Abstract
The Arabidopsis FRO2 gene encodes the low-iron-inducible ferric chelate reductase responsible for reduction of iron at the root surface. Here, we report that FRO2 and IRT1, the major transporter responsible for high-affinity iron uptake from the soil, are coordinately regulated at both the transcriptional and posttranscriptional levels. FRO2 and IRT1 are induced together following the imposition of iron starvation and are coordinately repressed following iron resupply. Steady-state mRNA levels of FRO2 and IRT1 are also coordinately regulated by zinc and cadmium. Like IRT1, FRO2 mRNA is detected in the epidermal cells of roots, consistent with its proposed role in iron uptake from the soil. FRO2 mRNA is detected at high levels in the roots and shoots of 35S-FRO2 transgenic plants. However, ferric chelate reductase activity is only elevated in the 35S-FRO2 plants under conditions of iron deficiency, indicating that FRO2 is subject to posttranscriptional regulation, as shown previously for IRT1. Finally, the 35S-FRO2 plants grow better on low iron as compared with wild-type plants, supporting the idea that reduction of ferric iron to ferrous iron is the rate-limiting step in iron uptake.
Iron is an essential element for plants. It functions to accept and donate electrons and thus serves as an important cofactor for a number of metalloenzymes involved in respiration and photosynthesis. Although iron is abundant in the earth's crust, it is often unavailable to plants because it tends to form insoluble ferric hydroxide complexes in aerobic environments at neutral or basic pH (Guerinot and Yi, 1994). In addition to the solubility problem, the chemical properties of iron require cells to place limitations on its accumulation. Fe(II) and Fe(III) act catalytically to generate hydroxyl radicals that can damage cellular constituents such as DNA and lipids (Halliwell and Gutteridge, 1992). Thus, iron uptake (Eide et al., 1996; Robinson et al., 1999; Connolly et al., 2002) and storage (Lescure et al., 1991; Briat and Lobréaux, 1997; Wei and Theil, 2000) are carefully regulated processes.
Dicots and non-grass monocots employ the strategy I response to mobilize iron from the soil (Marschner and Römheld, 1994). This response includes release of protons and subsequent acidification of the rhizosphere, which serves to drive more Fe(III) into solution, reduction of Fe(III) to Fe(II) at the root surface, and transport of Fe(II) across the root epidermal cell membrane. We have previously identified the genes in Arabidopsis that encode the iron-regulated ferric chelate reductase (FRO2) and the ferrous iron transporter (IRT1) that function to take up iron from the soil as part of the strategy I response (Eide et al., 1996; Yi and Guerinot, 1996; Robinson et al., 1999; Vert et al., 2002). FRO2 is predicted to encode a polypeptide of 725 amino acids, with conserved FAD- and NADPH-binding sites. FRO2 has six hydrophobic domains within its amino-terminal region along with two additional carboxyl-terminal hydrophobic domains, all of which are predicted to form trans-membrane helices. This suggests that an intracellular region of FRO2, which contains the deduced cofactor-binding sites, is anchored at both ends by membrane-spanning regions. Two pairs of His residues that are thought to coordinate heme in FRO2 lie on two similarly oriented trans-membrane helices and are in equivalent locations to His residues in superfamily members FRE1 and gp91phox (Robinson et al., 1999). The FRO2 gene belongs to an eight-member gene family in Arabidopsis; the other seven FRO genes have not yet been functionally characterized.
Here, we report the results of studies aimed at further characterizing expression of FRO2. Previously, we showed that IRT1 expression is induced when plants are transferred to iron-deficient medium and is repressed upon iron resupply (Connolly et al., 2002). In this report, we show coordinate control of the expression of FRO2 and IRT1. Because previous work suggested that reduction of ferric iron to ferrous iron by ferric chelate reductase is the rate-limiting step in iron uptake (Grusak et al., 1990), we hypothesized that overexpression of FRO2 in transgenic plants might lead to enhanced iron accumulation. 35S-FRO2 transgenic plants do not accumulate iron when compared with wild type; analysis of these plants revealed that FRO2 is subject to posttranscriptional regulation by iron, as previously shown for IRT1 (Connolly et al., 2002). However, 35S-FRO2 plants do display tolerance to growth on low-iron medium. This result suggests that expression of FRO2 in crop plants may yield lines with improved iron efficiency.
