Abstract
Caspase-3 and -7 are considered functionally redundant proteases with similar proteolytic specificities. We performed a proteome-wide screen on a mouse macrophage lysate using the N-terminal combined fractional diagonal chromatography technology and identified 46 shared, three caspase-3-specific, and six caspase-7-specific cleavage sites. Further analysis of these cleavage sites and substitution mutation experiments revealed that for certain cleavage sites a lysine at the P5 position contributes to the discrimination between caspase-7 and -3 specificity. One of the caspase-7-specific substrates, the 40 S ribosomal protein S18, was studied in detail. The RPS18-derived P6–P5′ undecapeptide retained complete specificity for caspase-7. The corresponding P6–P1 hexapeptide still displayed caspase-7 preference but lost strict specificity, suggesting that P′ residues are additionally required for caspase-7-specific cleavage. Analysis of truncated peptide mutants revealed that in the case of RPS18 the P4–P1 residues constitute the core cleavage site but that P6, P5, P2′, and P3′ residues critically contribute to caspase-7 specificity. Interestingly, specific cleavage by caspase-7 relies on excluding recognition by caspase-3 and not on increasing binding for caspase-7.
Caspases, a family of evolutionarily conserved proteases, mediate apoptosis, inflammation, proliferation, and differentiation by cleaving many cellular substrates (1–3). The apoptotic initiator caspases (caspase-8, -9, and -10) are activated in large signaling platforms and propagate the death signal by cleavage-induced activation of executioner caspase-3 and -7 (4, 5). Most of the cleavage events occurring during apoptosis have been attributed to the proteolytic activity of these two executioner caspases, which can act on several hundreds of proteins (2, 3, 6, 7). The substrate degradomes of the two main executioner caspases have not been determined but their identification is important to gaining greater insight in their cleavage specificity and biological functions.
The specificity of caspases was rigorously profiled by using combinatorial tetrapeptide libraries (8), proteome-derived peptide libraries (9), and sets of individual peptide substrates (10, 11). The results of these studies indicate that specificity motifs for caspase-3 and -7 are nearly indistinguishable with the canonical peptide substrate, DEVD, used to monitor the enzymatic activity of both caspase-3 and -7 in biological samples. This overlap in cleavage specificity is manifested in their generation of similar cleavage fragments from a variety of apoptosis-related substrates such as inhibitor of caspase-activated DNase, keratin 18, PARP,1 protein-disulfide isomerase, and Rho kinase I (for reviews, see Refs. 2, 3, and 7). This propagated the view that these two caspases have completely redundant functions during apoptosis. Surprisingly, mice deficient in one of these caspases (as well as mice deficient in both) have distinct phenotypes. Depending on the genetic background of the mice, caspase-3-deficient mice either die before birth (129/SvJ) or develop almost normally (C57BL/6J) (12–14). This suggests that dynamics in the genetic background, such as increased caspase-7 expression, compensate for the functional loss of caspase-3 (15). In the C57BL/6J background, caspase-7 single deficient mice are also viable, whereas caspase-3 and -7 double deficient mice die as embryos, further suggesting redundancy (12–14). However, because caspase-3 and -7 probably arose from gene duplication between the Cephalochordata-Vertebrata diversion (16), they might have acquired different substrate specificities during evolution. Caspase-3 and -7 do exhibit different activities on a few arbitrarily identified natural substrates, including BID, X-linked inhibitor of apoptosis protein, gelsolin, caspase-6, ataxin-7, and co-chaperone p23 (17–20). In addition, caspase-3 generally cleaves more substrates during apoptosis than caspase-7 and therefore appears to be the major executioner caspase. Moreover, a recent report describing caspase-1-dependent activation of caspase-7, but not of caspase-3, in macrophages in response to microbial stimuli supports the idea of a non-redundant function for caspase-7 downstream of caspase-1 (21).
Commercially available “caspase-specific” tetrapeptide substrates are widely used for specific caspase detection, but they display substantial promiscuity and cannot be used to monitor individual caspases in cells (22, 23). Detecting proteolysis by measuring the release of C-terminal fluorophores, such as 7-amino-4-methylcoumarin (amc), restricts the specificity of these peptide substrates to non-prime cleavage site residues, which may have hampered the identification of specific cleavage events. To address this limitation, a recently developed proteomics technique, called proteomic identification of protease cleavage sites, was used to map both non-prime and prime preferences for caspase-3 and -7 on a tryptic peptide library (9). However, no clear distinction in peptide recognition motifs between caspase-3 and -7 could be observed (9). Because not all classical caspase cleavage sites are processed (7), structural or post-translational higher order constraints are likely involved in steering the cleavage site selectivity. Peptide-based approaches generally overlook such aspects.
We made use of the COFRADIC N-terminal peptide sorting methodology (24–26) to profile proteolytic events of caspase-3 and -7 in a macrophage proteome labeled by triple stable isotope labeling by amino acids in cell culture (SILAC), which allowed direct comparison of peak intensities in peptide MS spectra and consequent quantification of N termini that are equally, preferably, or exclusively generated by the action of caspase-3 or -7 (26, 27). We identified 55 cleavage sites in 48 protein substrates, encompassing mutual, preferred, and unique caspase-3 and -7 cleavage sites.
EXPERIMENTAL PROCEDURES
Activity of Recombinant Caspases
Expression and purification of recombinant mouse caspase-3 and -7 have been described (28). Activity of both caspases was determined by active site titration using a serial dilution of zVAD-fmk (25 min at 37 °C) in cell-free system (CFS) buffer containing 220 mm mannitol, 170 mm sucrose, 5 mm NaCl, 5 mm MgCl2, 10 mm HEPES, pH 7.5, 2.5 mm KH2PO4, 2.1 μm leupeptin, 0.15 μm aprotinin, 100 μm PMSF, and 10 mm DTT. Proteolytic activity was quantified on Ac-DEVD-amc as described below (data not shown). Activity was also quantified on proendothelial monocyte-activating polypeptide II, the p43 component of the aminoacyl-tRNA complex as described previously (29) (data not shown). Recombinant mouse caspase-3 or -7 was incubated with 100 μm fluorogenic Ac-DEVD-amc peptide (Peptide Institute, Osaka, Japan) in 150 μl of CFS buffer. The generation of free amc was continuously monitored for 50 min in a fluorometer (CytoFluor, PerSeptive Biosystems) at 360/460-nm excitation/emission wavelengths. The linear rate of fluorophore generation was used to quantify caspase activity.
Synthesis of Ac-peptide-amc
Fmoc-Asp(OtBu)-OH was converted to the corresponding amide using amc. After side chain deprotection, the Fmoc-Asp-amc residue was attached to 2-chlorotrityl resin followed by automated peptide synthesis using an O-benzotriazol-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate/diisopropylethylamine protocol. Subsequently, following N-terminal acetylation, acidic cleavage was done from the solid support. Finally, purification by precipitation and lyophilization yielded the C-terminal labeled Ac-peptide-amc. Identical kinetic values were obtained with commercial or in-house synthesized Ac-DEVD-amc, illustrating sufficiently high quality of in-house made peptides (Table I).
Table I. Kinetic parameters of DEVD- and DVKD-based peptide cleavage by caspase-7 and -3.
The catalytic constant (kcat), the Michaelis-Menten constant (Km), and the specificity constant (kcat/Km) of Ac-DEVD-amc, Ac-XXDVKD-amc, and the internally quenched substrate Abz-QKDVKDGKYSQY(NO2) by caspase-7 and -3 were determined as described under “Experimental Procedures.”
| Substrate |
kcat |
Km |
kcat/Km |
|||
|---|---|---|---|---|---|---|
| C7 | C3 | C7 | C3 | C7 | C3 | |
| s−1 | μm | m−1 s−1 | ||||
| DEVDa | 0.17 ± 0.00 | 8.32 ± 0.32 | 9.0 ± 0.6 | 108 ± 14 | 18,900 ± 1,280 | 76,500 ± 10,800 |
| DEVDb | 0.14 ± 0.00 | 8.39 ± 0.47 | 7.9 ± 0.6 | 112 ± 11 | 17,800 ± 1,380 | 74,700 ± 8,700 |
| DVKDb | 0.23 ± 0.01 | 0.74 ± 0.03 | 53 ± 6 | 267 ± 27 | 4,330 ± 590 | 2,790 ± 312 |
| ADVKDb | 0.11 ± 0.00 | 0.77 ± 0.03 | 35 ± 4 | 243 ± 27 | 3,080 ± 437 | 3,180 ± 389 |
| KDVKDb | 0.59 ± 0.02 | 0.79 ± 0.03 | 133 ± 13 | 407 ± 36 | 4,390 ± 494 | 1,940 ± 188 |
| AKDVKDb | 0.63 ± 0.02 | 0.48 ± 0.01 | 129 ± 11 | 439 ± 27 | 4,840 ± 444 | 1,100 ± 77 |
| QKDVKDb | 0.43 ± 0.02 | 0.19 ± 0.01 | 92 ± 12 | 356 ± 39 | 4,690 ± 657 | 560 ± 69 |
| “Quencher”c | 0.25 ± 0.02 | 0.005 ± 0.001 | 173 ± 27 | 192 ± 68 | 1,450 ± 253 | 26 ± 10 |
a Commercial source.
b Synthesized in house.
c Abz-QKDVKDGKYSQY(NO2).
