- 31.Takada M., Kusunoki Y., Masuda N., Matui K., Yana T., Ushijima S., Iida K., Tamura K., Komiya T., Kawase I., Kikui N., Morino H., Fukuoka M. ( 1996) Pro-gastrin-releasing peptide (31–98) as a tumour marker of small-cell lung cancer: comparative evaluation with neuron-specific enolase. Br. J. Cancer 73, 1227– 1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharjee A., Richards W. G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., Loda M., Weber G., Mark E. J., Lander E. S., Wong W., Johnson B. E., Golub T. R., Sugarbaker D. J., Meyerson M. ( 2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U.S.A 98, 13790– 13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planque C., Li L., Zheng Y., Soosaipillai A., Reckamp K., Chia D., Diamandis E. P., Goodglick L. ( 2008) A multiparametric serum kallikrein panel for diagnosis of non-small cell lung carcinoma. Clin. Cancer Res 14, 1355– 1362 [DOI] [PubMed] [Google Scholar]
- 34.Planque C., Bléchet C., Ayadi-Kaddour A., Heuzé-Vourc'h N., Dumont P., Guyétant S., Diamandis E. P., El Mezni F., Courty Y. ( 2008) Quantitative RT-PCR analysis and immunohistochemical localization of the kallikrein-related peptidases 13 and 14 in lung. Biol. Chem 389, 781– 786 [DOI] [PubMed] [Google Scholar]
- 35.Lynch D. F., Jr., Hassen W., Clements M. A., Schellhammer P. F., Wright G. L., Jr. ( 1997) Serum levels of endothelial and neural cell adhesion molecules in prostate cancer. Prostate 32, 214– 220 [DOI] [PubMed] [Google Scholar]
- 36.Jaques G., Auerbach B., Pritsch M., Wolf M., Madry N., Havemann K. ( 1993) Evaluation of serum neural cell adhesion molecule as a new tumor marker in small cell lung cancer. Cancer 72, 418– 425 [DOI] [PubMed] [Google Scholar]
- 37.Ledermann J. A., Pasini F., Olabiran Y., Pelosi G. ( 1994) Detection of the neural cell adhesion molecule (NCAM) in serum of patients with small-cell lung cancer (SCLC) with “limited” or “extensive” disease, and bone-marrow infiltration. Int. J. Cancer Suppl.8, 49– 52 [DOI] [PubMed] [Google Scholar]
- 38.Magi B., Bargagli E., Bini L., Rottoli P. ( 2006) Proteome analysis of bronchoalveolar lavage in lung diseases. Proteomics 6, 6354– 6369 [DOI] [PubMed] [Google Scholar]
- 39.Issaq H. J. ( 2001) The role of separation science in proteomics research. Electrophoresis 22, 3629– 3638 [DOI] [PubMed] [Google Scholar]
- 40.Cho C. K., Shan S. J., Winsor E. J., Diamandis E. P. ( 2007) Proteomics analysis of human amniotic fluid. Mol. Cell. Proteomics 6, 1406– 1415 [DOI] [PubMed] [Google Scholar]
- 41.Shaw J. L., Smith C. R., Diamandis E. P. ( 2007) Proteomic analysis of human cervico-vaginal fluid. J. Proteome Res 6, 2859– 2865 [DOI] [PubMed] [Google Scholar]
- 42.Martin D. B., Gifford D. R., Wright M. E., Keller A., Yi E., Goodlett D. R., Aebersold R., Nelson P. S. ( 2004) Quantitative proteomic analysis of proteins released by neoplastic prostate epithelium. Cancer Res 64, 347– 355 [DOI] [PubMed] [Google Scholar]
- 43.Li C., Hong Y., Tan Y. X., Zhou H., Ai J. H., Li S. J., Zhang L., Xia Q. C., Wu J. R., Wang H. Y., Zeng R. ( 2004) Accurate qualitative and quantitative proteomic analysis of clinical hepatocellular carcinoma using laser capture microdissection coupled with isotope-coded affinity tag and two-dimensional liquid chromatography mass spectrometry. Mol. Cell. Proteomics 3, 399– 409 [DOI] [PubMed] [Google Scholar]
