Abstract
Acute myocardial infarction (AMI) is a common cause of death for which effective treatments are available provided that diagnosis is rapid. The current diagnostic gold standards are circulating cardiac troponins I and T. However, their slow release delays diagnosis, and their persistence limits their utility in the identification of reinfarction. The aim was to identify candidate biomarkers of AMI. Isolated mouse hearts were perfused with oxygenated protein-free buffer, and coronary effluent was collected after ischemia or during matched normoxic perfusion. Effluents were analyzed using proteomics approaches based on one- or two-dimensional initial separation. Of the 459 proteins identified after ischemia with one-dimensional separation, 320 were not detected in the control coronary effluent. Among these were all classic existing biomarkers of AMI. We also identified the cardiac isoform of myosin-binding protein C in its full-length form and as a 40-kDa degradation product. This protein was not detected in the other murine organs examined, increased markedly with even trivial myocardial infarction, and could be detected in the plasma after myocardial infarction in vivo, a profile compatible with a biomarker of AMI. Two-dimensional fluorescence DIGE of ischemic and control coronary effluents identified more than 200 asymmetric spots verified by swapping dyes. Once again existing biomarkers of injury were confirmed as well as posttranslational modifications of antioxidant proteins such as peroxiredoxins. Perfusing hearts with protein-free buffers provides a platform of graded ischemic injury that allows detailed analysis of protein release and identification of candidate cardiac biomarkers like myosin-binding protein C.
Acute myocardial infarction (AMI)1 is a common cause of death for which effective treatments are available provided that the condition is rapidly diagnosed. The modern diagnosis of AMI relies on the rise and fall of a specific serum biomarker accompanied by an appropriate circumstance such as chest pain or revascularization. In this accepted paradigm, the diagnosis cannot be ruled in or ruled out without the definite presence or definite absence of a serum biomarker. The ideal biomarker of cardiac injury should be cardiac specific and released rapidly after myocardial injury in direct proportion to the extent of damage. Furthermore, the biomarker should have a high sensitivity and specificity (1). Several biomarkers of AMI have been described in the literature, but only a few, none of which are ideal, have found their way into routine clinical practice. For example, CK-MB starts to increase 4–8 h after coronary artery occlusion and returns to base line within 2–3 days (2). However, its use is limited by its presence in skeletal muscle and normal serum and by sensitivity of the assay to interference, causing some to question its utility (3). Myoglobin is another cytoplasmic protein found in cardiac and skeletal, but not smooth, muscle. It is released even earlier within 1–2 h of AMI and peaks within 5–6 h (2). Unfortunately, any injury to skeletal muscle also causes elevated levels of myoglobin, reducing specificity. Fatty acid-binding proteins (FABPs) are small (15-kDa) cytoplasmic proteins expressed in all tissues with active fatty acid metabolism. Among the nine proteins, heart-specific FABP (H-FABP) is found in heart but also kidney, brain, skeletal muscle, and placenta (4). Following acute myocardial infarction, H-FABP can be detected within 20 min and peaks at 4 h, considerably faster even than CK/CK-MB in the same patient cohort. Although H-FABP concentrations in normal plasma are low, they are known to rise nonspecifically during physical exertion (without a troponin rise), kidney injury, and stroke (5).
The most specific and sensitive cardiac proteins released after acute myocardial infarction are cardiac troponins I and T. Both troponins I and T are released slowly, peaking ∼18 h after myocardial infarction, and remain elevated for 7–10 days (2). This slow release is likely the result of their relatively inaccessible cellular location compared with CK-MB, myoglobin, and H-FABP. Troponins regulate the physical interaction of actin and myosin and thus are found almost entirely associated within the crystalline structure of the sarcomere of striated muscle cells (6). The troponin complex is composed of three forms: I, T, and C. Troponins I and T exist as cardiac specific isoforms with epitopes that differ from the corresponding skeletal isoforms. In addition, the absent or extremely low normal circulating levels of troponin provide the greatest dynamic range of any of the currently available biomarkers (7). Although there is no doubt troponins have revolutionized the detection and management of patients with AMI (8), they do have disadvantages. The slow release of troponin delays diagnosis and the initiation of specific treatments that could salvage heart tissue in those in whom it is raised. Similarly, patients in whom it is absent and who are ultimately reassured and discharged are admitted to the hospital unnecessarily. Furthermore, the persistence of troponins limits their utility in the diagnosis of reinfarction.
It is therefore widely accepted that there is a need for new biomarkers that can diagnose AMI earlier during its natural history and/or that have a short plasma half-life, allowing use in diagnosis and quantification of reinfarction. The purpose of this study was to use the platform of the crystalloid perfused mouse hearts to perform a systematic proteomics analysis of the coronary effluent after minimal AMI to identify new potential biomarkers (9).
MATERIALS AND METHODS
Animals
All experiments were performed in accordance with the United Kingdom Home Office Guidance on the Operation of Animals (Scientific Procedures) Act 1986, published by Her Majesty's Stationary Office, London, UK and with the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH Publication No. 85-23, revised 1996).
Perfusion of Isolated Murine Hearts
Male C57BL/6 mice were anesthetized with pentobarbital (300 mg/kg) and heparin (150 units) intraperitoneally. Hearts were rapidly isolated, mounted onto a Langendorff apparatus, and retrogradely perfused at a constant pressure of 80 mm Hg with Krebs-Henseleit buffer (118.5 mmol/liter NaCl, 25.0 mmol/liter NaHCO3, 4.75 mmol/liter KCl, 0.18 mmol/liter KH2PO4, 1.19 mmol/liter MgSO4, 11.0 mmol/liter d-glucose, and 1.41 mmol/liter CaCl2) equilibrated with 95% O2 and 5% CO2 at 37 °C. Atrial pacing was performed at 580 beats/min.
Experimental Protocol for Isolated Murine Myocardial Infarction Studies
A detailed method has been published previously (10). Briefly, hearts were stabilized for 30 min after initiation of retrograde perfusion. All hearts underwent a period of global ischemia (as indicated in Fig. 1A) followed by 2 h of reperfusion. At the end of reperfusion, hearts were perfused with triphenyl tetrazolium chloride (TTC). Risk and infarct areas were calculated from surface area analysis of short axis slices of left ventricular myocardium. Infarct analysis, by TTC-negative myocardium, was performed in all cases by an investigator blinded to the group assignments.
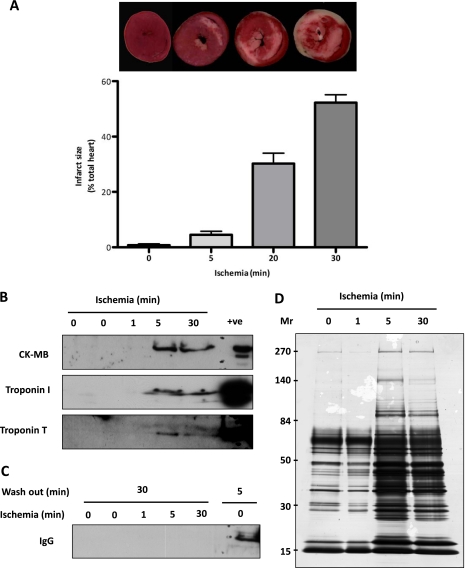
Fig. 1.
Characterization of myocardial infarction and coronary effluent protein content. A, relationship between the extent of infarction and duration of ischemia. Isolated retrogradely perfused mouse hearts were subjected to 0, 5, 20, or 30 min of global ischemia (no flow) followed by 2 h of reperfusion. Infarct size was demarcated by TTC staining. Infarct size expressed as mean ± S.E. B, coronary effluent analysis by immunoblotting. Coronary effluents were collected after the indicated duration of global ischemia and probed for CK-MB, troponin I, and troponin T. C, samples were probed for the presence of IgG after 30 or 5 min of perfusion (washout). D, estimation of coronary effluent protein content by silver staining. Coronary effluents (n = 2 per time point) collected at the onset of reperfusion after the indicated period of ischemia were concentrated and analyzed by a 1D gel (5–20%) and silver stained. All bands were excised, and after in gel tryptic digestion, peptides were identified by MS-MS (proteins are listed in supplemental Table 1). +ve, positive control.
Experimental Protocol for Coronary Effluent Studies
Hearts were perfused on a Langendorff apparatus as described previously (10) with the following modifications. To limit non-ischemic damage to the heart and thereby nonspecific protein release, an intraventricular balloon was omitted. Hearts were stabilized for 30 min after initiation of retrograde perfusion to allow a complete washout of blood and limit the contamination of coronary effluent by plasma proteins. Hearts underwent 5 min of global ischemia (no flow) unless otherwise specified, and the coronary effluent was collected at the onset of reperfusion in a 5-ml aliquot with Triton X-100 at a final concentration of 0.05% to reduce the absorption of protein (11). Aliquots were immediately frozen in liquid nitrogen. Time-matched control mouse hearts were identically perfused but not subjected to ischemia. Coronary effluents were thawed and kept at 4 °C at all times. Coronary effluent was concentrated using a Vivaspin column (with a cutoff of 3000 Da) from Sartorius. Subsequent studies were carried out on the same volume of coronary effluent (1D gel), reflecting the physiology of acute myocardial infarction, or on the same quantity of protein (2D DIGE gel) as measured by a modified Lowry assay to allow an unbiased quantification by fluorescence measurement.