Determination of Kinetic Constants
50 nm recombinant caspase-3 or -7 was incubated with a serial dilution of DEVD- or DVKD-based fluorogenic amc-linked substrates (concentration ranging between 1 and 1200 μm) in CFS buffer in a final volume of 80 μl. Similarly, 100 nm recombinant caspase-3 or -7 was incubated with a serial dilution of Abz-QKDVKDGKYSQY(NO2) (synthesized using the Fmoc chemistry on an Applied Biosystems 433A Peptide Synthesizer; Y(NO2) denotes a 3-nitrotyrosine residue). Generation of free amc or Abz was monitored as described above. The initial linear rates of fluorescence at all concentrations of the substrate were used to obtain plots of activity versus substrate concentration. The Michaelis-Menten (Km) and the catalytic constant (kcat) were determined from these plots. Absolute Km and kcat values were calculated using a standard curve determined with free amc.
In Vitro Transcription/Translation
pCMV-SPORT6-GSTP1, pCMV- SPORT6-HDGF, pCMV-SPORT6-MYBBP1a, pCMV-SPORT6-NUCKS1, pCMV-SPORT6-PKM2, pCMV-SPORT6-RPL28, pCMV-SPORT6.1-MKI67ip, pYX-ASC-FAM21 (all from the German Resource Center for Genome Research (RZPD), Berlin, Germany), pFLCI-ACTA2 (from Geneservice Ltd.), and the four pCR3-based plasmids described under “Plasmids” were used as templates for [35S]methionine (PerkinElmer Life Sciences)-radiolabeled in vitro coupled transcription/translation in a rabbit reticulocyte lysate system according to the manufacturer's instructions (Promega). Before caspase treatment, the translated protein samples were alkylated with 5 mm iodoacetamide (IAA) for 30 min at 30 °C in the dark. Excess IAA was removed by making use of protein desalting spin columns (Perbio Science). The desalted translated proteins (2 μl) were subsequently incubated for 1.5 h at 37 °C in CFS buffer with the indicated concentrations of recombinant caspases in a total volume of 30 μl. Next, samples were boiled for 10 min after addition of Laemmli buffer, separated by 10, 15, or 20% SDS-PAGE, and transferred to a nitrocellulose membrane (Schleicher & Schuell) by semidry blotting. The blotted membranes were sealed to keep them humid and exposed to a film (Amersham Biosciences Hyperfilm™ ECL) for radiography.
Plasmids
The cDNA encoding RPS18 was amplified by PCR from the pYX-ASC-RPS18 vector (RZPD) using the following primer pair: RPS18-forward, 5′-CTGAATTCGCCATGTCTCTAGTGATCCC-3′, and RPS18-reverse, 5′-ATATGCGGCCGCTCATTTCTTCTTGGATACACCC-3′. The amplified product was digested with EcoRI and NotI and cloned in a multiple cloning site-modified pCR3 vector (Invitrogen). The cDNA encoding the “ADVKD” mutant of RPS18 was generated by overlap PCR technology using the following primer pair: RPS18-K88A-forward, 5′-AACAGACAGGCGGATGTGAAGGATGGGAAG-3′, and RPS18-K88A-reverse, 5′-TTCACATCCGCCTGTCTGTTCAGGAACCAG-3′. The cDNA encoding PARP was amplified by PCR from the pGEM-PARP1 plasmid using the following primer pair: PARP-forward, 5′-ATCTGCGGCCGCTCATGGCGGAGTCTTCGGATAAGC-3′, and PARP-reverse, 5′-TATAGCGGCCGCTCCCAATTACCACAGGGAGGTC-3′. The amplified product was digested with NotI and cloned into a multiple cloning site-modified pCR3 vector (Invitrogen). The cDNA encoding the “KDEVD” mutant of PARP was generated by overlap PCR technology using the following primer pair: PARP-G210K-forward, 5′-AGAGAAAAAAGGATGAGGTGGATGGAGTGG-3′, and PARP-G210K-reverse, 5′-ACCTCATCCTTTTTTCTCTTTCCTTCACTCTTGC-3′. The cDNAs encoding caspase-3 and -7 devoid of their prodomain (i.e. caspase-p30) were cloned in the pLT10TH vector downstream of an N-terminal His6 tag as described (28). All plasmids were sequence-verified.
COFRADIC Setup
Mf4/4 (30) mouse macrophages were grown in lipopolysaccharide-free RPMI 1640 medium (Invitrogen) containing either l-[12C6]arginine, l-[13C6]arginine, or l-[13C6,15N4]arginine (Cambridge Isotope Laboratories, Andover, MA) at a concentration of 287 μm (i.e. 25% of the normal concentration in RPMI 1640 medium at which l-arginine to proline conversion was not observed). Cell populations were cultured at 37 °C in a humidified 5% CO2 atmosphere for at least five doublings for complete incorporation of the labeled l-arginine. Mf4/4 cells were detached using enzyme-free cell dissociation buffer (Invitrogen), washed in D-PBS (Dulbecco's PBS), resuspended in D-PBS supplemented with 2.1 μm leupeptin, 0.15 μm aprotinin, 100 μm PMSF, and 1 mm oxidized glutathione, and freeze-thawed three times. Lysates were cleared by centrifugation for 15 min at 20,000 × g. Proteins were alkylated with 5 mm iodoacetamide for 30 min at 30 °C in the dark, and to remove excess IAA they were separately desalted on NAP-10 columns in D-PBS supplemented with leupeptin, aprotinin, PMSF, and oxidized glutathione. Subsequently, the separate samples were supplemented with DTT (10 mm final concentration) and incubated with either 130 nm recombinant caspase-7 (l-[12C6]Arg) or -3 (l-[13C6]Arg) or left untreated (l-[13C6,15N4]Arg) for 1.5 h at 37 °C. Solid guanidinium hydrochloride was then added to a final concentration of 4 m followed by downstream analysis as described previously (24–26).
LC-MS/MS Analysis and Peptide Identification by Mascot
ESI LC-MS/MS analysis was performed as described before (26). ESI-Q-TOF MS/MS peptide fragmentation spectra were converted to pkl files using the MassLynx® software (version 4.1, Waters Corp.), and ESI ion trap MS/MS spectra were converted to mgf files using the Automation Engine software (version 3.2, Bruker). Peptides were identified using a locally installed version of the Mascot database search engine version 2.1 (Matrix Science) and the Swiss-Prot (version 53.2 of UniProtKB/Swiss-Prot protein database, containing 269,293 sequence entries of which 13,316 originated from mouse protein sequences) and TrEMBL databases (version 35.0 of UniProtKB/TrEMBL protein database, containing 3,874,166 sequence entries comprising 52,403 mouse protein entries) were searched with restriction to mouse proteins. Truncated peptide databases made by DBToolkit (31) were searched in parallel to pick up protein processing events more efficiently (e.g. Refs. 6 and 32). The following search parameters were used. Peptide mass tolerance was set at 0.2 Da, and peptide fragment mass tolerance was set at 0.1 Da with the “ESI-QUAD-TOF” as selected instrument for peptide fragmentation rules for the Q-TOF Premier data. For ion trap data, peptide mass tolerance was set at 0.5 Da, and peptide fragment mass tolerance was set at 0.5 Da with the “ESI-IT” as selected instrument for peptide fragmentation rules. Endoproteinase Arg-C/P was set as the enzyme with a maximum number of one missed cleavage. Peptide charge was set to 1+, 2+, and 3+. Variable modifications were set to methionine oxidation (to methionine sulfoxide), pyroglutamate formation of N-terminal glutamine, pyrocarbamidomethyl formation of N-terminal alkylated cysteine, deamidation of asparagine, acetylation of the N terminus, and trideuteroacetylation of the N terminus. Fixed modifications were trideuteroacetylation of lysine and S-carbamidomethylation of cysteine, and for identifying heavy labeled peptides, [13C6]arginine or [13C6,15N4]arginine were also set as additional fixed modifications. Only MS/MS spectra that exceeded the corresponding Mascot threshold score of identity (at 95% confidence level) and that were ranked first were withheld. These were further manually evaluated for the presence of sequence-specific fragment ions (typically half of all b- and y-type fragment ions were observed). In addition, spectra that received a low Mascot ion score (five or fewer points above the threshold for identity) were further interrogated, and only spectra that contained b- and y-fragment ions covering a stretch of three consecutive amino acids were considered identified (see supplemental Figs. 1 and 2 listing identified MS/MS spectra). The estimated false discovery rate by searching decoy databases was typically found to lie between 2 and 4% on the spectrum level (25). Whenever a peptide matched to multiple members of a protein family (redundancy), the protein entry reported (supplemental Tables 2, 3, and 4, column “Accession”) was according to its alphabetical description ranking or, in the case when multiple peptides could be assigned to a specific entry, the one with the highest number of matching peptides was reported.