RESULTS
Coordinate Regulation of FRO2 and IRT1 by Iron and Zinc
FRO2 mRNA is detected in the roots of Arabidopsis plants after 3 d of growth on iron-deficient medium (Robinson et al., 1999). Previously, we examined the induction of IRT1 expression over time and showed that IRT1 mRNA is detectable in roots 24 h after plants are transferred to iron-deficient media (Connolly et al., 2002). IRT1 mRNA levels peak 3 d after plants are transferred to iron-deficient media. Because strategy I plants require reduction of iron before transport, we reasoned that expression of FRO2 and IRT1 would either be coordinately controlled or that the induction of FRO2 expression would precede the induction of IRT1 expression. We performed RNA gel-blot analysis using the FRO2 cDNA as a hybridization probe; this analysis revealed that the induction of FRO2 expression exactly parallels that of IRT1 (Fig. 1). Moreover, after the transfer of plants from iron-deficient medium to iron-sufficient medium, both FRO2 and IRT1 mRNA levels decline rapidly (Fig. 2). The two transcripts are undetectable 24 h after iron resupply.
Figure 1.
Time course of FRO2 and IRT1 mRNA abundance patterns in roots exposed to iron deficiency. Wild-type plants were grown for 14 d on standard medium and transferred to iron-deficient medium. Roots were harvested 0, 12, 24, 36, 48, 72, and 144 h after the transfer to iron-deficient medium. RNA was isolated from the samples and used to prepare RNA gel blots. The blots were hybridized with radiolabeled FRO2 and IRT1 cDNA probes. Ethidium bromide-stained rRNA is shown as a control for loading.
Figure 2.
Time course of FRO2 and IRT1 mRNA abundance patterns in roots after iron resupply. Wild-type plants were grown for 14 d on standard medium, transferred to iron-deficient medium for 3 d, and then transferred a second time to iron-sufficient medium. Roots were harvested 0, 12, 24, 36, 48, 72, and 144 h after the second transfer. RNA was isolated from the samples and used to prepare RNA gel blots. The blots were hybridized with radiolabeled FRO2 and IRT1 cDNA probes. Ethidium bromide-stained rRNA is shown as a control for loading.
In addition, we previously showed that IRT1 transcript abundance is affected by zinc (Connolly et al., 2002). It is logical that IRT1 expression is repressed when high amounts of zinc are present in the medium, because accumulation of zinc is toxic and IRT1 mediates zinc transport (Eide et al., 1996; Korshunova et al., 1999; Vert et al., 2002). FRO2 is not known to play a role in zinc uptake or homeostasis. However, because FRO2 and IRT1 are coordinately regulated by iron, we wondered whether the coordinate control would extend to other metals. Both FRO2 and IRT1 were detected in the roots of plants grown for 3 d on either iron-deficient medium or iron-deficient medium supplemented with 100 μm zinc (Fig. 3). However, neither transcript was detectable in roots when plants were grown on iron-deficient medium supplemented with 500 μm zinc, confirming that zinc mediates coordinate control of FRO2 and IRT1 transcript abundance. Although 500 μm zinc does cause zinc toxicity, previous work with IRT1 showed that the decrease in IRT1 mRNA transcript abundance after zinc exposure is dependent upon the IRT1 promoter (Connolly et al., 2002). Similar experiments with cadmium confirm that high levels of cadmium in the growth medium also mediate coordinate repression of FRO2 and IRT1 expression (data not shown). Together, these results show that the expression of two genes that are required for iron uptake from the soil are coordinately regulated by metals.
Figure 3.