Experimental Protocol for Left Anterior Descending Coronary Artery Ligation in Vivo
Coronary artery occlusion was achieved using the hanging weight system as described previously (12, 13). Briefly, mice were anesthetized by isoflurane inhalation and ventilated by tracheal intubation during the ischemia/reperfusion period. A left thoracotomy was performed, and the left anterior descending coronary artery occlusion was achieved using the hanging weight system. Mice were subjected to 30 min of ischemia and 2 h of reperfusion, and blood was taken by cardiac puncture at the end of reperfusion. Blood was centrifuged at 10,000 × g for 15 min at 4 °C, and plasma aliquots were stored at −80 °C.
Immunoblotting
Concentrated coronary effluents were separated by SDS-PAGE and immunoblotted using previously described protocols (10). The following primary antibodies were used against creatine kinase M/B (catalogue number sc-28898, Santa Cruz Biotechnology), troponin T (catalogue number ab10214, Abcam), troponin I (catalogue number 4002, Cell Signaling Technology), peroxiredoxin 6 (catalogue number LF-PA0011), peroxiredoxin 6-SO3 (catalogue number LF-PA0005), and peroxiredoxin-SO3 (catalogue number LF-PA0004) (all from Labfrontier). Antibody directed against the C0-C1 region of cardiac myosin-binding protein C was a gift from Prof. M. Gautel (Randall Institute, King's College London, London, UK) (14). For immunoblotting of 2D gels, concentrated coronary effluents from control or ischemic mouse hearts were pooled and concentrated further before using the ReadyPrep 2D cleanup kit from Bio-Rad following the manufacturer's instructions. 30 μg of protein extracts were loaded on an IPG strip (18 cm, pH 4–7, nonlinear; GE Healthcare) and then on an SDS-polyacrylamide large gradient gel (12–20%) using a stacking gel.
One-dimensional Gradient Gel Electrophoresis
For prefractionation, concentrated coronary effluents were reconstituted in Laemmli buffer and separated by SDS-PAGE. Large format gradient gels (5–20%) were cast using the a2DEoptimizer (NextGen Sciences, Huntingdon, UK). After the gels were overlaid with water-saturated butanol (2:3) and left to polymerize overnight, the stacking gel containing 4–5% acrylamide weakly buffered at pH 9.0 was cast over the already set resolving gel. Once samples were loaded, a constant 50 mA current was applied as proteins migrated down the stacking gel; at the stacking gel/resolving gel boundary, the current was increased and maintained at 75 mA until the dye front reached the end of the gel. After silver staining (PlusOne silver staining kit, GE Healthcare) with slight modifications to ensure compatibility with subsequent mass spectrometry analysis (15), all gel bands were excised, digested, and subjected to LC-MS/MS analysis.
Two-dimensional Gel Electrophoresis
Protein extracts prepared using the ReadyPrep 2D cleanup kit (Bio-Rad) were resuspended in lysis buffer (8 m urea, 4% (w/v) CHAPS, and 30 mm Tris-Cl, pH 8.5) compatible with DIGE labeling (GE Healthcare). After centrifugation at 13,000 × g for 10 min, the supernatant containing soluble proteins was harvested, and the protein concentration was determined using a modified Bradford method. The fluorescence dye labeling reaction was carried out at a dye/protein ratio of 400 pmol/100 μg. After incubation on ice for 30 min in the dark, the labeling reaction was stopped by scavenging non-bound dye with 10 mm l-lysine (catalogue number L8662, Sigma) for 15 min. For two-dimensional gel electrophoresis, 20 μg of proteins from each group were mixed with the same volume of DIGE 2× buffer (8 m urea, 4% (w/v) CHAPS, 2% (w/v) DTT, and 2% (v/v) Pharmalytes pH 3–10 for IEF), and 20 μg per sample were diluted in rehydration solution (8 m urea, 0.5% (w/v) CHAPS, 0.2% (w/v) DTT, and 0.2% (v/v) Pharmalytes pH 3–10) and loaded on IPG strips (18 cm, pH 3–10, nonlinear, GE Healthcare). After rehydration overnight, strips were focused at 60 kV-h at 20 °C (Multiphor II, GE Healthcare). Once IEF was complete, the strips were equilibrated in 6 m urea containing 30% (v/v) glycerol, 2% (w/v) SDS, and 0.01% (w/v) bromphenol blue with addition of 1% (w/v) DTT for 15 min followed by the same buffer without DTT but with the addition of 4.8% (w/v) iodoacetamide for 15 min. SDS-PAGE was performed using 12% T (total acrylamide concentration), 2.6% C (degree of cross-linking) polyacrylamide gels without a stacking gel using the Ettan DALT system (GE Healthcare). The second dimension was terminated when the bromphenol blue dye front had migrated off the lower end of the gels. Fluorescence images were acquired using the Ettan DIGE imager (GE Healthcare). Finally, gels were fixed overnight in methanol:acetic acid:water solution (4:1:5, v/v/v). Protein profiles were visualized by silver staining using the PlusOne silver staining kit (GE Healthcare) with slight modifications to ensure compatibility with subsequent mass spectrometry analysis (15). For documentation, silver-stained gels were scanned in transmission scan mode using a calibrated scanner (GS-800, Bio-Rad). DIGE gels were analyzed using the DIA module of the DeCyder software (version 6.5, GE Healthcare). A detailed methodology is available upon request.
LC-MS/MS
For MS/MS, in-gel digestion with trypsin was performed according to published methods (16) modified for use with an Investigator ProGest (Genomic Solutions) robotic digestion system. Tryptic peptides were separated by nanoflow liquid chromatography on a reverse phase column (C18 PepMap100 (3 μm, 100 Å, 25 cm), Dionex) and applied to an LTQ Orbitrap XL mass spectrometer (Thermo Scientific). Spectra were collected from the mass analyzer using full ion scan mode over the m/z range 300–2000. Six dependent MS/MS scans were performed on each ion using dynamic exclusion. Database searches were performed using the SEQUEST v.28 (revision 13) (BioWorks version 3.3.1, Thermo Scientific) against mouse UniProt/Swiss-Prot database (release version 13.5, 15,702 protein entries) and X! Tandem (version 2007.01.01.2). Carboxyamidomethylation of cysteine was used as a fixed modification, and oxidation of methionine was used as a variable modification. Mass tolerance for precursor ions was 50 ppm, and mass tolerance for fragment ions was 0.8 Da. Two missed cleavages were allowed. Scaffold (version 2.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (17, 18). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides with ±10-ppm mass accuracy.
RESULTS
Model of Langendorff Apparatus-perfused Murine Hearts
To mimic myocardial ischemia, we used Langendorff apparatus-perfused mouse hearts. In this model, the heart is retrogradely perfused with a standard protein-free buffer (Krebs-Henseleit buffer) at a constant pressure. Before the index ischemia, all hearts were perfused for 30 min to allow recovery of normal function (19) and to avoid a carryover of plasma proteins. Compared with shorter stabilization/washout periods, there was significantly less IgG in the coronary effluent (Fig. 1C). The time-dependent effect of ischemia on infarct size is shown in Fig. 1A: the infarct size increased from 4.5 to 50% with 5–30 min of global ischemia. The coronary effluent was collected after 1, 5, and 30 min of ischemia and examined for the release of known biomarkers of tissue damage by immunoblotting. Fig. 1B shows the release of creatine kinase (a cytoplasmic protein) and of troponin I and troponin T (myofilament proteins) after a 1-, 5-, or 30-min duration of ischemia. It is important to note that these markers of myocardial injury are not present in the control hearts, confirming that cardiac excision and perfusion, in the absence of ischemia, do not cause discernable damage. We were unable to detect the release of markers of myocardial injury with the shortest duration of ischemia (1 min), suggesting that detectable injury occurs between 1 and 5 min in this model. The protein release profile in isolated perfused heart was greatly accelerated compared with that occurring in vivo, and a similar rapid release of CK has been described previously (20, 21). This is likely the result of relative hyperperfusion caused by the low viscosity of the perfusate. The protein release in the coronary effluent was monitored by reverse phase HPLC (supplemental Fig. 1). Consistent with a previous report (21), we observed a predominant peak eluting around 23 min using a C8 Zorbax column. As expected, the protein release increased with the duration of ischemia.