- 44.Yocum A. K., Busch C. M., Felix C. A., Blair I. A. ( 2006) Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations. J. Proteome Res 5, 2743– 2753 [DOI] [PubMed] [Google Scholar]
- 45.Kapp E. A., Schütz F., Connolly L. M., Chakel J. A., Meza J. E., Miller C. A., Fenyo D., Eng J. K., Adkins J. N., Omenn G. S., Simpson R. J. ( 2005) An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5, 3475– 3490 [DOI] [PubMed] [Google Scholar]
- 46.Domon B., Aebersold R. ( 2006) Challenges and opportunities in proteomics data analysis. Mol. Cell. Proteomics 5, 1921– 1926 [DOI] [PubMed] [Google Scholar]
- 47.Kagohashi K., Satoh H., Kurishima K., Kadono K., Ishikawa H., Ohtsuka M., Sekizawa K. ( 2008) Squamous cell carcinoma antigen in lung cancer and nonmalignant respiratory diseases. Lung 186, 323– 326 [DOI] [PubMed] [Google Scholar]
- 48.Vassilakopoulos T., Troupis T., Sotiropoulou C., Zacharatos P., Katsaounou P., Parthenis D., Noussia O., Troupis G., Papiris S., Kittas C., Roussos C., Zakynthinos S., Gorgoulis V. ( 2001) Diagnostic and prognostic significance of squamous cell carcinoma antigen in non-small cell lung cancer. Lung Cancer 32, 137– 144 [DOI] [PubMed] [Google Scholar]
- 49.Schneider J., Velcovsky H. G., Morr H., Katz N., Neu K., Eigenbrodt E. ( 2000) Comparison of the tumor markers tumor M2-PK, CEA, CYFRA 21-1, NSE and SCC in the diagnosis of lung cancer. Anticancer Res 20, 5053– 5058 [PubMed] [Google Scholar]
- 50.Liotta L. A., Ferrari M., Petricoin E. ( 2003) Clinical proteomics: written in blood. Nature 425, 905. [DOI] [PubMed] [Google Scholar]
- 51.Santiago-Josefat B., Esselens C., Bech-Serra J. J., Arribas J. ( 2007) Post-transcriptional up-regulation of ADAM17 upon epidermal growth factor receptor activation and in breast tumors. J. Biol. Chem 282, 8325– 8331 [DOI] [PubMed] [Google Scholar]
- 52.McGowan P. M., Ryan B. M., Hill A. D., McDermott E., O'Higgins N., Duffy M. J. ( 2007) ADAM-17 expression in breast cancer correlates with variables of tumor progression. Clin. Cancer Res 13, 2335– 2343 [DOI] [PubMed] [Google Scholar]
- 53.McGowan P. M., McKiernan E., Bolster F., Ryan B. M., Hill A. D., McDermott E. W., Evoy D., O'Higgins N., Crown J., Duffy M. J. ( 2008) ADAM-17 predicts adverse outcome in patients with breast cancer. Ann. Oncol 19, 1075– 1081 [DOI] [PubMed] [Google Scholar]
- 54.Zhou B. B., Peyton M., He B., Liu C., Girard L., Caudler E., Lo Y., Baribaud F., Mikami I., Reguart N., Yang G., Li Y., Yao W., Vaddi K., Gazdar A. F., Friedman S. M., Jablons D. M., Newton R. C., Fridman J. S., Minna J. D., Scherle P. A. ( 2006) Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 10, 39– 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breviario F., d'Aniello E. M., Golay J., Peri G., Bottazzi B., Bairoch A., Saccone S., Marzella R., Predazzi V., Rocchi M., Della Valle G., Dejana E., Mantovani A., Introna M. ( 1992) Interleukin-1-inducible genes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J. Biol. Chem 267, 22190– 22197 [PubMed] [Google Scholar]
- 56.He X., Han B., Liu M. ( 2007) Long pentraxin 3 in pulmonary infection and acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol 292, L1039– L1049 [DOI] [PubMed] [Google Scholar]
- 57.Thomas T. Z., Wang H., Niclasen P., O'Bryan M. K., Evans L. W., Groome N. P., Pedersen J., Risbridger G. P. ( 1997) Expression and localization of activin subunits and follistatins in tissues from men with high grade prostate cancer. J. Clin. Endocrinol. Metab 82, 3851– 3858 [DOI] [PubMed] [Google Scholar]