Analysis of Synthetic Undecapeptide Cleavage
The undecapeptides containing the P6–P5′ cleavage sites (wild type RPS18 (RPS18-wt), RPS18-P5, PARP-wt, and PARP-P5) and the C-terminal amino acid deletion mutants derived from RPS18 (P6–P4′, P6–P3′, P6–P2′, and P6–P1′) were synthesized using the Fmoc chemistry on an Applied Biosystems 433A Peptide Synthesizer. Substrate peptides were used at 100 μm, and proteases were used at concentrations of 5–1000 nm. The reactions proceeded at 37 °C for 1.5 h in CFS buffer and were then quenched by the addition of trifluoroacetic acid to 1% final concentration. Cleavage by the caspases was assessed by monitoring UV absorbance (214 nm) of the precursor peptide and its fragments during RP-HPLC separation. The extent of hydrolysis was determined from the intensity of the product peak(s) compared with their unprocessed precursor.
Heat Maps and Statistics
The heat maps were generated as follows. For n substrates, n amino acids were picked at random from the mouse Swiss-Prot 56.0 database, and their frequencies were calculated. This random picking was repeated 100 times, and the standard deviation for each amino acid frequency was calculated. In this way, a statistical value was obtained for the random occurrence of each amino acid for a sample size of n in a theoretical mouse proteome. Finally, the amino acid frequencies of the identified cleavage sites were compared with these calculated values. The cleavage site-specific divergence of the positional amino acid distribution opposed to the random frequency of occurrence was calculated, and the number of standard deviations were plotted in color-coded heat maps.
Molecular Modeling
Human caspase-3 (Protein Data Bank code 2H65 (33)) and human caspase-7 (Protein Data Bank code 1F1J (34)) were selected as templates for modeling of their respective mouse homologues because both of them are in complex with a peptide sequence similar to KDVKD. After aligning all sequences, the FoldX force field (35) was applied to model the new side chains. The peptide VDVAD in the model of mouse caspase-3 was substituted by KDVKD by using FoldX followed by an unrestrained energy minimization using the YASARA2 force field (36) to optimize the interactions between the KDVKD peptide and the binding pocket of mouse caspase-3. For mouse caspase-7, N-terminal extension of the DEVD tetrapeptide to the KDVKD pentapeptide was enabled by extracting pentapeptide fragments from the BriX library (37). Each of the 194 matching pentapeptide fragments was fitted into the caspase binding cleft by structural superposition onto the tetrapeptide backbone and mutated to the sequence KDVKD in the model of mouse caspase-7 using FoldX. To release strain from the introduced P5 residue in the binding cleft, a first energy minimization was run. During this minimization, only the caspase side chains in the binding cleft (defined by a 5-Å threshold from the ligand), the ligand side chains, and the complete P5 residue were allowed to move. The second round of energy minimization was carried out without restraints to permit accommodation of the pentapeptide in the binding pocket. A round of side chain optimization with FoldX produced the final models. The caspase-7 model with the best binding energy, as calculated with FoldX, was selected as the final structural model of caspase-7 complexed to the peptide KDVKD.
RESULTS
Identification of Mouse Caspase-7 and/or -3 Cleavage Events
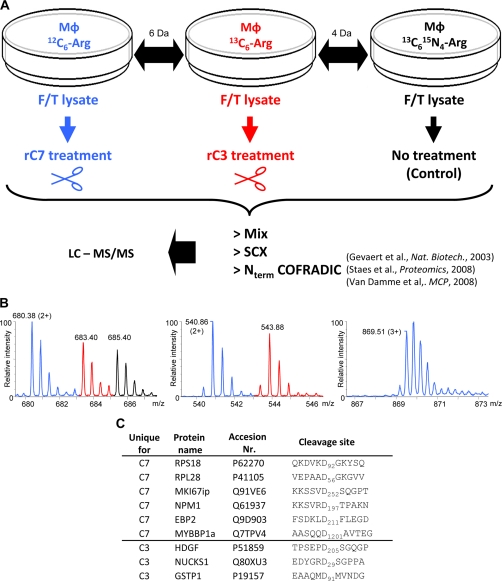
Mouse macrophages were metabolically labeled by SILAC using l-[12C6]arginine, l-[13C6]arginine, or l-[13C6,15N4]arginine (27) and lysed by freeze-thawing. Lysates were treated with iodoacetamide to inactivate unwanted downstream cysteine protease cascades during incubation with recombinant caspases. Next, the extracts were either left untreated (13C6,15N4) or incubated with identical amounts of active recombinant mouse caspase-3 (13C6) or -7 (12C6) as determined by active site titration assays (data not shown). Finally, equal amounts of the three SILAC samples were mixed and subjected to differential analysis using gel-free COFRADIC N-terminal peptide sorting (24–26) (Fig. 1, A and B). Following LC-MS/MS analysis, 795 unique peptide sequences (from 1634 MS/MS spectra) were identified using the Mascot algorithm (38). These represented 629 unique protein accessions. Overall, in 48 of these proteins 55 aspartic acid-specific cleavage sites were found (supplemental Table 1). Six of the 55 cleavage sites (11%) were found only in the caspase-7-treated lysate (unique caspase-7 (C7)), whereas three sites (5%) were retrieved only from the caspase-3-treated lysate (unique caspase-3 (C3)) (Fig. 1C and supplemental Table 2). The remaining 46 sites (84%) are shared by both caspases and were ranked according to the ratio of cleavage efficiency of caspase-7 to -3 (see supplemental Table 3). Of the 55 cleavage events, six match those previously reported in mouse substrates, 13 match those previously reported in human orthologs, and 36 have not been reported (supplemental Table 4).
Fig. 1.
Experimental setup for identifying caspase-7 and -3 substrates. A, SILAC was used to compare three differently treated macrophage (MΦ) cell lysates. 12C6- and 13C6-labeled lysates were treated with recombinant mouse caspase-7 (rC7) or -3 (rC3), respectively, whereas a 13C6,15N4-labeled lysate served as non-treated control. After caspase treatment, equal amounts of the respective proteome preparations were mixed, and N-terminal peptides were isolated by the N-terminal COFRADIC procedure using an enriched sample of α-amino-blocked peptides obtained after strong cation exchange (SCX) prefractionation step. LC-MS/MS was used to identify the isolated peptides. Gevaert et al., Ref. 24; Staes et al., Ref. 25; Van Damme et al., Ref. 26. B, representative MS spectra of the different categories of isolated N-terminal peptides. The color code refers to the different isotopes used for SILAC labeling (see A). Left, unaltered protein-N termini present in all three samples analyzed. These peptides were identified as Acd3-2GVQVETISPGDGR14 (Acd3 denotes an α-trideuteroacetylated amino group) from the FK506-binding protein 1A. Middle, neo-N termini generated equally well, based on equal ion intensities, by caspase-3 and -7. These neo-N termini correspond to the cleavage of the TAR DNA-binding protein 43 at DETD↓Acd3-90ASSAVKVKR98. Right, neo-N termini uniquely generated by mouse recombinant caspase-7. This unique neo-N terminus corresponds to cleavage at SVRD↓Acd3-198TPAKNAQKSNQNGKDLKPSTPR214 in nucleophosmin. C, list of the COFRADIC-identified unique C7 and C3 cleavage sites in the different substrates.