Coronary Effluent Analysis by 1D Gel-LC-MS/MS
To reduce the complexity of the sample, the coronary effluents were first separated on large format gradient gels (5–20%) and stained with silver (Fig. 1D). Notably, the shortest ischemia duration at which there was a detectable difference in the coronary effluent compared with base-line protein release was 5 min. Although this time point caused only minor infarction (see Fig. 1A), it was used for further proteomics analysis. All bands were excised from the gel, subjected to tryptic digestion, and identified by nanoflow reverse phase LC-MS/MS. In total, 459 proteins were identified in the ischemic coronary effluent compared with only 139 in the control samples (see supplemental Table 1 for the complete list of proteins). Thus, 320 proteins were unique to ischemia/reperfusion samples. Moreover, all known biomarkers of myocardial injury, such as troponins I, C, and T; creatine kinase; lactate dehydrogenase; myoglobin; heart-specific fatty acid-binding protein; aspartate aminotransferase; etc., were among the proteins identified (Table I). Relative quantification of the protein release after ischemia was obtained by using spectral counts computed using the Scaffold software (version 2.0, Proteome Software Inc.). Whereas troponin I, troponin T, and troponin C were only detected in the coronary effluent after ischemia but not at base line, other markers such as CK, FABP, LDH, etc. were present under both conditions (Table I). However, the number of peptides identified for CK increased by 8-fold, those identified for FABP increased by 11-fold, and those identified for lactate dehydrogenase increased by 6-fold in ischemic compared with normoxic hearts, and their appearance/enrichment underscores their potential as clinical biomarkers for myocardial injury.
Table I. Known biomarkers of myocardial infarction identified by one-dimensional electrophoresis and LC-MS/MS of coronary effluent.
The normalized spectral count was computed using Scaffold software (version 2.0, Proteome Software Inc.), and protein release was expressed as a ratio using the control count as the denominator and ischemic count as the numerator. “+++” indicates proteins identified in coronary effluent from ischemic but not control hearts (denominator is zero).
| Protein name | Accession number | Molecular mass | Unique peptides in control | Unique peptides in ischemia | Ratio ischemia/control |
|---|---|---|---|---|---|
| kDa | |||||
| Troponin I, cardiac muscle | TNNI3_MOUSE | 24 | 0 | 66 | +++ |
| Troponin T, cardiac muscle | TNNT2_MOUSE | 36 | 0 | 5 | +++ |
| Troponin C, slow skeletal and cardiac muscles | TNNC1_MOUSE | 18 | 0 | 11 | +++ |
| Creatine kinase B-type | KCRB_MOUSE | 43 | 6 | 24 | 4.0 |
| Creatine kinase M-type | KCRM_MOUSE | 43 | 37 | 306 | 8.3 |
| Fatty acid-binding protein, heart | FABPH_MOUSE | 15 | 6 | 68 | 11.3 |
| l-Lactate dehydrogenase A chain | LDHA_MOUSE | 36 | 31 | 205 | 6.6 |
| l-Lactate dehydrogenase B chain | LDHB_MOUSE | 37 | 56 | 349 | 6.2 |
| Myoglobin | MYG_MOUSE | 17 | 45 | 237 | 5.3 |
| Myosin light chain 3 | MYL3_MOUSE | 22 | 5 | 30 | 6.0 |
| α-Enolase | ENOA_MOUSE | 47 | 39 | 49 | 1.3 |
| β-Enolase | ENOB_MOUSE | 47 | 59 | 206 | 3.5 |
Coronary Effluent Analysis by DIGE
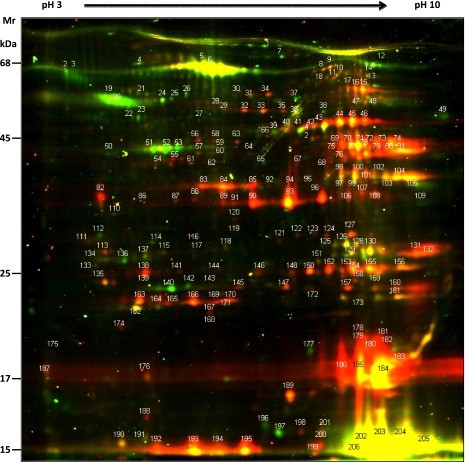
Posttranslational modifications of albumin caused by oxidative stress during ischemia/reperfusion (known as ischemia-modified albumin) are already used clinically as a biomarker of ischemia. To determine whether similarly useful posttranslational modifications occurred in proteins in the coronary effluent, control and ischemic samples were analyzed by 2D DIGE (Fig. 2). The fluorescence intensity was quantified for more than 200 spots. Results were reproduced by using a dye swap (supplemental Fig. 2). After silver staining, all spots were excised and identified by tandem mass spectrometry. The list of proteins identified is presented in supplemental Table 2. Quantitative values were obtained by using the DeCyder software (GE Healthcare). The DIGE approach confirmed the quantitation based on spectral counting in that known markers of cardiac infarction, such as troponin T, troponin C, CK-MB isoforms, FABP, LDH, and myoglobin, were present in higher abundance in ischemic than control samples (Table II and Fig. 3).
Fig. 2.
Representative DIGE 2D gel of proteins appearing in coronary effluents after 5 min of global ischemia (Cy5; red) and matched control perfusion (Cy3; green). Coronary effluents from control and ischemic hearts (n = 3–4) were concentrated, and the same amount of protein was CyDye-labeled before separation by 2D gel electrophoresis. The annotated spots were excised and identified by LC-MS/MS (see supplemental Table 2 for the complete list of proteins).
Table II. Known biomarkers of myocardial infarction identified by two-dimensional electrophoresis.
The fluorescence intensity (Cy3 and Cy5) for each spot was measured, and the ratio was calculated using Cy5 as the numerator and Cy3 as the denominator to express release after ischemia as a ratio of that with matched control perfusion.
| Spot identification | Protein name | Accession number | Molecular mass | Ischemia/control-fold |
|---|---|---|---|---|
| kDa | ||||
| 55 | Troponin T | TNNT2_MOUSE | 35 | 4.5 |
| 187 | Troponin C | TNNC1_MOUSE | 18 | 4.23 |
| 56 | Creatine kinase B-type | KCRB_MOUSE | 42 | 4.5 |
| 69 | Creatine kinase M-type | KCRM_MOUSE | 43 | 15.3 |
| 71 | Creatine kinase M-type | KCRM_MOUSE | 43 | 15.1 |
| 83 | l-Lactate dehydrogenase B chain | LDHB_MOUSE | 36 | 8.2 |
| 84 | l-Lactate dehydrogenase B chain | LDHB_MOUSE | 36 | 17.9 |
| 107 | l-Lactate dehydrogenase A chain | LDHA_MOUSE | 36 | 7.0 |
| 108 | l-Lactate dehydrogenase A chain | LDHA_MOUSE | 36 | 4.8 |
| 138 | Myosin light polypeptide 3 | MYL3_MOUSE | 22 | 10.1 |
| 184 | Myoglobin | MYG_MOUSE | 17 | 20.7 |
| 185 | Myoglobin | MYG_MOUSE | 17 | 22.7 |
| 192 | Fatty acid-binding protein, heart | FABPH_MOUSE | 14 | 9.4 |
| 193 | Fatty acid-binding protein, heart | FABPH_MOUSE | 14 | 14.2 |
| 194 | Fatty acid-binding protein, heart | FABPH_MOUSE | 14 | 11.1 |
| 195 | Fatty acid-binding protein, heart | FABPH_MOUSE | 14 | 20.4 |
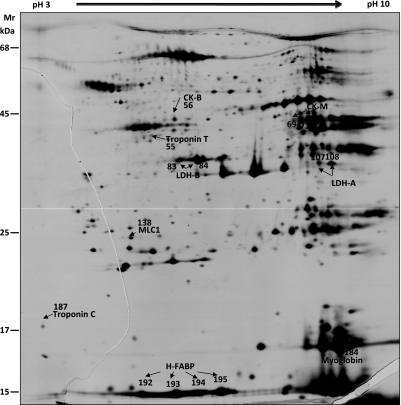
Fig. 3.
Representative silver-stained DIGE 2D gel of proteins appearing in coronary effluents after 5 min of global ischemia and matched control perfusion. The known biomarkers identified by mass spectrometry are indicated.