Coordinate control of FRO2 and IRT1 expression by zinc. Wild-type plants were grown for 14 d on standard medium. Plants were subsequently transferred and grown for 3 d on iron-sufficient medium (lane 1), iron-deficient medium (lane 2), iron-sufficient medium supplemented with 500 μm zinc (lane 3), iron-deficient medium supplemented with 100 μm zinc (lane 4), or iron-deficient medium supplemented with 500 μm zinc (lane 5). RNA was isolated from roots and used to prepare RNA gel blots. The blots were hybridized with radiolabeled FRO2 and IRT1 cDNA probes. Ethidium bromide-stained rRNA is shown as a control for loading.
FRO2 Is Expressed in the Root Epidermis and Flowers
Next, we investigated the localization of FRO2 mRNA in the roots of iron-deficient plants. Previous studies suggested that FRO2 functions in iron uptake from the rhizosphere (Robinson et al., 1999; Yi and Guerinot, 1996). IRT1 is known to be expressed in epidermal root cells of iron-deficient plants (Vert et al., 2002). For these reasons, we hypothesized that FRO2 expression would be observed in the root epidermis. In situ hybridization analysis with a FRO2 antisense probe on longitudinal sections of iron-deficient roots showed that FRO2 mRNA is present in the epidermal layer (Fig. 4A). Hybridization of a sense probe to similar sections yielded no detectable signal (Fig. 4B). These results are consistent with the proposed role of FRO2 in iron uptake from the soil and further support coordinate regulation of IRT1 and FRO2.
Figure 4.
Localization of the FRO2 transcript in longitudinal sections of iron-deficient roots. A, In situ hybridization experiments were performed on longitudinal sections (10 μm) of iron-deficient roots using a FRO2 antisense probe. B, In situ hybridization experiments were performed on longitudinal sections (10 μm) of iron-deficient roots using a FRO2 sense probe.
We constructed transgenic plants in which the FRO2 promoter drives expression of the uidA gene, which encodes β-glucuronidase (GUS). Histochemical staining of 10 independent transgenic lines demonstrated that the FRO2 promoter is much more active in iron-deficient roots than in iron-sufficient roots and is highest in lateral roots of iron-deficient plants (data not shown). GUS activity was observed in the outer tissues of iron-deficient roots including root hairs and epidermal cells (data not shown); this data confirms the results obtained with in situ hybridization.
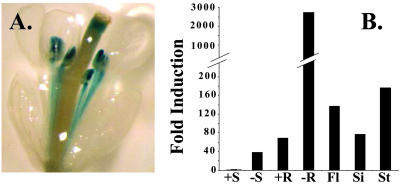
The FRO2 promoter is active in flowers (Fig. 5A). Histochemical staining of FRO2-GUS flowers revealed GUS activity in the anther filaments (Fig. 5A). The IRT1 promoter also is active in filaments (Vert et al., 2002). In addition, we observed FRO2 promoter activity in the anthers and the style (Fig. 5A). To confirm FRO2 expression in flowers, we examined FRO2 transcript abundance in various tissues using real time reverse transcriptase (RT)-PCR. FRO2 transcript abundance is greatest in iron-deficient roots, but FRO2 mRNA also is detected in other tissues tested (Fig. 5B). Our results suggest a role for FRO2 in the delivery of iron to developing pollen grains.
Figure 5.
FRO2 is expressed in flowers. A, Histochemical staining of flowers from FRO2-GUS transgenic plants grown in soil. A representative individual is shown. No significant GUS activity was detected in wild-type plants (data not shown). B, Real time RT-PCR analysis of FRO2 expression in various tissues. Plants were grown for 14 d on standard medium and then transferred to either iron-deficient or -sufficient medium as indicated or were grown in soil (flowers, siliques, and stems). FRO2 values were normalized and expressed as -fold induction relative to +Fe shoots. Due to the ratio calculation, sds are not shown in this figure.