Validation of Identified Cleavage Sites
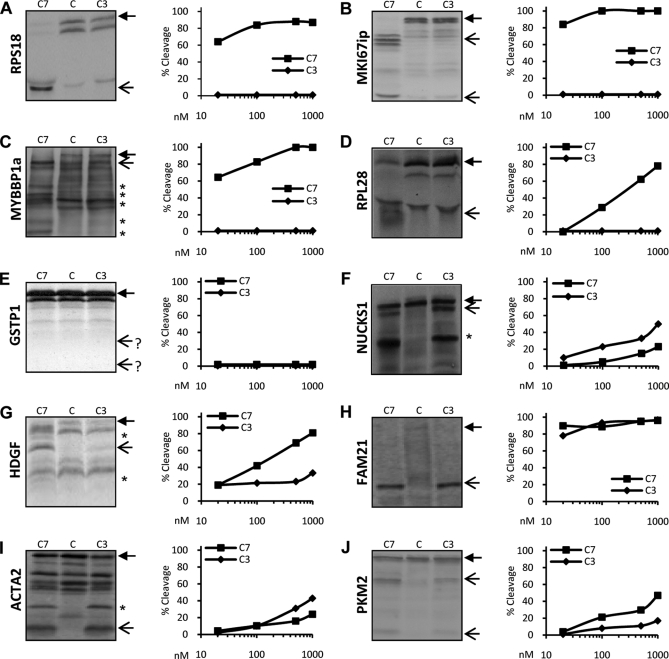
To further validate and probe for the extent of identified proteolytic cleavage events, in vitro translated and [35S]methionine-radiolabeled substrates were incubated with recombinant mouse caspase-7 or -3, separated by SDS-PAGE, analyzed by autoradiography, and quantified by densitometry. These experiments confirmed that RPS18 (Fig. 2A), MKI67ip (Fig. 2B), and MYBBP1a (Fig. 2C) were efficiently and uniquely cleaved by caspase-7 at its physiological concentrations (10–100 nm range), whereas caspase-3 concentrations up to 1 μm were unable to cleave these substrates. Also, RPL28 was uniquely processed by caspase-7, although less efficiently (Fig. 2D). In contrast, the COFRADIC-identified caspase-3-specific cleavage events in GSTP1, NUCKS1, and HDGF could not be confirmed by the in vitro cleavage assays. The precursor of GSTP1 was apparently unaffected by either caspase (Fig. 2E) and was probably identified in the proteomics screen because of the reported higher sensitivity of mass spectrometry for the identification of protease cleavage sites compared with gel-based methods (39). According to mass spectrometry data, NUCKS1 is cleaved into fragments of 22.9 and 3.5 kDa. In the caspase-3-treated samples, the 22.9-kDa band is visible just below the 26.3-kDa band of the full-length protein (Fig. 2F). However, another cleavage product is clearly observed in samples treated with caspase-3 or caspase-7, suggesting the presence of a second NUCKS1 caspase-3/-7 cleavage that remained undetected in the COFRADIC analysis. HDGF was also cleaved by both caspase-3 and -7, suggesting the presence of additional redundant caspase cleavage sites close in position to the one identified by COFRADIC; these additional sites generated fragments that are nearly indistinguishable by electrophoresis from the fragments generated by cleavage at the identified site (Fig. 2G). Hence, the COFRADIC-identified unique caspase-3 cleavage site in HDGF might be inefficiently processed by caspase-3 and masked by more efficient cleavage by both caspases at a neighboring aspartic acid, which was missed in the COFRADIC analysis. Validation of some shared substrates revealed that FAM21 (Fig. 2H), ACTA2 (Fig. 2I), and PKM2 (Fig. 2J) are indeed cleaved by both caspases. Combined, our data demonstrate the identification and in vitro validation of unique caspase-7 cleavage events. In contrast, the unique caspase-3 cleavage events based on COFRADIC technology could not be validated probably because the identified sites display low cleavage susceptibility or because of the proximity of yet unidentified redundant cleavage sites.
Fig. 2.
Caspase-7 and -3 cleavage efficiency of COFRADIC-identified substrates. RPS18 (A), MKI67ip (B), MYBBP1a (C), RPL28 (D), GSTP1 (E), NUCKS1 (F), HDGF (G), FAM21 (H), ACTA2 (I), and PKM2 (J) were transcribed-translated in vitro and labeled with [35S]methionine. Translated proteins were left untreated (C) or were treated with 10, 100, or 1000 nm recombinant C7 or C3 for 1.5 h at 37 °C and separated using SDS-PAGE, and the dried gels were exposed to autoradiograms (left panels; only samples treated with 1000 nm are shown). Densitometric analysis of the autoradiogram of each protein substrate was performed to determine the percent cleavage efficiency. For each caspase concentration, the percent of cleavage was calculated as the sum of the densities of the expected cleavage fragments divided by the sum of the densities of these fragments and the full-length protein. A closed arrow denotes the full-length protein. Open arrows indicate the expected caspase-mediated cleavage fragments. A star on the autoradiogram indicates cleavage fragments generated from cleavage sites not identified in the COFRADIC analysis.
Profiling Amino Acid Specificities of Caspase-7 and/or -3 Proteolytic Events
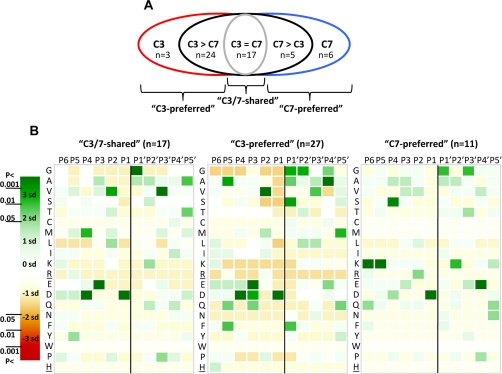
To compare the preference or aversion of caspase-7 and -3 for certain amino acid residues near the P1 cleavage site, we performed a statistical analysis on the occurrence of the amino acids at positions P6–P5′ and their deviation from a random distribution. The results were plotted in heat maps with a color code reflecting the level of preference or aversion. We grouped the cleavage sites identified by COFRADIC into three groups (Fig. 3A): sites that were cleaved equally by caspase-3 and -7 (called “C3/7-shared”; n = 17), sites that were more than 2-fold better cleaved by caspase-3 (“C3-preferred”; n = 27), and those that were cleaved more than 2-fold better by caspase-7 (“C7-preferred”; n = 11).
Fig. 3.
P6–P5′ cleavage site specificities of caspase-7 and -3. A, a diagram representing the five different groups of COFRADIC-identified caspase-3 and -7 cleavage events. “C3” and “C7” group the unique caspase-3 and -7 cleavage sites, respectively. “C3 > C7” contains the targets cleaved at least 2 times more efficiently by caspase-3, and “C7 > C3” contains the sites cleaved more than 2 times more efficiently by caspase-7. “C3 = C7” shows the sites cleaved equally by caspase-7 and -3 (C3/7-shared). The C3-preferred cleavage sites combine the C3 and C3 > C7 groups, and the C7-preferred cleavage sites combine the C7 and C7 > C3 cleavage sites. B, heat maps plotting the number of standard deviations (sd) that the frequency of an amino acid at each position in the C3/7-shared (n = 17) (left), C3-preferred (n = 27) (middle), or C7-preferred (n = 11) (right) cleavage sites differs from the frequency of randomly taken amino acids at a particular position (detailed under “Experimental Procedures”). The 95% (p < 0.05 = <1.96 × SD), 99% (p < 0.01 = <2.58 × SD), and 99.9% (p < 0.001 = <3.39 × SD) confidence thresholds are color-coded. Green indicates overrepresentation and red indicates underrepresentation of a particular amino acid at a certain position (P6–P5′ positions were analyzed). Arg and His residues (underlined) are not found at P′ positions as an artifact of the technique used. The scissile bond between P1 and P1′ is indicated by a vertical black line.
In general, the P1′ position of the cleavage products was enriched for Gly, Ala, or Ser residues, which corresponds to the N-end rule of stabilizing residues (40), suggesting that most of the generated C-terminal fragments would be stable. Statistical analysis of the C3/7-shared cleavage sites revealed that the most frequent residues at the P4 to P1′ positions are DEVD↓G (Fig. 3B, left panel) as reported previously (8, 10, 11). The same P4–P1′ profile was also found in the caspase-3-preferred cleavage sites (Fig. 3B, middle panel). Moreover, a slight preference for alanine and phenylalanine was noticed at the P5 position. This agrees with reports indicating that peptide substrates with a hydrophobic residue in the P5 position are better substrates for caspase-3 than peptide substrates lacking such a residue (33, 41). However, statistical analysis of the P6–P1′ positions revealed a different profile for the caspase-7-preferred cleavage sites, namely a preference for a KKSXXD↓G primary sequence (Fig. 3B, right panel). Four of the 11 caspase-7-preferred sites have a lysine at the P5 position, whereas none of the 44 caspase-3-preferred or shared cleavage sites contain a lysine at that position, so a lysine at position P5 might generate incompatibility constraints for caspase-3 specificity (see below).