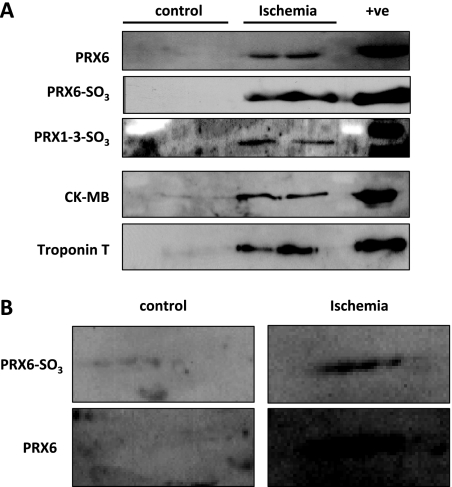
Verification by Immunoblotting of Proteomics Identification of Oxidative Stress-related Proteins
Oxygen and other free radicals are generated in the ischemic and reperfused heart and are important mediators of postischemic injury (for a review, see Ref. 22). Our proteomics analysis revealed the release of numerous proteins involved in neutralizing free radicals following ischemia/reperfusion, including thioredoxin reductase, peroxiredoxins, etc. (see Table III). Peroxiredoxins (Prdxs) are a family of antioxidant proteins, which are highly abundant in the heart, that have a cytoprotective role in the myocardium and vasculature. Myocardial peroxiredoxins undergo a complex series of redox-dependent structural changes in response to oxidant stress (23). Prdx3 confers protection against oxidative stress within the mitochondria of aortic cells and within the ischemic myocardium (24) and also prevents left ventricular remodeling and failure after myocardial infarction (25). Their functional role is indicated by Prdx6-null mice, which are more susceptible to ischemia/reperfusion injury (26). The separation by 2D gel electrophoresis revealed that ischemic injury induced an acidic shift in the isoelectric point of proteins containing redox-active cysteines as illustrated for peroxiredoxin 6 (supplemental Fig. 3B) (27, 28). This finding is consistent with previous reports confirming sulfoxidation as a common posttranslational modification in redox-sensitive proteins (23). To confirm the presence of these potentially cardioprotective proteins in ischemic coronary effluents, we validated our findings by immunoblotting. The release of peroxiredoxins 1–3 and 6 was not only significantly higher in ischemic compared with control samples (Fig. 4A) but antibodies against oxidized peroxiredoxin 6 also confirmed the presence of the oxidized isoform after myocardial ischemia by immunoblotting on a 1D gel (Fig. 4A) and on a 2D gel (Fig. 4B). Thus, the proteomics analysis revealed evidence for posttranslational modifications of antioxidant defense proteins in ischemic coronary effluents. Furthermore, in combination with the literature (26), our findings suggest that the hyperoxidized form of peroxiredoxin identified in the coronary effluent is not only a marker of myocardial ischemic injury but may be part of a defense mechanism to limit oxidative stress-induced damage.
Table III. Proteins involved in oxidative stress response that were identified in coronary effluents using 1D or 2D gel-based approach.
The -fold variation was calculated as described in Tables I and II.
| Protein name | Accession number | Molecular mass | Ischemia/control-fold |
|---|---|---|---|
| kDa | |||
| 1D gel | |||
| Aflatoxin B1 aldehyde reductase member 2 | ARK72_MOUSE | 41 | +++ |
| Catalase | CATA_MOUSE | 60 | 0.6 |
| Extracellular superoxide dismutase (Cu,Zn) | SODE_MOUSE | 27 | 4.0 |
| Glutaredoxin 3 | GLRX3_MOUSE | 38 | +++ |
| Glutaredoxin 3 | GLRX3_MOUSE | 38 | +++ |
| Glutathione peroxidase 1 | GPX1_MOUSE | 22 | 0.7 |
| Glutathione peroxidase 3 | GPX3_MOUSE | 25 | 1.9 |
| Glutathione reductase | GSHR_MOUSE | 54 | 2.3 |
| Glutathione S-transferase A4 | GSTA4_MOUSE | 26 | 3.6 |
| Glutathione S-transferase Mu 1 | GSTM1_MOUSE | 26 | 5.2 |
| Glutathione S-transferase Mu 2 | GSTM2_MOUSE | 26 | 5.7 |
| Glutathione S-transferase Mu 5 | GSTM5_MOUSE | 27 | +++ |
| Glutathione S-transferase Mu 7 | GSTM7_MOUSE | 26 | 2.0 |
| Glutathione S-transferase P 1 | GSTP1_MOUSE | 24 | 4.5 |
| Glutathione synthetase | GSHB_MOUSE | 52 | +++ |
| Methionine-R-sulfoxide reductase B3 | MSRB3_MOUSE | 20 | +++ |
| Peptide methionine sulfoxide reductase | MSRA_MOUSE | 26 | 9.0 |
| Peroxiredoxin 1 | PRDX1_MOUSE | 22 | 4.3 |
| Peroxiredoxin 2 | PRDX2_MOUSE | 22 | 3.9 |
| Peroxiredoxin 5 | PRDX5_MOUSE | 22 | 3.8 |
| Peroxiredoxin 6 | PRDX6_MOUSE | 25 | 5.0 |
| Superoxide dismutase (Cu,Zn) | SODC_MOUSE | 16 | 1.7 |
| Thioredoxin | THIO_MOUSE | 12 | 2.0 |
| Thioredoxin domain-containing protein 17 | TXD17_MOUSE | 14 | +++ |
| Thioredoxin reductase 1, cytoplasmic | TRXR1_MOUSE | 67 | +++ |
| Xanthine dehydrogenase/oxidase | XDH_MOUSE | 147 | 1.4 |
| 2D gel | |||
| Glutaredoxin 3 | GLRX3_MOUSE | 38 | 2.7 |
| Hydroxyacylglutathione hydrolase | GLO2_MOUSE | 29 | 2.4 |
| Peroxiredoxin 1 | PRDX1_MOUSE | 22 | 3.8 |
| Peroxiredoxin 2 | PRDX2_MOUSE | 22 | 8.0 |
| Peroxiredoxin 5, mitochondrial precursor | PRDX5_MOUSE | 22 | 20.7 |
| Peroxiredoxin 6 | PRDX6_MOUSE | 25 | 16.2 |
| Peroxiredoxin 6 | PRDX6_MOUSE | 25 | 7.5 |
| Peroxiredoxin 6 | PRDX6_MOUSE | 25 | 6.4 |
| Superoxide dismutase (Cu,Zn) | SODC_MOUSE | 16 | 8.1 |
| Superoxide dismutase (Cu,Zn) | SODC_MOUSE | 16 | 4.8 |
| Thioredoxin | THIO_MOUSE | 12 | 2.0 |
Fig. 4.
Verification by immunoblotting of oxidative stress-related proteins appearing in coronary effluent after ischemia. A, analyses of coronary effluents collected after 5 min of ischemia or matching control perfusion by 1D electrophoresis. Peroxiredoxins and their posttranslationally modified oxidized forms (PRX-SO3) were detected only after ischemia. Immunoblots for CK and troponin were used as controls to indicate ischemia-selective protein release. B, analyses of coronary effluents collected after 5 min of ischemia or matching control perfusion by 2D electrophoresis. Immunoblots of a 2D gel (18-cm strip, pH 4–7, 12–20% gradient gel) for peroxiredoxin 6 total and oxidized forms are shown. Peroxiredoxin 6 is present only in ischemic coronary effluent, and the charge train of the oxidized peroxiredoxin-SO3 can be visualized. +ve, positive.
Myosin-binding Protein C as Candidate Biomarker of Myocardial Injury
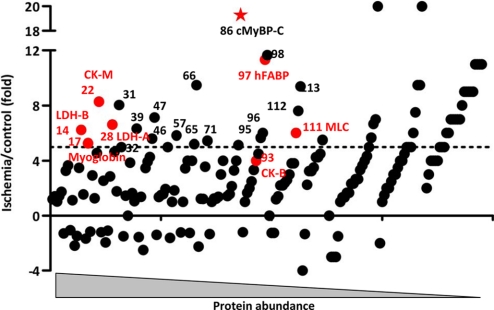
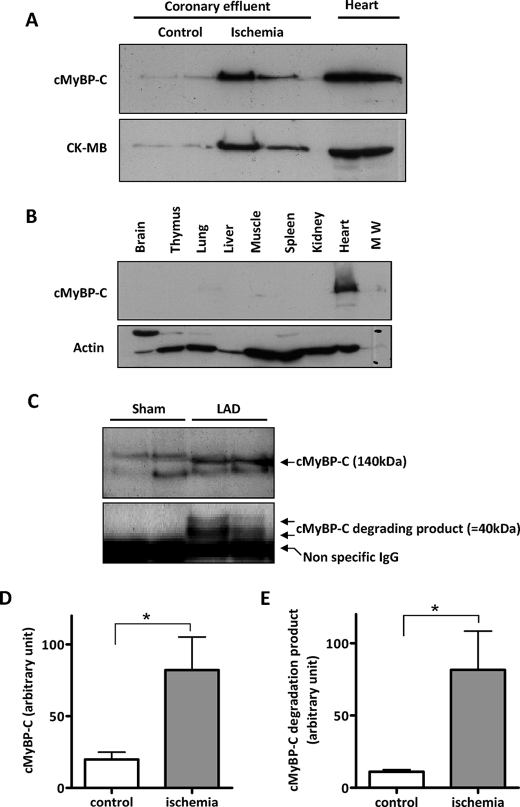
In Fig. 5, proteins in the postischemic coronary effluent are plotted according to their estimated abundance based on the normalized spectral count. The -fold change compared with control hearts is plotted on the y axis, and those proteins with markedly increased release after ischemia are listed in Table IV. Troponins are not present in this table as they were detected only in ischemic, and not in control, coronary effluent, confirming their very high specificity for cardiac damage. Most known biomarkers (CK, LDH, etc.) are in a cluster at the top left, depicting highly abundant proteins with a pronounced propensity to leak into the coronary effluent after ischemia. Interestingly, cardiac myosin-binding protein C (cMyBP-C) appeared in a similar region (Fig. 5), and its -fold change (19-fold) exceeded that of many established biomarkers. We subsequently verified the release of cMyBP-C in coronary effluent by immunoblotting (Fig. 6A) using an antibody directed against the C0-C1 region of cMyBP-C, which is only present in the cardiac isoform (Fig. 6B) (for a review, see Ref. 29). We further confirmed ischemia-selective cMyBP-C release into plasma after regional myocardial ischemia and reperfusion in vivo. Interestingly, we were not only able to detect a band at 140 kDa, corresponding to the full-length cMyBP-C, but also a degradation product at 40 kDa (Fig. 6, C, D, and E), which has previously been noted in postischemic myocardium (30, 31). Thus, our proteomics approach has validated a number of existing biomarkers and revealed a cardiac specific potential new marker detectable with only minor myocardial injury.