Characterization of 35S-FRO2 Plants
Previous studies suggested that reduction of ferric iron to ferrous iron, as opposed to transport of ferrous iron, is the rate-limiting step for iron uptake (Grusak et al., 1990). Thus, we reasoned that overexpression of FRO2 in transgenic plants would lead to enhanced iron uptake and accumulation. We used the cauliflower mosaic virus 35S promoter to drive expression of FRO2 in Arabidopsis and identified six independent, homozygous transgenic lines. We examined FRO2 steady-state RNA levels in all six lines; the results for three lines are shown in Figure 6. RNA was isolated from the roots and shoots of T5 seedlings grown for 3 d on either iron-sufficient or -deficient medium. In wild-type plants, FRO2 mRNA was detected only in the roots of iron-deficient plants, as has been shown previously (Fig. 6). In contrast, all three transgenic lines showed altered accumulation of FRO2 mRNA. Lines 11E and 15G display high levels of FRO2 mRNA in roots and shoots of iron-deficient and -sufficient plants. Line 5A accumulates lower, but detectable, levels of FRO2 mRNA in iron-sufficient and -deficient shoots as well as in iron-sufficient roots. Quantification of FRO2 mRNA levels in iron-deficient roots showed that lines 5A, 11E, and 15G have 2.1, 1.9, and 2.3 times more FRO2 mRNA, respectively, than wild type. Three additional 35S-FRO2 transgenic lines examined contained detectable levels of FRO2 mRNA only in iron-deficient roots and thus resembled wild-type plants. None of the lines displayed a visible phenotype when grown under normal growth conditions (data not shown).
Figure 6.
Northern-blot analysis of wild-type and transgenic 35S-FRO2 plants. RNA was prepared from the roots and shoots of 17-d-old plants grown for 3 d on either iron-sufficient or -deficient medium. The radiolabeled FRO2 cDNA was used as a probe. Ethidium bromide-stained rRNA is shown as a control for loading.
It is interesting to note that FRO2 mRNA levels are higher in iron-deficient roots than in iron-sufficient roots of 35S-FRO2 transgenic plants. Lines 5A, 11E, and 15G have 13, 1.3, and 2.7 times more FRO2 mRNA in iron-deficient roots than iron-sufficient roots, respectively. This result indicates that the FRO2 transcript may be more stable when plants are iron deficient. However, at least for lines 11E and 15G, FRO2 mRNA levels in iron-deficient roots can be explained by expression of FRO2 from the endogenous gene and the transgene resulting in elevated levels in iron-deficient roots as compared with iron-sufficient roots (compare wild-type –Fe root with 15G –Fe root). Because line 5A is the only line obtained thus far that contains disproportionately high levels of FRO2 mRNA in iron-deficient roots, whether the FRO2 transcript is stabilized under conditions of iron deficiency is unclear and warrants further investigation.
Next, we examined ferric chelate reductase activity in the transgenic plants as compared with wild type. Plants were grown for 2 weeks on standard medium and then transferred to iron-deficient or -sufficient medium for 3 d. Ferric chelate reductase assays showed that the transgenic plants have elevated ferric chelate reductase activity only when plants are grown on iron-deficient medium (Fig. 7). Lines 5A, 11E, and 15G displayed 1.9, 1.7, and 1.7 times higher activity than wild type, respectively. These values correspond well with the mRNA levels (Fig. 6). These results indicate that expression of FRO2 is controlled at the posttranscriptional level. Previously, we showed that IRT1 is subject to posttranscriptional regulation; IRT1 protein only accumulates in the roots of iron-deficient plants even when IRT1 expression is driven from the cauliflower mosaic virus 35S promoter. We have been unable to generate antibodies that recognize FRO2 protein, despite numerous attempts, and thus are unable to determine whether FRO2 protein levels correlate with activity levels. Regardless, it is clear that FRO2 and IRT1 are coordinately regulated at the posttranscriptional level, as they are at the level of transcript accumulation.
Figure 7.
Assays of ferric chelate reductase activity in wild-type and 35S-FRO2 transgenic lines. Plants were grown on standard medium for 14 d and then transferred to either iron-deficient (open bars) or -sufficient (filled bars) medium for 3 d before the assay. Values are the mean of five assays. The means of wild-type and 35S-FRO2 (lines 5A and 15G) plants grown on iron-deficient medium are statistically different at P < 0.05.