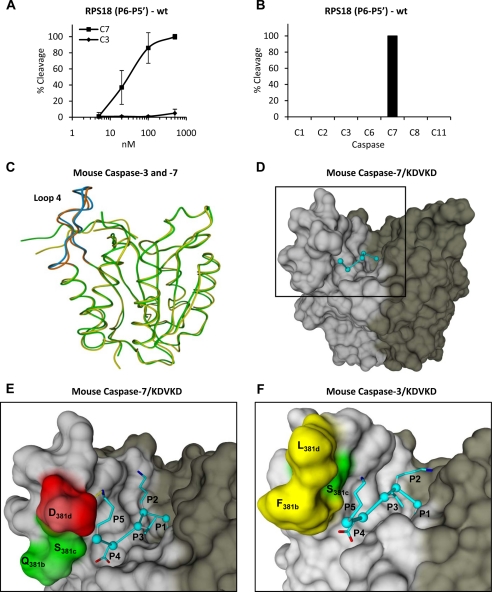
Caspase-7 Specificity Is Preserved on RPS18-derived P6–P5′ Undecapeptide
The differences in the P6–P5′ region for caspase-7- and caspase-3-preferred cleavage sites (Fig. 3B) may explain the different specificities for some substrates. Therefore, the P6–P5′ region of the caspase-7-specific protein substrate RPS18 was synthesized as an undecapeptide, incubated with recombinant caspase-3 and -7, and analyzed by reverse phase chromatography for cleavage efficiency. Interestingly, similar to its natural protein counterpart, the RPS18-derived undecapeptide substrate was cleaved efficiently by caspase-7 but only marginally by caspase-3 even at high concentrations (Fig. 4A). Furthermore, the RPS18-derived undecapeptide was not cleaved by any of the other tested apoptotic (caspase-2, -6, and -8) or inflammatory (caspase-1 and -11) caspases (Fig. 4B). Because caspases show hardly any specificity for tetrapeptides (23), these results indicate that additional residues in the P6–P5′ cleavage site region other than P4–P1 may determine the specific recognition of RPS18 by caspase-7. In silico molecular modeling was used to evaluate molecular interactions distinguishing between caspase-3 and -7 cleavage vulnerability. Modeling of the complete P6–P5′ undecapeptide on mouse caspase-7 or -3 structural models would be unreliable because of the many degrees of freedom that each additional residue can acquire when compared with the currently known caspase-3 and -7 crystal structures in complex with a penta- (VDVAD) and tetrapeptide (DEVD), respectively. Therefore, we modeled the RPS18-derived KDVKD sequence on mouse caspase-7 or -3 structural models. In general, no major differences in the interactions of P1, P2, P3, and P4 with their respective S pocket are seen between mouse caspase-7 and -3 in complex with KDVKD. However, the amino acids constituting the S5 pocket, which reside in loop 4 (Fig. 4C), are hydrophilic (Gln276, Ser277, and Asp278) in caspase-7 (Fig. 4E), whereas two of three are hydrophobic (Phe250, Ser251, and Leu252) in caspase-3 (Fig. 4F). These observations indicate that cleavage sites with a hydrophilic lysine at the P5 position will probably be less tolerated by caspase-3 as compared with caspase-7.
Fig. 4.
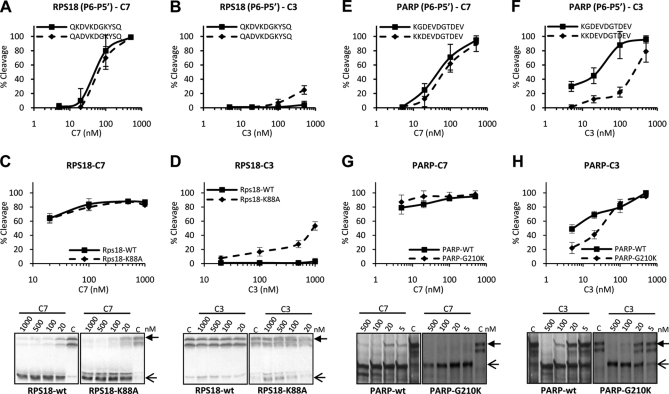
Caspase-7-specific cleavage of RPS18-delineated P6–P5′ undecapeptide and modeling of P5–P1 pentapeptide in substrate binding cleft of caspase-7 and -3. A, the P6–P5′ undecapeptide overlapping the caspase-7 cleavage site in RPS18 (RPS18(P6–P5′)-wt) was left untreated or treated with the indicated concentration of C7 or C3 for 1.5 h at 37 °C, and the generated fragments were analyzed by RP-HPLC for cleavage efficiency. B, RPS18(P6–P5′)-wt was incubated with a 250 nm concentration of the indicated caspases and analyzed by RP-HPLC. The activity of the recombinant caspase fractions was checked on their respective optimal tetrapeptide substrates (8) (data not shown). C, superposition of the structural model of mouse caspase-7 (green) and -3 (yellow) in ribbon representation. Loop 4 is in brown for caspase-7 and in blue for caspase-3. D, molecular model of KDVKD (cyan) in the substrate binding cleft of caspase-7 in surface representation. The black square delineates the active site of the caspase monomer and is enlarged for caspase-7 (E) and -3 (F) to indicate the S5-P5 lysine interactions. The dotted yellow line indicates a possible ion bridge between Asp278 and P5 lysine in caspase-7 (E). In D, E, and F, the p20 and p10 subunits are colored light and dark gray, respectively. Red, green, and yellow indicate negatively charged, uncharged polar, and hydrophobic residues, respectively. Molecular graphics were generated with YASARA and PovRay. The caspase-1 numbering convention is used (2, 43). Error bars indicate standard deviation of at least three independent experiments.
Lysine at P5 Position Is Involved in Determining Caspase-7 and -3 Specificity
To verify the contribution of the P5 lysine in distinguishing between caspase-7 and -3 cleavage susceptibility, we compared the sensitivity of the RPS18-derived undecapeptide with its P5-K/A substitution counterpart for cleavage by both caspases. Substitution of the lysine by an alanine at the P5 position did not significantly alter the efficiency of cleavage by caspase-7 (Fig. 5A). However, a P5-K/A substitution made the undecapeptide susceptible to cleavage by caspase-3 (Fig. 5B). To determine whether the role of the lysine at P5 is the same in natural proteins, we measured the caspase-7-specific cleavage of the natural RPS18 protein and its P5-K/A substitution mutant. As with the undecapeptides, the mutant RPS18-K88A protein was cleaved by caspase-7 to the same extent as wild type RPS18 (Fig. 5C) and became susceptible to caspase-3 cleavage (Fig. 5D). Thus, the lysine at position P5 probably does not affect cleavage of this substrate by caspase-7 but excludes it from the caspase-3 substrate binding cleft. To verify this hypothesis, we introduced a P5-G/K substitution in the caspase cleavage site of PARP (a C3/7-shared substrate) and into its thereof derived undecapeptide. We found that caspase-7-mediated cleavage of the P5-modified PARP-G210K protein and its undecapeptide was similar to cleavage of their wild type counterparts (Fig. 5, E and G). In contrast, caspase-3-mediated cleavage of PARP-G210K was less efficient than that of wild-type PARP (Fig. 5, F and H). These data argue for a contribution of the P5 lysine in determining caspase-7 and -3 cleavage site recognition, mainly by reducing recognition by caspase-3.
Fig. 5.
Lysine at P5 position is involved in distinguishing caspase-7 and -3 cleavage. RPS18-wt, PARP-wt, and the respective mutants with modified cleavage sites, RPS18-K88A and PARP-G210K, were transcribed and translated in vitro in the presence of [35S]methionine. The translated proteins and their delineated P6–P5′ undecapeptide (RPS18-wt, QKDVKDGKYSQ; RPS18-K/A, QADVKDGKYSQ; PARP-wt, KGDEVDGTDEV; PARP-G/K, KKDEVDGTDEV) were left untreated or treated with the indicated concentrations of C7 or C3 for 1.5 h at 37 °C and analyzed by autoradiography (lower panels in C, D, G, and H) and densitometry or by RP-HPLC. Cleavage efficiencies of the wild type and K/A-substituted P6–P5′ undecapeptides of RPS18 by caspase-7 (A) or -3 (B), cleavage efficiencies of the wild type and K88A-substituted mutant of RPS18 protein by caspase-7 (C) or -3 (D), cleavage efficiencies of the wild type and G/K-substituted P6–P5′ undecapeptides of PARP by caspase-7 (E) or -3 (F), and cleavage efficiencies of the wild type and G210K mutant PARP proteins by caspase-7 (G) or -3 (H) are shown. A closed arrow indicates the full-length protein. Open arrows indicate the expected caspase-mediated cleavage fragments. Error bars indicate standard deviation of at least three independent experiments.