Fig. 5.
Proteins released into coronary effluent. Protein release after 5 min of ischemia (identified on the 1D gel) is plotted as -fold change (unique peptide in ischemia/unique peptide in control ratio) on the ordinate against absolute abundance on the abscissa. Abundance is estimated by the obtained spectral count normalized to the molecular weight of the individual protein. The known biomarkers of myocardial infarction are highlighted in red, and proteins in the coronary effluent with a -fold change greater than 5 are numbered and listed in Table IV. MLC, myosin light chain. Asterisk highlights MyBP-C.
Table IV. Proteins released into coronary effluent at abundance and with ischemia selectivity similar to those of established biomarkers (see Fig. 5).
Coronary effluents from control and 5-min ischemia were separated on a 1D gel and identified by LC-MS/MS, and abundance was estimated based on the normalized spectral count. A cutoff of a 5-fold enrichment in postischemic effluent was used (see Fig. 5). Known biomarkers appear in italic text, and potential candidate biomarkers are in non-italic text.
| Protein identification | Protein name | Accession number | Molecular mass | Unique peptides in control | Unique peptides in ischemia | Ratio ischemia/control |
|---|---|---|---|---|---|---|
| kDa | ||||||
| 14 | l-Lactate dehydrogenase B chain | LDHB_MOUSE | 37 | 56 | 349 | 6.2 |
| 17 | Myoglobin | MYG_MOUSE | 17 | 45 | 237 | 5.3 |
| 21 | Phosphoglycerate kinase 1 | PGK1_MOUSE | 45 | 39 | 178 | 4.6 |
| 22 | Creatine kinase M-type | KCRM_MOUSE | 43 | 37 | 306 | 8.3 |
| 28 | l-Lactate dehydrogenase A chain | LDHA_MOUSE | 36 | 31 | 205 | 6.6 |
| 29 | Heat shock cognate 71-kDa protein | HSP7C_MOUSE | 71 | 28 | 130 | 4.6 |
| 31 | Malate dehydrogenase, cytoplasmic | MDHC_MOUSE | 37 | 26 | 209 | 8.0 |
| 32 | Fructose-bisphosphate aldolase A | ALDOA_MOUSE | 39 | 25 | 125 | 5.0 |
| 39 | Triose-phosphate isomerase | TPIS_MOUSE | 27 | 18 | 114 | 6.3 |
| 47 | Vinculin | VINC_MOUSE | 117 | 15 | 107 | 7.1 |
| 46 | Phosphoglucomutase 1 | PGM1_MOUSE | 62 | 15 | 84 | 5.6 |
| 45 | Peroxiredoxin 1 | PRDX1_MOUSE | 22 | 15 | 64 | 4.3 |
| 53 | Peroxiredoxin 6 | PRDX6_MOUSE | 25 | 14 | 70 | 5.0 |
| 57 | Elongation factor 1-α1 | EF1A1_MOUSE | 50 | 12 | 70 | 5.8 |
| 62 | Epoxide hydrolase 2 | HYES_MOUSE | 63 | 11 | 44 | 4.0 |
| 66 | Selenium-binding protein 1 | SBP1_MOUSE | 53 | 10 | 95 | 9.5 |
| 65 | Glutathione S-transferase Mu 1 | GSTM1_MOUSE | 26 | 10 | 52 | 5.2 |
| 64 | UTP-glucose-1-phosphate uridylyltransferase | UGPA_MOUSE | 57 | 10 | 40 | 4.0 |
| 71 | Adenosylhomocysteinase | SAHH_MOUSE | 48 | 9 | 49 | 5.4 |
| 78 | Glyoxylate reductase/hydroxypyruvate reductase | GRHPR_MOUSE | 35 | 8 | 32 | 4.0 |
| 86 | Myosin-binding protein C, cardiac type | MYPC3_MOUSE | 141 | 7 | 135 | 19.3 |
| 85 | Lactoylglutathione lyase | LGUL_MOUSE | 21 | 7 | 36 | 5.1 |
| 84 | Isocitrate dehydrogenase (NADP) cytoplasmic | IDHC_MOUSE | 47 | 7 | 28 | 4.0 |
| 98 | Elongation factor 2 | EF2_MOUSE | 95 | 6 | 70 | 11.7 |
| 97 | Fatty acid-binding protein, heart | FABPH_MOUSE | 15 | 6 | 68 | 11.3 |
| 96 | Heat shock-related 70-kDa protein 2 | HSP72_MOUSE | 70 | 6 | 36 | 6.0 |
| 95 | Glutathione S-transferase Mu 2 | GSTM2_MOUSE | 26 | 6 | 34 | 5.7 |
| 94 | Phosphatidylethanolamine-binding protein 1 | PEBP1_MOUSE | 21 | 6 | 27 | 4.5 |
| 93 | Creatine kinase B-type | KCRB_MOUSE | 43 | 6 | 24 | 4.0 |
| 113 | WD repeat-containing protein 1 | WDR1_MOUSE | 66 | 5 | 47 | 9.4 |
| 112 | Proteasome subunit β type-5 precursor | PSB5_MOUSE | 29 | 5 | 38 | 7.6 |
| 111 | Myosin light chain 3 | MYL3_MOUSE | 22 | 5 | 30 | 6.0 |
| 125 | Transitional endoplasmic reticulum ATPase | TERA_MOUSE | 89 | 4 | 56 | 14.0 |
| 124 | Adenylate kinase isoenzyme 1 | KAD1_MOUSE | 22 | 4 | 54 | 13.5 |
| 123 | 6-Phosphogluconate dehydrogenase, decarboxylating | 6PGD_MOUSE | 53 | 4 | 22 | 5.5 |
| 122 | Glutathione S-transferase P 1 | GSTP1_MOUSE | 24 | 4 | 18 | 4.5 |
| 120 | Peptidyl-prolyl cis-trans isomerase A | PPIA_MOUSE | 18 | 4 | 17 | 4.3 |
| 121 | Ferritin light chain 1 | FRIL1_MOUSE | 21 | 4 | 17 | 4.3 |
| 148 | Aldose reductase | ALDR_MOUSE | 36 | 3 | 60 | 20.0 |
| 147 | Apolipoprotein A-I precursor | APOA1_MOUSE | 31 | 3 | 21 | 7.0 |
| 146 | 14-3-3 protein ζ/δ | 1433Z_MOUSE | 28 | 3 | 20 | 6.7 |
| 145 | Kynurenine-oxoglutarate transaminase 1 | KAT1_MOUSE | 48 | 3 | 17 | 5.7 |
| 144 | 14-3-3 protein γ | 1433G_MOUSE | 28 | 3 | 15 | 5.0 |
| 143 | Proteasome subunit β type-6 precursor | PSB6_MOUSE | 25 | 3 | 13 | 4.3 |
| 142 | Serpin B6 | SPB6_MOUSE | 43 | 3 | 12 | 4.0 |
| 169 | Phosphoglycerate mutase 1 | PGAM1_MOUSE | 29 | 2 | 50 | 25.0 |
| 168 | Phosphoglycerate mutase 2 | PGAM2_MOUSE | 29 | 2 | 40 | 20.0 |
| 166 | Cytosolic nonspecific dipeptidase | CNDP2_MOUSE | 53 | 2 | 19 | 9.5 |
| 167 | Protein-disulfide isomerase A3 precursor | PDIA3_MOUSE | 57 | 2 | 19 | 9.5 |
| 165 | Aminoacylase 1 | ACY1_MOUSE | 46 | 2 | 15 | 7.5 |
| 164 | Protein NDRG2 | NDRG2_MOUSE | 41 | 2 | 12 | 6.0 |
| 161 | Destrin | DEST_MOUSE | 19 | 2 | 10 | 5.0 |
| 162 | GMP reductase 1 | GMPR1_MOUSE | 37 | 2 | 10 | 5.0 |
| 163 | Tubulin β-2C chain | TBB2C_MOUSE | 50 | 2 | 10 | 5.0 |
| 160 | Pyridoxal kinase | PDXK_MOUSE | 35 | 2 | 9 | 4.5 |
| 158 | Extracellular superoxide dismutase (Cu,Zn) precursor | SODE_MOUSE | 27 | 2 | 8 | 4.0 |