FRO2 is known to function in the reduction of copper; frd1-1 mutants lack low-iron-inducible copper chelate reductase activity. However, frd1-1 mutants do not show alterations in copper accumulation, making the role of FRO2 in copper mobilization and uptake unclear (Yi and Guerinot, 1996). We measured copper chelate reductase activity in the 35S-FRO2 transgenic plants and wild-type plants. Copper chelate reductase activity is elevated in the roots of iron-deficient 35S-FRO2 plants but not in roots of iron-sufficient 35S-FRO2 plants (Fig. 8). Lines 5A, 11E, and 15G show levels that are 2.6, 1.3, and 2.5 times that of wild-type iron-deficient roots, respectively. Thus, this result also is consistent with posttranscriptional regulation of FRO2. To determine whether copper chelate reduction by FRO2 is physiologically relevant, we measured root growth of wild-type and 35S-FRO2 plants grown vertically on plates that were iron-deficient and supplemented with varying concentrations of copper. We reasoned that, if reduction of copper by FRO2 is physiologically important, then the 35S-FRO2 lines might show sensitivity to growth on high-copper medium, but only when plants were grown on iron-deficient medium, because this is the only growth condition that permits higher copper chelate reductase activity. We were unable to detect any differences in root growth between wild-type and 35S-FRO2 plants over a range of copper concentrations (iron-deficient minimal medium supplemented with 0, 1, 10, 50, 75, 100, 150, and 200 μm CuSO4; data not shown). Root growth of all plants was inhibited by copper concentrations greater than 100 μm. This result suggests either that reduction of copper by FRO2 is not physiologically relevant or that copper, like iron, causes posttranscriptional regulation of FRO2 when present at high concentrations.
Figure 8.
Assays of copper chelate reductase activity in wild-type and 35S-FRO2 transgenic plants. Plants were grown on standard medium for 14 d and then transferred to either iron-deficient (open bars) or -sufficient (filled bars) medium for 4 d before the assay. Values are the mean of five assays. The means of wild-type and 35S-FRO2 (lines 5A and 15G) plants grown on iron-deficient medium are statistically different at P < 0.05.
We also examined metal levels in the 35S-FRO2 transgenic plants. Elemental analysis was performed on 13 individual plants each of wild type and 35S-FRO2 lines 5A, 11E, and 15G grown in soil. No significant trends were observed when the three transgenic lines were compared with wild type (data not shown). Elemental analysis also was performed on roots and shoots of wild type and 35S-FRO2 transgenic lines 5A, 11E, and 15G grown on iron-deficient and -sufficient medium. We reasoned that because FRO2 activity is elevated only in the roots of iron-deficient plants, we might detect differences in metal ion content only when plants were grown on iron-deficient medium. We did not detect any differences in metal accumulation between wild type and the three 35S-FRO2 transgenic lines grown on iron-deficient medium (data not shown). It is not surprising that we were unable to detect elevated iron levels because iron is not available for reduction and uptake when the plants are grown on iron-deficient media. The elemental analysis data is consistent with posttranscriptional regulation of FRO2; elevated FRO2 mRNA levels result in elevated FRO2 activity only when plants are starved for iron. Thus, posttranscriptional regulation of FRO2 may serve to protect against the accumulation of potentially toxic amounts of iron.
We germinated and grew wild type and the transgenic lines on media that contained varying amounts of iron. First, we grew plants on minimal media that was not supplemented with iron, but that also did not contain an iron chelator. Thus, the small amount of iron that remains following water purification or that contaminates the agar may be available to seedlings growing in the media. Each of the three transgenic lines displayed an enhanced ability to tolerate growth on low-iron media with line 15G showing the greatest tolerance (Fig. 9). The enhanced growth of 35S-FRO2 plants on low-iron medium is pH dependent, consistent with the pH optima of ferric chelate reductase (data not shown; Susin et al., 1996). Presumably, the 35S-FRO2 plants show enhanced growth on low-iron medium due to enhanced iron reduction leading to higher rates of uptake. This result, therefore, supports previous work that showed that iron reduction is the rate-limiting step for iron uptake (Grusak et al., 1990).
Figure 9.
35S-FRO2 plants (line 15G) tolerate growth on low-iron medium. Seedlings were germinated on B5 medium and then were transferred and grown for 2 weeks on low-iron medium before being photographed.