P6, P2′, and P3′ Residues Are Involved in Specific Caspase-7 and -3 Cleavage Site Recognition
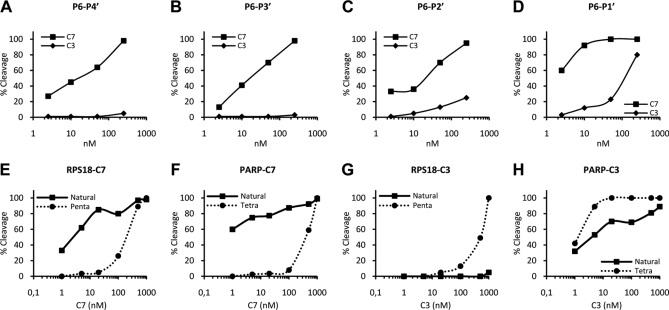
The specific cleavage of the RPS18-derived P6–P5′ undecapeptide by caspase-7 and the role of the P5 lysine in the substrate specificity prompted us to synthesize the P5–P1 acetyl-KDVKD-amc. We compared the catalytic activities of recombinant mouse caspase-7 and -3 on the Ac-KDVKD-amc pentapeptide and three control peptides: Ac-ADVKD-amc, Ac-DVKD-amc, and Ac-DEVD-amc (Table I). Consistent with previous reports (10, 11), the kcat/Km ratio on Ac-DEVD-amc was about 4 times higher for caspase-3 than for caspase-7. The kcat/Km ratio on Ac-DVKD-amc for caspase-3 and -7 were 26- and 4-fold lower than on Ac-DEVD-amc, respectively (Table I), resulting in a relative enhanced proteolytic specificity of 1.5-fold in favor of caspase-7. Addition of a lysine at the P5 position of Ac-DVKD-amc did not alter the specificity by caspase-7 but further reduced the proteolytic activity of caspase-3 (Table I). Despite the relative increased specificity of caspase-7 for Ac-KDVKD-amc this substrate was still cleaved by caspase-3, indicating that additional residues in the P6–P5′ undecapeptide may determine specificity. Addition of a glutamine at P6 of Ac-KDVKD-amc additionally reduced the specificity by caspase-3 (Table I), whereas the specificity by caspase-7 remained unaffected. This observation shows that the P6 glutamine in the P6–P1 Ac-QKDVKD-amc peptide further excludes cleavage site recognition by caspase-3 but still does not result in unique caspase-7 proteolytic specificity, suggesting that the P′ residues further define exclusive specificity. To study the role of the P′ residues in P6–P5′ specificity we made primed site deletion peptides for comparative analysis. Similar to the P6–P5′ undecapeptide, the P6–P4′ and P6–P3′ peptides were efficiently cleaved by caspase-7 and hardly cleaved by caspase-3 (Fig. 6, A and B). However, the P6–P2′ peptide became susceptible to caspase-3 but did not alter the cleavage efficiency by caspase-7 (Fig. 6C). Further truncation to the P6–P1′ peptide additionally increased the cleavage efficiency by caspase-3 but also fairly affected caspase-7 cleavage efficiency (Fig. 6D). Taken together, these findings show for the first time that P6, P5, and primed residues such as P2′ and P3′ are involved in determining cleavage site recognition by caspase-7 and -3, mainly by excluding recognition by caspase-3. Analogous to a previous report (11), the P6–P5′ undecapeptide-based internal quencher substrate Abz-QKDVKDGKYSQY(NO2) was generated as a reporter substrate, taking P′ residues for caspase cleavage into account. The kinetic constants on the internal quencher substrate confirm that the RPS18-derived P6–P5′ undecapeptide was far more specifically cleaved by caspase-7 than by caspase-3 (Table I).
Fig. 6.
Lysine at P2′ and tyrosine at P3′ are involved in distinguishing cleavage by caspase-7 and -3. A–D, the RPS18-delineated truncated peptides (P6–P1′, QKDVKDG; P6–P2′, QKDVKDGK; P6–P3′, QKDVKDGKY; P6–P4′, QKDVKDGKYS) were left untreated or treated with the indicated concentrations of C7 or C3 for 1.5 h at 37 °C and analyzed for cleavage efficiency. Caspase-7 and -3-mediated cleavage efficiencies of the P6–P4′ (A), P6–P3′ (B), P6–P2′ (C), and P6–P1′ (D) peptides are shown. E–H, RPS18 and PARP were transcribed and translated in vitro in the presence of [35S]methionine. The translated proteins and their derived penta- or tetrapeptide caspase substrates were left untreated or treated with the indicated concentrations of caspase-7 or -3 for 1.5 h at 37 °C and analyzed for cleavage efficiency. Caspase-7-mediated cleavage efficiencies of RPS18 and its KDVKD pentapeptide (E) or PARP and its DEVD tetrapeptide (F) and caspase-3-mediated cleavage efficiencies of RPS18 and its KDVKD pentapeptide (G) or PARP and its DEVD tetrapeptide (H) are shown.
Caspase-7 and -3 Activity on Peptides Does Not Reflect Efficiency of Protein Substrate Cleavage
Caspase-7 and -3 cleaved natural proteins (e.g. RPS18 and PARP) and their derived penta- or tetrapeptides with different efficiencies. Caspase-7 cleaved natural RPS18 more efficiently than the P5–P1 pentapeptide substrate (Fig. 6E). Also, caspase-7 cleaved natural PARP better than the thereof derived tetrapeptide (Fig. 6F). In contrast, caspase-3 processed the small pentapeptide better than natural RPS18 (Fig. 6G) and the PARP-derived tetrapeptide more efficiently than natural PARP (Fig. 6H). Altogether, these findings indicate that determining caspase activities in cellular lysates based on short peptide substrates may not reflect their actual in vivo activity on natural substrates.
DISCUSSION
The specificity of caspases for the P4–P2 cleavage site positions has been extensively documented by peptide-based approaches such as combinatorial tetrapeptide screenings (8, 10, 11) and proteome-derived peptide libraries (9). The value of such peptide-based methods is demonstrated by the fact that the protein-based COFRADIC analysis revealed the same specificity profile for the shared and caspase-3-preferred cleavage site motifs. However, some constraints limit the global applicability of synthetic peptide-based techniques, such as limited library size for hexa- and heptapeptides and restriction to non-prime cleavage specificities because of C-terminal labeling with fluorophores. Libraries of fluorescence resonance energy transfer peptides and the proteomic identification of protease cleavage sites technique have overcome most of the limitations of library size and non-prime motifs (9, 42). Peptide-based studies have yielded valuable insight into the sequence specificity of caspases, although interpretation of the biological significance remains difficult because protease-peptide interactions differ substantially from natural protease-protein interactions (2). To address this limitation, we applied the COFRADIC N-terminal peptide sorting proteomics technology (24–26) to macrophage lysates incubated with recombinant caspase-3 or -7. It is noteworthy that the preclusion of downstream caspase activation by iodoacetamide modification of free cysteines (which blocks the catalytic cysteine of the caspases) was a prerequisite to unambiguously assign and delineate caspase-3 and -7 substrate specificity rules. Consequently, this alkylation step might also have altered or prohibited substrate recognition to a certain extent. However, with respect to caspase-3- and -7-mediated substrate processing, we believe this to be of minor influence given the large overlap (16 of the 55 identified unique sites or 29%) between the cleavage events reported in our study and those reported in earlier studies using apoptotic cells in which no iodoacetamide pretreatment step was involved (supplemental Tables 2, 3, and 4).
COFRADIC has the advantage of identifying protein cleavage events within the complexity of a native proteome, but it also has limitations. One bias of the current COFRADIC analysis design is that neo-N-terminal peptides carrying histidine or non-C-terminal arginine will most likely be missed due to the use of a cation exchange step to pre-enrich N-terminal peptides (25). Also, peptides that are too small, too large, too hydrophilic, or too hydrophobic will be missed due to traditional LC-MS/MS techniques for peptide identification. Moreover, incomplete proteomic coverage occurs due to the stochastic nature of peptide sampling. These biases may explain the modest overlap observed between the 55 identified mouse caspase-3 and -7 cleavage sites in our proteomics experiment and the 57 previously reported mouse caspase processing sites (3, 21). Nevertheless, our study doubles the number of known caspase cleavage sites in mouse.
A critical step in our approach is combining COFRADIC with differential SILAC labeling, enabling comparative and quantitative analysis of the relative sensitivity of each cleavage site to caspase-3 and -7 proteolysis as deduced from the ion intensity peaks of the N-terminal peptides. Our study revealed three caspase-3-specific, six caspase-7-specific, and 46 shared cleavage sites, indicating that both caspases, although most of their cleavage sites are redundant, also have some non-overlapping cleavage sites. In agreement with our findings, Walsh et al. (17) recently showed that caspase-3 and -7 may have non-redundant roles in the cell death machinery as concluded from the different activities of these caspases on different arbitrarily identified protein substrates. Noteworthy is that whether cleavage occurs by caspase-7 or -3 or by both the resulting N-terminal peptide is generally consistent with the N-end rule (40), suggesting that both caspases generate stable fragments.