| 159 | 3,2-trans-Enoyl-CoA isomerase, mitochondrial precursor | D3D2_MOUSE | 32 | 2 | 8 | 4.0 |
| 213 | Glycogen phosphorylase, brain form | PYGB_MOUSE | 97 | 1 | 89 | 89.0 |
| 212 | Glycogen phosphorylase, muscle form | PYGM_MOUSE | 97 | 1 | 62 | 62.0 |
| 211 | Inositol monophosphatase | IMPA1_MOUSE | 30 | 1 | 39 | 39.0 |
| 208 | 14-3-3 protein β/α | 1433B_MOUSE | 28 | 1 | 27 | 27.0 |
| 209 | Ferritin heavy chain | FRIH_MOUSE | 21 | 1 | 27 | 27.0 |
| 210 | Phosphoglucomutase-like protein 5 | PGM5_MOUSE | 62 | 1 | 27 | 27.0 |
| 207 | Moesin | MOES_MOUSE | 68 | 1 | 23 | 23.0 |
| 206 | Alanine aminotransferase 1 | ALAT1_MOUSE | 55 | 1 | 22 | 22.0 |
| 205 | Cytoplasmic aconitate hydratase | ACOC_MOUSE | 98 | 1 | 18 | 18.0 |
| 204 | Aconitate hydratase, mitochondrial precursor | ACON_MOUSE | 85 | 1 | 17 | 17.0 |
| 202 | Dihydropteridine reductase | DHPR_MOUSE | 26 | 1 | 16 | 16.0 |
| 203 | Isocitrate dehydrogenase (NADP), mitochondrial precursor | IDHP_MOUSE | 51 | 1 | 16 | 16.0 |
| 196 | Bifunctional purine biosynthesis protein PURH | PUR9_MOUSE | 64 | 1 | 14 | 14.0 |
| 197 | Secernin 3 | SCRN3_MOUSE | 48 | 1 | 14 | 14.0 |
| 198 | Leukotriene A-4 hydrolase | LKHA4_MOUSE | 69 | 1 | 14 | 14.0 |
| 199 | 6-Phosphogluconolactonase | 6PGL_MOUSE | 27 | 1 | 14 | 14.0 |
| 200 | Aspartyl aminopeptidase | DNPEP_MOUSE | 52 | 1 | 14 | 14.0 |
| 201 | Nicotinamide phosphoribosyltransferase | NAMPT_MOUSE | 55 | 1 | 14 | 14.0 |
| 192 | Carboxymethylenebutenolidase homolog | CMBL_MOUSE | 28 | 1 | 11 | 11.0 |
| 193 | Catechol O-methyltransferase | COMT_MOUSE | 29 | 1 | 11 | 11.0 |
| 194 | Ribosyldihydronicotinamide dehydrogenase (quinone) | NQO2_MOUSE | 26 | 1 | 11 | 11.0 |
| 195 | Inorganic pyrophosphatase | IPYR_MOUSE | 33 | 1 | 11 | 11.0 |
| 187 | UPF0587 protein C1orf123 homolog | CA123_MOUSE | 18 | 1 | 9 | 9.0 |
| 188 | Fructose-1,6-bisphosphatase isozyme 2 | F16P2_MOUSE | 37 | 1 | 9 | 9.0 |
| 189 | ATP synthase subunit α, mitochondrial precursor | ATPA_MOUSE | 60 | 1 | 9 | 9.0 |
| 190 | Peptide methionine sulfoxide reductase | MSRA_MOUSE | 26 | 1 | 9 | 9.0 |
| 191 | Glycyl-tRNA synthetase | SYG_MOUSE | 82 | 1 | 9 | 9.0 |
| 186 | Hypoxanthine-guanine phosphoribosyltransferase | HPRT_MOUSE | 25 | 1 | 8 | 8.0 |
| 181 | Alcohol dehydrogenase class-3 | ADHX_MOUSE | 40 | 1 | 7 | 7.0 |
| 182 | Translationally controlled tumor protein | TCTP_MOUSE | 19 | 1 | 7 | 7.0 |
| 183 | Guanine nucleotide-binding protein subunit β-2-like 1 | GBLP_MOUSE | 35 | 1 | 7 | 7.0 |
| 184 | Hydroxyacyl-coenzyme A dehydrogenase | HCDH_MOUSE | 34 | 1 | 7 | 7.0 |
| 185 | Purine-nucleoside phosphorylase | PNPH_MOUSE | 32 | 1 | 7 | 7.0 |
| 179 | Inter-α-trypsin inhibitor heavy chain H1 precursor | ITIH1_MOUSE | 101 | 1 | 6 | 6.0 |
| 180 | DNA damage-binding protein 1 | DDB1_MOUSE | 127 | 1 | 6 | 6.0 |
| 176 | α2-HS-glycoprotein precursor | FETUA_MOUSE | 37 | 1 | 5 | 5.0 |
| 177 | Dihydropyrimidinase-related protein 2 | DPYL2_MOUSE | 62 | 1 | 5 | 5.0 |
| 178 | Actin-related protein 2/3 complex subunit 2 | ARPC2_MOUSE | 34 | 1 | 5 | 5.0 |
| 172 | Clathrin light chain B | CLCB_MOUSE | 25 | 1 | 4 | 4.0 |
| 173 | Stress 70 protein, mitochondrial precursor | GRP75_MOUSE | 74 | 1 | 4 | 4.0 |
| 174 | Heat shock protein β-2 | HSPB2_MOUSE | 20 | 1 | 4 | 4.0 |
| 175 | NADP-dependent malic enzyme | MAOX_MOUSE | 64 | 1 | 4 | 4.0 |
Fig. 6.
Verification and validation of cMyBP-C as potential new biomarker of acute myocardial infarction. A, 1D separation and immunoblotting of proteins in coronary effluent. Coronary effluents were collected after 5 min of ischemia or matching control perfusion. B, cMyBP-C content among mouse organs. The anti-cMyBP-C antibody reacted only with cardiac tissue. Anti-actin was used as loading control. C, detection of cMyBP-C in the plasma of mice subjected to acute myocardial infarction. Mice were subjected to temporary ligation of the left anterior descending (LAD) coronary artery (30 min) followed by reperfusion (2 h). Blood was collected, and plasma was immunoblotted for cMyBP-C. The full-length cMyBP-C at 140 kDa and the degradation product at 40 kDa were both detected selectively in plasma of mice subjected to temporary, but not sham, coronary artery ligation. D and E, quantification of full-length and short forms of cMyBP-C. Data are derived from n = 6 mice per group and are expressed as mean ± S.E. *, p < 0.05.
DISCUSSION
The aim of this study was to identify candidate biomarkers by comprehensively analyzing the proteins released into the coronary effluent after a duration of myocardial ischemia chosen to cause mild injury. Our extensive proteomics analysis confirmed the presence of all existing biomarkers of AMI but also revealed posttranslational modifications of antioxidant proteins and identified the cardiac isoform of cMyBP-C as a candidate biomarker. Furthermore, using an in vivo murine model of AMI, we could detect cMyBP-C in the plasma. These promising early results have yet to be supported by a detailed time course of MyBP-C release/clearance in serum and further confirmed by clinical studies where the magnitude and profile of release and ultimately the predictive performance are compared with those of the established biomarkers. Without such validation it is not possible to progress from the current status of promising candidate to bona fide biomarker.