We also grew wild type and the transgenic lines on high-iron media. Germination and growth of plants on minimal media supplemented with 100, 250, or 500 μm Fe(III)-EDTA did not reveal any difference in tolerance between wild type and the transgenic plants. Both wild-type and 35S-FRO2 plants displayed iron toxicity symptoms when grown in the presence of 250 or 500 μm Fe(III)-EDTA (data not shown). Moreover, we observed no difference in growth between the 35S-FRO2 transgenic plants and wild type when plants were grown on standard medium, which contains 50 μm Fe(III)-EDTA. These results are consistent with posttranscriptional regulation of FRO2.
DISCUSSION
The data presented here provide evidence for coordinate regulation of two of the major components of the strategy I response used by dicots and non-grass monocots, FRO2 and IRT1. Our results suggest that expression of FRO2 and IRT1 is coordinately regulated at the level of transcript accumulation by iron, zinc, and cadmium. It makes sense that induction of FRO2 and IRT1 would be coordinately controlled, because FRO2 activity is required to provide the ferrous iron that is a substrate for IRT1. This regulatory mechanism may be acting either at transcription initiation or at the transcript stability level; at this time, we are unable to eliminate either one of these possibilities.
Coordinate control of transcription initiation likely would be mediated through common cis-acting elements and trans-acting proteins. Recently, the tomato (Lycopersicon esculentum) FER gene was isolated; FER encodes a putative transcription factor that controls iron uptake responses in roots (Ling et al., 2002). Both root ferric chelate reductase activity and LeIRT1 expression are altered in the fer mutant. A FER ortholog is present in the Arabidopsis genome and may be responsible for coordinate regulation of FRO2 and IRT1.
IRT1 mediates uptake of both zinc and cadmium (Eide et al., 1996; Korshunova et al., 1999; Connolly et al., 2002; Vert et al., 2002), and both metals can cause toxicity when they accumulate to high levels. Therefore, repression of IRT1 expression when plants are grown in the presence of high amounts of zinc and cadmium would avoid accumulation of either metal to toxic levels (Connolly et al., 2002). However, it is surprising that FRO2 expression is controlled by zinc and cadmium because there is no evidence that FRO2 functions in the uptake of either of these metals. One possibility is that FRO2 and IRT1 form a complex that is only stable when both proteins are present in the membrane. Thus, when a cell represses IRT1 expression, it would make sense that FRO2 expression would be repressed as well. In frd1-1, the FRO2 gene encodes a protein with a stop codon at position 21; western analysis of these plants showed that IRT1 protein is detected at normal levels (data not shown). In addition, inducible root ferric chelate reductase activity is elevated in an IRT1 KO line (Vert et al., 2002). These results suggest that the accumulation of neither IRT1 nor FRO2 protein is dependent upon the presence of the other protein.
In situ hybridization with a FRO2 antisense probe showed that FRO2 mRNA is detected at high levels in the epidermal cells of the root. This pattern is consistent with the proposed role of FRO2 in iron reduction at the root surface (Yi and Guerinot, 1996; Robinson et al., 1999) and again is consistent with the idea that FRO2 and IRT1 are coordinately regulated. IRT1 also is expressed in root epidermal cells (Vert et al., 2002). In tomato, FER mRNA is detected in root epidermal cells and in the outer cortical cell layers of roots (Ling et al., 2002). However, FER mRNA also is detected in the vascular cylinder of the mature roothair zone of the root. A gene encoding ferric chelate reductase has been cloned from pea (Pisum sativum), and in situ hybridization experiments have shown that it is expressed throughout the root, including the epidermis, cortex, and vascular cylinder (Waters et al., 2002). Pea FRO1 also is expressed in nodules as well as leaf mesophyll and parenchyma cells. In situ hybridization failed to show that FRO2 and IRT1 mRNA are present in the vascular cylinder of Arabidopsis roots (this study; Vert et al., 2002). However, staining of FRO2-GUS plants did reveal FRO2 promoter activity in the central cylinder (data not shown). Our real time RT-PCR results show that FRO2 is expressed at relatively low levels in tissues other than iron-deficient roots and support the results obtained with FRO2-GUS plants. However, because histochemical staining of promoter-GUS plants is unreliable and because we could not confirm FRO2 expression in the central cylinder using in situ hybridization, a role for FRO2 in the vasculature remains unsubstantiated.