Unexpectedly, the identified caspase-7-preferred cleavage sites did not exhibit the documented optimal DEVD tetrapeptide motif. Bioinformatics profiling of the P6–P5′ positions in the identified caspase-3 and -7 cleavage sites revealed enrichment of lysine at the P5 position in caspase-7-preferred substrates. Because no statistically significant enrichment of arginine or histidine was found at the P5 position in caspase-7 substrates, the presence of the lysine itself rather than a positive charge seems important. In addition, amino acid substitution analysis indicated that a lysine at P5 is not selected by caspase-7 but disfavored by caspase-3. However, such a mechanism does not explain the specific enrichment of a lysine at this position in caspase-7-prefered cleavage sites. In silico molecular modeling indicates that the hydrophobic S5 pocket residues of mouse caspase-3 disfavor interactions with the hydrophilic P5 lysine compared with the hydrophilic S5 pocket of caspase-7. In agreement with the modeling data, we observed a slight preference for hydrophobic amino acids at the P5 position of caspase-3-preferred cleavage sites. Indeed, most caspase-3-preferred substrates favored an alanine at the P5 position. In line with this observation, the P5-K/A substitution in the caspase-7-specific substrate RPS18 made it susceptible to caspase-3 but did not increase its proteolysis by caspase-7. Similarly, the reverse P5-G/K substitution in PARP, a C3/7-shared substrate, reduced cleavage by caspase-3 but did not increase caspase-7 cleavage efficiency. Previous reports demonstrated the kinetic importance of hydrophobic P5 residues, showing that caspase-3 hydrolyzed the substrates Ac-VDVAD-pNA and Ac-LDVAD-pNA more efficiently than Ac-DVAD-pNA and hydrolyzed Ac-LDEVD-pNA more efficiently than Ac-DEVD-pNA (33, 41). This underscores the significant contribution of the P5 position in caspase-3 specificity. Our results add to this knowledge by demonstrating that, at least for RPS18, a lysine at the P5 position contributes to caspase-7 specificity by reducing caspase-3 recognition.
Although caspase-3-mediated cleavage was increased in efficiency by the P5-K/A mutation in RPS18, it still remained less efficient than caspase-7 cleavage. On the other hand, caspase-3-mediated cleavage of PARP was decreased by the P5-G/K mutation, but still P5-modified PARP was not a unique caspase-7 substrate. These results indicate that the P5 lysine is not the sole determining factor in the RPS18 P6–P5′ undecapeptide distinguishing between caspase-3 and -7 cleavage. Indeed, the glutamine on the P6 position also contributes to caspase-7-specific cleavage by further excluding caspase-3 recognition. Analysis of the P′ positions further revealed that addition of the P2′ lysine and P3′ tyrosine also strongly reduced caspase-3 susceptibility without affecting caspase-7 reactivity, demonstrating the involvement of these positions in determining specificity between caspase-7 and -3. Together, our observations indicate that specific cleavage site recognition by caspase-7 primarily relies on disfavoring the cleavage site for caspase-3 and not on increased binding specificity of the substrate by caspase-7. These data also indicate that the RPS18 P6–P3′ nonapeptide is required and sufficient to distinguish cleavage site recognition by caspase-7 and -3. Because the P1–P5′, P2–P5′, and P3–P5′ peptides were not cleaved by caspase-3 and -7 (data not shown), our combined data indicate that the DVKD tetrapeptide “core” roughly steers recognition but that the N-terminal P6 and P5 residues together with the C-terminal P2′ and P3′ residues are required to infer caspase-7-specific cleavage site recognition. Interestingly, the internally quenched substrate Abz-QKDVKDGKYSQY(NO2) was about 55 times more selective for caspase-7 than for caspase-3, suggesting that it is likely far better suited to monitor caspase-7-specific activity than Ac-DEVD-amc.
Comparative analysis of the cleavage efficiencies of proteins and peptides by caspase-7 and -3 revealed that caspase-7 cleaves the PARP- and RPS18-derived tetra- or pentapeptide, respectively, less efficiently than their protein counterparts. The far more efficient cleavage of the RPS18 protein by caspase-7 illustrates that the P6–P3′ region might determine its selectivity, whereas other determinants on the native protein might serve to optimize cleavage by caspase-7. In this context, it has been reported that interactions distant from the catalytic site, at so-called exosites (2, 43), and specific higher order structures of the cleavage site (6, 44) might help to optimize recognition and cleavage. In contrast, caspase-3 cleaved the natural proteins PARP and RPS18 less efficiently than the thereof derived small tetra- or pentapeptides, respectively, suggesting that optimal cleavage by caspase-3 is less dependent on additional native protein interactions. In line with these findings, we observed that the cleavage of DEVD- and DVKD-based peptides by caspase-3 is mainly driven by its strong catalytic activity, compensating for its lower affinity toward these peptides. Caspase-7 has lower kcat values for DEVD- and DVKD-based peptides but higher Km values. In functional terms, for certain cleavage sites caspase-7 is a more selective but less active specialist, whereas caspase-3 is a less selective but very active generalist. This is further supported by the observation that most identified C3/7-shared cleavage sites (33 of 46 or 72%) were more efficiently cleaved by caspase-3. These data also indicate that the cleavage efficiency of tetra- or pentapeptide substrates by caspase-7 or -3 does not reflect the in vivo cleavage efficiency of their corresponding natural proteins. Our data further indicate that using small peptide substrates in cellular lysates overestimates caspase-3 activity, whereas caspase-7 activity is likely underestimated.
Although in vivo relevance of the caspase-3- or -7-specific proteolytic events has yet to be proven, the actual cleavage of some of these substrates, e.g. cleavage of the caspase-7-specific substrate nucleophosmin (one of the six caspase-7-specific substrates identified in this study) and the caspase-3-specific substrate nuclear ubiquitous casein and cyclin-dependent kinase substrate (one of the three caspase-3-specific substrates) was already documented for their human orthologs in apoptotic dying cells (6, 45, 46) (supplemental Tables 2, 3, and 4). This suggests that at least some of the reported cleavage events in this study occur in cellular conditions, although one cannot a priori discriminate between signaling and bystander cleavage events in the context of apoptotic dying cells. However, the major aim of our study was to document possible cleavage specificities of the highly redundant executioner caspases. In that respect, we were able to delineate a caspase-7-specific RPS18-derived P6–P5′ undecapeptide, which was refractory to cleavage by all other caspases tested. This emphasizes the potential relevance and existence of caspase-7-specific proteolytic events in vivo as suggested by a previous report (17).
To conclude, our data show that caspase-3- and -7-specific cleavage events are probably scarce but that identifying them is of crucial importance to determine their individual specificities. Moreover, in-depth functional analysis of the identified caspase-3 and -7 proteolytic events will help to unravel the in vivo functional role of both caspases and might lead to novel specific biomarker discovery. Importantly, our analysis shows for the first time that the P4–P1 residues form the core for cleavage site recognition but that the P6, P5, P2′, and P3′ positions are essential for fine tuning caspase-7-specific cleavage, principally by excluding caspase-3 recognition. Whether the recognition of a P6–P3′ nonapeptide is a general concept for caspase-specific cleavage site recognition remains to be evaluated.
Supplementary Material
Acknowledgments
We thank Amin Bredan for editing the manuscript, Marc Goethals for synthesizing the undecapeptide substrates, and Marc Mirande (CNRS-UPR 9063, Gif-sur-Yvette, France) for the generous supply of recombinant p43 (proendothelial monocyte-activating polypeptide II) protein.
Footnotes
* This work was supported in part by the Flanders Institute for Biotechnology (VIB) and several grants from the European Union (European Commission (EC) Marie Curie Training and Mobility Program, FP6, ApopTrain, Grant MRTN-CT-035624; EC Research and Technology Development Integrated Project, FP6, Epistem, Grant LSHB-CT-2005-019067; and APO-SYS, FP7, Grant HEALTH-F4-2007-200767), the Interuniversity Poles of Attraction-Belgian Science Policy (Grant IAP6/18), the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (Grants G.0133.05 and 3G.0218.06), and the Special Research Fund of Ghent University (Geconcerteerde Onderzoekstacties Grant 12.0505.02). This work was also supported by research grants from the Fund for Scientific Research-Flanders (Belgium) (Projects G.0156.05, G.0077.06, and G.0042.07), the Concerted Research Actions (Project BOF07/GOA/012) from Ghent University, the Interuniversity Attraction Poles (Grant IUAP06), and the European Union Interaction Proteome (6th Framework Program) (to the Department of Medical Protein Research).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–4 and Figs. 1 and 2.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–4 and Figs. 1 and 2.
1 The abbreviations used are:
- PARP
- poly(ADP-ribose) polymerase
- amc
- 7-amino-4-methylcoumarine
- Abz
- 2-amino-benzoic acid
- CFS
- cell-free system
- COFRADIC
- combined fractional diagonal chromatography
- IAA
- iodoacetamide
- kcat
- catalytic constant
- Km
- Michaelis-Menten constant
- Y(NO2)
- 3-nitrotyrosine
- RP
- reverse phase
- SILAC
- stable isotope labeling by amino acids in cell culture
- zVAD-fmk
- benzyloxycarbonyl-valine-alanine-aspartic acid(OMe)fluoromethyl ketone
- pNA
- p-nitroanilide
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- wt
- wild type
- C3
- caspase-3
- C7
- caspase-7
- HDGF
- hepatoma-derived growth factor
- OtBu
- tert-butyl ester.