Langendorff apparatus-perfused mouse hearts formed the basis of our discovery platform. This model recapitulates true ischemic injury and allows a careful titration of the duration of ischemia to cause minimal, but definite, AMI. Furthermore, the model has the advantage of limiting contamination of the coronary effluent by plasma proteins. The choice of this model was vindicated by its ability to identify all the commonly used existing biomarkers of AMI (troponin I, troponin T, creatine kinase, FABP, LDH, and myoglobin). An extensive proteomics analysis of the coronary effluent identified more than 450 proteins using a 1D gel-LC-MS/MS approach and around 200 proteins using the DIGE platform. There was a large overlap in the proteins identified by these two methods, and only 1.1% of proteins were identified solely on 2D gels. Nevertheless, the 2D approach gave useful complementary information regarding posttranslational modifications and in particular identified changes in proteins involved in redox defense such as the peroxiredoxins. The proteomics analysis revealed that the troponins, the current gold standard to assess AMI, are among the most abundant proteins in ischemic coronary effluents and show minimal leakage from normoxic myocardium (Table I).
Over the last few years, proteomics has been used extensively for the discovery of potential biomarkers for cancer (32), atherosclerosis (33), and cardiovascular disease (for reviews, see Refs. 9 and 34). Plasma and serum were routinely used for biomarker discovery, although the dynamic range of plasma proteins spans 10 orders of magnitude, and just five proteins typically constitute more than 90% of the total protein mass. This complexity exceeds the analytical capabilities of most proteomics approaches. Despite these evident difficulties, several proteomics studies have been undertaken to discover predictive cardiovascular biomarkers, but most studies failed to even detect the existing biomarkers and revealed only changes in high abundance plasma proteins (35). In an alternative approach, we analyzed the coronary effluent in an ex vivo model where the heart was perfused with a crystalloid buffer without proteins. This approach has been used previously by Koomen et al. (36) in a model of ischemia/reperfusion injury in rat hearts, but they encountered a high contamination with plasma proteins (30% of the 342 proteins identified were of plasma origin), which minimizes the chance of detecting potential cardiac specific biomarkers. By extending the washout period and perhaps by using smaller hearts, we managed to substantially reduce the contamination by plasma proteins. Moreover, we used the latest ion trap technology (LTQ Orbitrap XL) and used a 1D gel-LC/MS/MS approach besides 2D gel electrophoresis. This allowed us to detect substantially more proteins, including all the standard biomarkers of myocardial injury, unlike previous publications (36). We identified more than 450 proteins by the 1D gel ranging from 11 to 272 kDa and nearly 200 proteins by the 2D gel ranging from 11 to 72 kDa. This difference in mass range was due to the use of a variable 5–20% gradient in our 1D gel compared with the fixed 12% concentration of our 2D gel. The 1D approach was better for protein identification in this simple protein mixture and allowed the detection of MyBP-C, which was not revealed on the 2D gel. The 2D gel coupled with fluorescence labeling of samples provided the advantage of comparative protein quantification and allowed the identification of relevant posttranslation modifications such as the oxidization of the peroxiredoxins by ischemia. Thus the 1D and 2D gel-based approaches are best viewed as complementary, each having unique characteristics that have contributed to our findings.
The detection of many oxidative stress-related proteins in the coronary effluent may reflect a mechanism of defense of the heart to eliminate hydrogen peroxide formed during ischemia/reperfusion. Indeed, peroxiredoxin, catalase, and glutathione peroxidase are the predominant enzymes responsible for the elimination of H2O2 in cells. In addition, the catalytic activities of these enzymes are extensively regulated by posttranslational modifications such as sulfinylation (37). Further studies will be needed to understand why and how these protective enzymes are released after myocardial ischemia/reperfusion stress.
A methodological question arose during this study as to whether proteomics analysis should be done on the same volume of coronary effluent or on the same quantity of protein released. It is well established that during ischemia/reperfusion there is an increased protein release by the myocardial tissue (14 ng/ml in control and 90 ng/ml after 5 min of ischemia). To reflect this pathophysiological phenomenon, we compared the same volume of coronary effluents from control and ischemic hearts in the 1D gel-LC/MS-MS experiment and then ranked the proteins according to their biggest increase in spectral counts. To identify ischemia-specific posttranslational modifications, however, we compared the same quantity of proteins in the DIGE experiment. Although fewer proteins were detected in the 2D gel-based approach compared with the 1D gel-LC/MS/MS approach, it is noteworthy that the known biomarkers of cardiac injury were identified in both experimental approaches.
Among the numerous proteins identified in ischemic coronary effluent, there were only rare examples that had cardiac specific tissue expression, an essential criterion for biomarker specificity, and were therefore studied in more detailed. The identification of the cardiac isoform of myosin-binding protein C in ischemic coronary effluents fulfilled some of the criteria of good biomarker candidate: namely a cardiac specific protein with a pronounced change in response to ischemic injury. Notably, the presence of cMyBP-C and a 40-kDa degradation product was only found in the plasma of mice with AMI. The later fragment corresponds to the N-terminal part of cMyBP-C and was described previously in mouse hearts subjected to ischemia/reperfusion (30) and in dog hearts subjected to low flow ischemia (31). However, its release into the circulation after myocardial injury has not been reported previously, and it may provide physiological circumstance-specific information. An analysis of the relative time courses of release and clearance of MyBP-C and troponin is being undertaken in an in vivo murine model of AMI as part of our ongoing investigation as to whether MyBP-C is a valid biomarker.
This information together with the quantification of MyBP-C by a proteomics approach (38) in timed collections of patients' plasma should provide an indication of the potential of MyBP-C as a novel biomarker of AMI and is mandatory to the clinical translation of our findings. Such information may reveal that cMyBP-C is released more quickly and has a shorter circulating half-life than troponin. If this is the case, it would have the advantage of allowing more rapid ruling in and ruling out of AMI in patients with chest pain. A further advantage is that it could allow the diagnosis and the quantification of reinfarction in patients in whom AMI is confirmed. This would be of particular advantage in trials of interventional devices that are thought to diminish periprocedural AMI (39).
In summary, the present study aimed to identify potential new biomarkers of AMI. We performed a comprehensive identification of proteins released in coronary effluent during myocardial ischemia that revealed the cardiac isoform of myosin-binding protein C as a candidate biomarker. Further studies in patients will be necessary to corroborate these findings and assess the clinical advantage, if any, of cMyBP-C over existing biomarkers of AMI.
Supplementary Material
Acknowledgments
We thank Prof. M. Gautel (Randall Institute, King's College London, London, UK) for the kind gift of the cMyBP-C antibody.
Footnotes
* This work was supported by project Grant R060701 from the Guys and St. Thomas' Charity and by the Department of Health via the National Institute for Health Research comprehensive Biomedical Research Centre award to Guy's and St. Thomas' National Health Service (NHS) Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
1 The abbreviations used are:
- AMI
- acute myocardial infarction
- CK
- creatine kinase
- FABP
- fatty acid-binding protein
- H-FABP
- heart-specific FABP
- TTC
- triphenyl tetrazolium chloride
- 1D
- one-dimensional
- 2D
- two-dimensional
- LTQ
- linear trap quadrupole
- LDH
- lactate dehydrogenase
- Prdx
- peroxiredoxin
- MyBP-C
- myosin-binding protein C
- cMyBP-C
- cardiac myosin-binding protein C.