- 58.McPherson S. J., Mellor S. L., Wang H., Evans L. W., Groome N. P., Risbridger G. P. ( 1999) Expression of activin A and follistatin core proteins by human prostate tumor cell lines. Endocrinology 140, 5303– 5309 [DOI] [PubMed] [Google Scholar]
- 59.Nakagawa H., Liyanarachchi S., Davuluri R. V., Auer H., Martin E. W., Jr., de la Chapelle A., Frankel W. L. ( 2004) Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene 23, 7366– 7377 [DOI] [PubMed] [Google Scholar]
- 60.Di Simone N., Crowley W. F., Jr., Wang Q. F., Sluss P. M., Schneyer A. L. ( 1996) Characterization of inhibin/activin subunit, follistatin, and activin type II receptors in human ovarian cancer cell lines: a potential role in autocrine growth regulation. Endocrinology 137, 486– 494 [DOI] [PubMed] [Google Scholar]
- 61.Ogino H., Yano S., Kakiuchi S., Muguruma H., Ikuta K., Hanibuchi M., Uehara H., Tsuchida K., Sugino H., Sone S. ( 2008) Follistatin suppresses the production of experimental multiple-organ metastasis by small cell lung cancer cells in natural killer cell-depleted SCID mice. Clin. Cancer Res 14, 660– 667 [DOI] [PubMed] [Google Scholar]
- 62.Tomita Y., Yang X., Ishida Y., Nemoto-Sasaki Y., Kondo T., Oda M., Watanabe G., Chaldakov G. N., Fujii C., Mukaida N. ( 2004) Spontaneous regression of lung metastasis in the absence of tumor necrosis factor receptor p55. Int. J. Cancer 112, 927– 933 [DOI] [PubMed] [Google Scholar]
- 63.Lipton A., Ali S. M., Leitzel K., Chinchilli V., Witters L., Engle L., Holloway D., Bekker P., Dunstan C. R. ( 2002) Serum osteoprotegerin levels in healthy controls and cancer patients. Clin. Cancer Res 8, 2306– 2310 [PubMed] [Google Scholar]
- 64.Mizutani Y., Matsubara H., Yamamoto K., Nan Li Y., Mikami K., Okihara K., Kawauchi A., Bonavida B., Miki T. ( 2004) Prognostic significance of serum osteoprotegerin levels in patients with bladder carcinoma. Cancer 101, 1794– 1802 [DOI] [PubMed] [Google Scholar]
- 65.Nikliñski J., Furman M., Palynyczko Z., Laudañski J., Bułatowicz J. ( 1991) Carcinoembryonic antigen, neuron-specific enolase and creatine kinase-BB as tumor markers for carcinoma of the lung. Neoplasma 38, 645– 651 [PubMed] [Google Scholar]
- 66.Nikliñski J., Furman M., Laudañski J., Pałynyczko Z., Welk M. ( 1991) Evaluation of carcinoembryonic antigen (CEA) and brain-type creatine kinase (CK-BB) in serum from patients with carcinoma of the lung. Neoplasma 38, 129– 135 [PubMed] [Google Scholar]
- 67.Chen Y., Zhang H., Xu A., Li N., Liu J., Liu C., Lv D., Wu S., Huang L., Yang S., He D., Xiao X. ( 2006) Elevation of serum l-lactate dehydrogenase B correlated with the clinical stage of lung cancer. Lung Cancer 54, 95– 102 [DOI] [PubMed] [Google Scholar]
- 68.Sun T., Gao Y., Tan W., Ma S., Zhang X., Wang Y., Zhang Q., Guo Y., Zhao D., Zeng C., Lin D. ( 2006) Haplotypes in matrix metalloproteinase gene cluster on chromosome 11q22 contribute to the risk of lung cancer development and progression. Clin. Cancer Res 12, 7009– 7017 [DOI] [PubMed] [Google Scholar]
- 69.Kim J. H., Bogner P. N., Baek S. H., Ramnath N., Liang P., Kim H. R., Andrews C., Park Y. M. ( 2008) Up-regulation of peroxiredoxin 1 in lung cancer and its implication as a prognostic and therapeutic target. Clin. Cancer Res 14, 2326– 2333 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.