REFERENCES
- 1.Lamkanfi M., Festjens N., Declercq W., Vanden Berghe T., Vandenabeele P. ( 2007) Caspases in cell survival, proliferation and differentiation. Cell Death Differ 14, 44– 55 [DOI] [PubMed] [Google Scholar]
- 2.Timmer J. C., Salvesen G. S. ( 2007) Caspase substrates. Cell Death Differ 14, 66– 72 [DOI] [PubMed] [Google Scholar]
- 3.Lüthi A. U., Martin S. J. ( 2007) The CASBAH: a searchable database of caspase substrates. Cell Death Differ 14, 641– 650 [DOI] [PubMed] [Google Scholar]
- 4.Boatright K. M., Salvesen G. S. ( 2003) Mechanisms of caspase activation. Curr. Opin. Cell Biol 15, 725– 731 [DOI] [PubMed] [Google Scholar]
- 5.Riedl S. J., Salvesen G. S. ( 2007) The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol 8, 405– 413 [DOI] [PubMed] [Google Scholar]
- 6.Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. ( 2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771– 777 [DOI] [PubMed] [Google Scholar]
- 7.Fischer U., Jänicke R. U., Schulze-Osthoff K. ( 2003) Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ 10, 76– 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. ( 1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem 272, 17907– 17911 [DOI] [PubMed] [Google Scholar]
- 9.Schilling O., Overall C. M. ( 2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol 26, 685– 694 [DOI] [PubMed] [Google Scholar]
- 10.Talanian R. V., Quinlan C., Trautz S., Hackett M. C., Mankovich J. A., Banach D., Ghayur T., Brady K. D., Wong W. W. ( 1997) Substrate specificities of caspase family proteases. J. Biol. Chem 272, 9677– 9682 [DOI] [PubMed] [Google Scholar]
- 11.Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. ( 2000) Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J 350, 563– 568 [PMC free article] [PubMed] [Google Scholar]
- 12.Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. ( 2006) Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311, 847– 851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuida K., Zheng T. S., Na S., Kuan C., Yang D., Karasuyama H., Rakic P., Flavell R. A.Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. ( 1996) Nature 384, 368– 372 [DOI] [PubMed] [Google Scholar]
- 14.Woo M., Hakem R., Soengas M. S., Duncan G. S., Shahinian A., Kägi D., Hakem A., McCurrach M., Khoo W., Kaufman S. A., Senaldi G., Howard T., Lowe S. W., Mak T. W. ( 1998) Essential contribution of caspase 3/CPP32 to apoptosis and its associated nuclear changes. Genes Dev 12, 806– 819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houde C., Banks K. G., Coulombe N., Rasper D., Grimm E., Roy S., Simpson E. M., Nicholson D. W. ( 2004) Caspase-7 expanded function and intrinsic expression level underlies strain-specific brain phenotype of caspase-3-null mice. J. Neurosci 24, 9977– 9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayascas J. R., Yuste V. J., Benito E., Garcia-Fernàndez J., Comella J. X. ( 2002) Isolation of AmphiCASP-3/7, an ancestral caspase from amphioxus (Branchiostoma floridae). Evolutionary considerations for vertebrate caspases. Cell Death Differ 9, 1078– 1089 [DOI] [PubMed] [Google Scholar]
- 17.Walsh J. G., Cullen S. P., Sheridan C., Lüthi A. U., Gerner C., Martin S. J. ( 2008) Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. U.S.A 105, 12815– 12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young J. E., Gouw L., Propp S., Sopher B. L., Taylor J., Lin A., Hermel E., Logvinova A., Chen S. F., Chen S., Bredesen D. E., Truant R., Ptacek L. J., La Spada A. R., Ellerby L. M. ( 2007) Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J. Biol. Chem 282, 30150– 30160 [DOI] [PubMed] [Google Scholar]
- 19.Ju W., Valencia C. A., Pang H., Ke Y., Gao W., Dong B., Liu R. ( 2007) Proteome-wide identification of family member-specific natural substrate repertoire of caspases. Proc. Natl. Acad. Sci. U.S.A 104, 14294– 14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J. J., Lee Y. J. ( 2008) Differential cleavage of Mst1 by caspase-7/-3 is responsible for TRAIL-induced activation of the MAPK superfamily. Cell. Signal 20, 892– 906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., Núñez G. ( 2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350– 2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agniswamy J., Fang B., Weber I. T. ( 2007) Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis. FEBS J 274, 4752– 4765 [DOI] [PubMed] [Google Scholar]
- 23.McStay G. P., Salvesen G. S., Green D. R. ( 2008) Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ 15, 322– 331 [DOI] [PubMed] [Google Scholar]
- 24.Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. ( 2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol 21, 566– 569 [DOI] [PubMed] [Google Scholar]
- 25.Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. ( 2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362– 1370 [DOI] [PubMed] [Google Scholar]
- 26.Van Damme P., Maurer-Stroh S., Plasman K., Van Durme J., Colaert N., Timmerman E., De Bock P. J., Goethals M., Rousseau F., Schymkowitz J., Vandekerckhove J., Gevaert K. ( 2009) Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol. Cell. Proteomics 8, 258– 272 [DOI] [PubMed] [Google Scholar]
- 27.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376– 386 [DOI] [PubMed] [Google Scholar]
- 28.Van de Craen M., Declercq W., Van den brande I., Fiers W., Vandenabeele P. ( 1999) The proteolytic procaspase activation network: an in vitro analysis. Cell Death Differ 6, 1117– 1124 [DOI] [PubMed] [Google Scholar]
- 29.Behrensdorf H. A., van de Craen M., Knies U. E., Vandenabeele P., Clauss M. ( 2000) The endothelial monocyte-activating polypeptide II (EMAP II) is a substrate for caspase-7. FEBS Lett 466, 143– 147 [DOI] [PubMed] [Google Scholar]
- 30.Desmedt M., Rottiers P., Dooms H., Fiers W., Grooten J. ( 1998) Macrophages induce cellular immunity by activating Th1 cell responses and suppressing Th2 cell responses. J. Immunol 160, 5300– 5308 [PubMed] [Google Scholar]
- 31.Martens L., Vandekerckhove J., Gevaert K. ( 2005) DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21, 3584– 3585 [DOI] [PubMed] [Google Scholar]
- 32.Vande Walle L., Van Damme P., Lamkanfi M., Saelens X., Vandekerckhove J., Gevaert K., Vandenabeele P. ( 2007) Proteome-wide identification of HtrA2/Omi substrates. J. Proteome Res 6, 1006– 1015 [DOI] [PubMed] [Google Scholar]
- 33.Fang B., Boross P. I., Tozser J., Weber I. T. ( 2006) Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition. J. Mol. Biol 360, 654– 666 [DOI] [PubMed] [Google Scholar]
- 34.Wei Y., Fox T., Chambers S. P., Sintchak J., Coll J. T., Golec J. M., Swenson L., Wilson K. P., Charifson P. S. ( 2000) The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem. Biol 7, 423– 432 [DOI] [PubMed] [Google Scholar]
- 35.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. ( 2005) The FoldX web server: an online force field. Nucleic Acids Res 33, W382– W388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krieger E., Darden T., Nabuurs S. B., Finkelstein A., Vriend G. ( 2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57, 678– 683 [DOI] [PubMed] [Google Scholar]
- 37.Baeten L., Reumers J., Tur V., Stricher F., Lenaerts T., Serrano L., Rousseau F., Schymkowitz J. ( 2008) Reconstruction of protein backbones from the BriX collection of canonical protein fragments. PLoS Comput. Biol 4, e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. ( 1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551– 3567 [DOI] [PubMed] [Google Scholar]
- 39.Enoksson M., Li J., Ivancic M. M., Timmer J. C., Wildfang E., Eroshkin A., Salvesen G. S., Tao W. A. ( 2007) Identification of proteolytic cleavage sites by quantitative proteomics. J. Proteome Res 6, 2850– 2858 [DOI] [PubMed] [Google Scholar]
- 40.Varshavsky A. ( 1992) The N-end rule. Cell 69, 725– 735 [DOI] [PubMed] [Google Scholar]
- 41.Fu G., Chumanevich A. A., Agniswamy J., Fang B., Harrison R. W., Weber I. T. ( 2008) Structural basis for executioner caspase recognition of P5 position in substrates. Apoptosis 13, 1291– 1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alves F. M., Hirata I. Y., Gouvea I. E., Alves M. F., Meldal M., Brömme D., Juliano L., Juliano M. A. ( 2007) Controlled peptide solvation in portion-mixing libraries of FRET peptides: improved specificity determination for Dengue 2 virus NS2B-NS3 protease and human cathepsin S. J. Comb. Chem 9, 627– 634 [DOI] [PubMed] [Google Scholar]
- 43.Fuentes-Prior P., Salvesen G. S. ( 2004) The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J 384, 201– 232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. ( 2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866– 876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Impens F., Van Damme P., Demol H., Van Damme J., Vandekerckhove J., Gevaert K. ( 2008) Mechanistic insight into Taxol-induced cell death. Oncogene 27, 4580– 4591 [DOI] [PubMed] [Google Scholar]
- 46.Thiede B., Treumann A., Kretschmer A., Söhlke J., Rudel T. ( 2005) Shotgun proteome analysis of protein cleavage in apoptotic cells. Proteomics 5, 2123– 2130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.