REFERENCES
- 1.Rajappa M., Sharma A. ( 2005) Biomarkers of cardiac injury: an update. Angiology 56, 677– 691 [DOI] [PubMed] [Google Scholar]
- 2.Apple F. S., Wu A. H., Mair J., Ravkilde J., Panteghini M., Tate J., Pagani F., Christenson R. H., Mockel M., Danne O., Jaffe A. S. ( 2005) Future biomarkers for detection of ischemia and risk stratification in acute coronary syndrome. Clin. Chem 51, 810– 824 [DOI] [PubMed] [Google Scholar]
- 3.Saenger A. K., Jaffe A. S. ( 2008) Requiem for a heavyweight: the demise of creatine kinase-MB. Circulation 118, 2200– 2206 [DOI] [PubMed] [Google Scholar]
- 4.Pelsers M. M., Hermens W. T., Glatz J. F. ( 2005) Fatty acid-binding proteins as plasma markers of tissue injury. Clin. Chim. Acta 352, 15– 35 [DOI] [PubMed] [Google Scholar]
- 5.Wunderlich M. T., Hanhoff T., Goertler M., Spener F., Glatz J. F., Wallesch C. W., Pelsers M. M. ( 2005) Release of brain-type and heart-type fatty acid-binding proteins in serum after acute ischaemic stroke. J. Neurol 252, 718– 724 [DOI] [PubMed] [Google Scholar]
- 6.Parmacek M. S., Solaro R. J. ( 2004) Biology of the troponin complex in cardiac myocytes. Prog. Cardiovasc. Dis 47, 159– 176 [DOI] [PubMed] [Google Scholar]
- 7.French J. K., White H. D. ( 2004) Clinical implications of the new definition of myocardial infarction. Heart 90, 99– 106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antman E. M., Tanasijevic M. J., Thompson B., Schactman M., McCabe C. H., Cannon C. P., Fischer G. A., Fung A. Y., Thompson C., Wybenga D., Braunwald E. ( 1996) Cardiac-specific troponin I levels to predict the risk of mortality in patients with acute coronary syndromes. N. Engl. J. Med 335, 1342– 1349 [DOI] [PubMed] [Google Scholar]
- 9.Mayr M., Zhang J., Greene A. S., Gutterman D., Perloff J., Ping P. ( 2006) Proteomics-based development of biomarkers in cardiovascular disease: mechanistic, clinical, and therapeutic insights. Mol. Cell. Proteomics 5, 1853– 1864 [DOI] [PubMed] [Google Scholar]
- 10.Jacquet S., Nishino Y., Kumphune S., Sicard P., Clark J. E., Kobayashi K. S., Flavell R. A., Eickhoff J., Cotten M., Marber M. S. ( 2008) The role of RIP2 in p38 MAPK activation in the stressed heart. J. Biol. Chem 283, 11964– 11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löwbeer C., Kawakami T., Tähepĵld P., Gustafsson S. A., Vaage J., Valen G. ( 2002) Importance of preanalytical handling of samples for measurement of cardiac troponin T in coronary effluent from isolated rat hearts. Scand J. Clin. Lab. Invest 62, 255– 262 [DOI] [PubMed] [Google Scholar]
- 12.Clark J. E., Kottam A., Motterlini R., Marber M. S. ( 2009) Measuring left ventricular function in the normal, infarcted and CORM-3-preconditioned mouse heart using complex admittance-derived pressure volume loops. J. Pharmacol. Toxicol. Methods 59, 94– 99 [DOI] [PubMed] [Google Scholar]
- 13.Eckle T., Grenz A., Köhler D., Redel A., Falk M., Rolauffs B., Osswald H., Kehl F., Eltzschig H. K. ( 2006) Systematic evaluation of a novel model for cardiac ischemic preconditioning in mice. Am. J. Physiol. Heart Circ. Physiol 291, H2533– H2540 [DOI] [PubMed] [Google Scholar]
- 14.Gautel M., Fürst D. O., Cocco A., Schiaffino S. ( 1998) Isoform transitions of the myosin binding protein C family in developing human and mouse muscles: lack of isoform transcomplementation in cardiac muscle. Circ. Res 82, 124– 129 [DOI] [PubMed] [Google Scholar]
- 15.Yan J. X., Wait R., Berkelman T., Harry R. A., Westbrook J. A., Wheeler C. H., Dunn M. J. ( 2000) A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21, 3666– 3672 [DOI] [PubMed] [Google Scholar]
- 16.Shevchenko A., Wilm M., Vorm O., Mann M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850– 858 [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646– 4658 [DOI] [PubMed] [Google Scholar]
- 18.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383– 5392 [DOI] [PubMed] [Google Scholar]
- 19.Jacquet S., Zarrinpashneh E., Chavey A., Ginion A., Leclerc I., Viollet B., Rutter G. A., Bertrand L., Marber M. S. ( 2007) The relationship between p38 mitogen-activated protein kinase and AMP-activated protein kinase during myocardial ischemia. Cardiovasc. Res 76, 465– 472 [DOI] [PubMed] [Google Scholar]
- 20.Marber M. S., Mestril R., Chi S. H., Sayen M. R., Yellon D. M., Dillmann W. H. ( 1995) Overexpression of the rat inducible 70-kD heat stress protein in a transgenic mouse increases the resistance of the heart to ischemic injury. J. Clin. Investig 95, 1446– 1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Eyk J. E., Powers F., Law W., Larue C., Hodges R. S., Solaro R. J. ( 1998) Breakdown and release of myofilament proteins during ischemia and ischemia/reperfusion in rat hearts: identification of degradation products and effects on the pCa-force relation. Circ. Res 82, 261– 271 [DOI] [PubMed] [Google Scholar]
- 22.Zweier J. L., Talukder M. A. ( 2006) The role of oxidants and free radicals in reperfusion injury. Cardiovasc. Res 70, 181– 190 [DOI] [PubMed] [Google Scholar]
- 23.Schröder E., Brennan J. P., Eaton P. ( 2008) Cardiac peroxiredoxins undergo complex modifications during cardiac oxidant stress. Am. J. Physiol. Heart Circ. Physiol 295, H425– H433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araki M., Nanri H., Ejima K., Murasato Y., Fujiwara T., Nakashima Y., Ikeda M. ( 1999) Antioxidant function of the mitochondrial protein SP-22 in the cardiovascular system. J. Biol. Chem 274, 2271– 2278 [DOI] [PubMed] [Google Scholar]
- 25.Matsushima S., Ide T., Yamato M., Matsusaka H., Hattori F., Ikeuchi M., Kubota T., Sunagawa K., Hasegawa Y., Kurihara T., Oikawa S., Kinugawa S., Tsutsui H. ( 2006) Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation 113, 1779– 1786 [DOI] [PubMed] [Google Scholar]
- 26.Nagy N., Malik G., Fisher A. B., Das D. K. ( 2006) Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol 291, H2636– H2640 [DOI] [PubMed] [Google Scholar]
- 27.Mayr M., Chung Y. L., Mayr U., McGregor E., Troy H., Baier G., Leitges M., Dunn M. J., Griffiths J. R., Xu Q. ( 2004) Loss of PKC-delta alters cardiac metabolism. Am. J. Physiol. Heart Circ Physiol 287, H937– H945 [DOI] [PubMed] [Google Scholar]
- 28.Mayr M., Chung Y. L., Mayr U., Yin X., Ly L., Troy H., Fredericks S., Hu Y., Griffiths J. R., Xu Q. ( 2005) Proteomic and metabolomic analyses of atherosclerotic vessels from apolipoprotein E-deficient mice reveal alterations in inflammation, oxidative stress, and energy metabolism. Arterioscler. Thromb. Vasc. Biol 25, 2135– 2142 [DOI] [PubMed] [Google Scholar]
- 29.Flashman E., Redwood C., Moolman-Smook J., Watkins H. ( 2004) Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res 94, 1279– 1289 [DOI] [PubMed] [Google Scholar]
- 30.Sadayappan S., Osinska H., Klevitsky R., Lorenz J. N., Sargent M., Molkentin J. D., Seidman C. E., Seidman J. G., Robbins J. ( 2006) Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc. Natl. Acad. Sci. U.S.A 103, 16918– 16923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan C., Guo Y., Ravi R., Przyklenk K., Shilkofski N., Diez R., Cole R. N., Murphy A. M. ( 2006) Myosin binding protein C is differentially phosphorylated upon myocardial stunning in canine and rat hearts—evidence for novel phosphorylation sites. Proteomics 6, 4176– 4186 [DOI] [PubMed] [Google Scholar]
- 32.Diamandis E. P. ( 2004) Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol. Cell. Proteomics 3, 367– 378 [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Pinna R., Martin-Ventura J. L., Mas S., Blanco-Colio L. M., Tuñon J., Egido J. ( 2008) Proteomics in atherosclerosis. Curr. Atheroscler. Rep 10, 209– 215 [DOI] [PubMed] [Google Scholar]
- 34.Edwards A. V., White M. Y., Cordwell S. J. ( 2008) The role of proteomics in clinical cardiovascular biomarker discovery. Mol. Cell. Proteomics 7, 1824– 1837 [DOI] [PubMed] [Google Scholar]
- 35.Mateos-Cáceres P. J., García-Méndez A., López Farré A., Macaya C., Núñez A., Gómez J., Alonso-Orgaz S., Carrasco C., Burgos M. E., de Andrés R., Granizo J. J., Farré J., Rico L. A. ( 2004) Proteomic analysis of plasma from patients during an acute coronary syndrome. J. Am. Coll. Cardiol 44, 1578– 1583 [DOI] [PubMed] [Google Scholar]
- 36.Koomen J. M., Wilson C. R., Guthrie P., Androlewicz M. J., Kobayashi R., Taegtmeyer H. ( 2006) Proteome analysis of isolated perfused organ effluent as a novel model for protein biomarker discovery. J. Proteome Res 5, 177– 182 [DOI] [PubMed] [Google Scholar]
- 37.Rhee S. G., Yang K. S., Kang S. W., Woo H. A., Chang T. S. ( 2005) Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid. Redox Signal 7, 619– 626 [DOI] [PubMed] [Google Scholar]
- 38.Kuhn E., Addona T., Keshishian H., Burgess M., Mani D. R., Lee R. T., Sabatine M. S., Gerszten R. E., Carr S. A. ( 2009) Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin. Chem 55, 1108– 1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan M., Rihal C., Holmes D. R., Prasad A. ( 2009) Adjunctive thrombectomy and distal protection in primary percutaneous coronary intervention: impact on microvascular perfusion and outcomes. Circulation 119, 1311– 1319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.