The Arabidopsis genome contains eight putative FRO genes; it is possible that other FROs function in the vascular cylinder and/or in the aerial portions of the plant. It is thought that iron is transported in the xylem as a Fe(III)-citrate complex (Tiffin, 1966) and that ferric chelate reductase is required in the aerial portions of the plant to reduce ferric iron before transport into the leaf cells. Ferric chelate reductase activity has been detected in leaves of sunflower (de la Guardia and Alcantara, 1996) and Vigna unguiculata (Brüggemann et al., 1993). Ferric chelate reductase activity also appears to be developmentally regulated, with ferric chelate reductase activity increasing during seed fill to supply the developing seeds (Grusak, 1995). The Arabidopsis FRO3 gene is expressed in both root and leaf cells in response to low iron (Robinson et al., 1997).
Our data suggest that, like IRT1, expression of FRO2 is controlled posttranscriptionally; IRT1 protein accumulates only in the roots of iron-deficient 35S-IRT1 lines despite the accumulation of high levels of IRT1 mRNA in all tissues (Connolly et al., 2002). Here, we show that FRO2 mRNA accumulates in roots and shoots of iron-sufficient and iron-deficient 35S-FRO2 transgenic plants, yet root ferric chelate reductase activity is elevated only when plants are starved for iron. In addition, 35S-FRO2 plants do not accumulate more metals than wild-type plants.
There certainly are precedents for metal control of gene expression. Zinc uptake in yeast is tightly regulated in response to changing zinc levels (Guerinot and Eide, 1999). Expression of ZRT1, the high-affinity zinc transporter in yeast, is controlled both at the level of transcription, via the ZAP1 transcription factor (Zhao et al., 1998; Zhao and Eide, 1997) and posttranslationally, via zinc-mediated ubiquitination and protein turnover (Gitan et al., 1998; Gitan and Eide, 2000). This system allows the rapid turnover of ZRT1 when cells come in contact with high zinc, thus preventing the accumulation of toxic amounts of this metal. Arabidopsis IRT1 and yeast ZRT1 belong to the ZIP family of metal transporters and thus share a high degree of sequence similarity; in addition, the two proteins share the same predicted membrane topology. For these reasons, it seems possible that IRT1 is regulated posttranslationally via ubiquitination and turnover, like ZRT1. It is harder to predict why FRO2 is regulated at the posttranscriptional level. FRO2 functions to reduce iron at the cell surface before transport. Thus, overexpression of FRO2 should not lead directly to the accumulation of toxic amounts of iron because it does not transport iron itself. Because posttranscriptional regulation of IRT1 expression prevents the accumulation of the transporter when iron and zinc are present, it seems unnecessary to regulate FRO2 posttranscriptionally as well. However, it would be inefficient to continue to express FRO2 protein if the major ferrous transporter is not present.
35S-FRO2 plants display elevated copper chelate reductase activity when plants are grown on iron-deficient medium. However, the 35S-FRO2 plants did not display enhanced sensitivity to growth on iron-deficient medium supplemented with copper, nor did 35S-FRO2 plants accumulate copper when grown on iron-deficient medium. Together, these data suggest that either copper reduction is not required before transport, or that copper reduction is not the rate-limiting step in copper uptake. Although loss-of-function mutations in FRO2 result in a loss of low-iron-inducible copper chelate reductase activity, FRO2 mutant plants do not show reduced copper accumulation. Moreover, FRO2 transcript abundance does not increase when plants are deprived of copper (Robinson et al., 1999). The yeast FRE1 and FRE2 proteins are known to function in the reduction of iron and copper (Georgatsou and Alexandraki, 1994; Georgatsou et al., 1997). FRE1 and FRE2 belong to family of genes in yeast that are thought to encode metal reductases. Two of these, FRE1 and FRE7, are regulated by copper through the Mac1 transcriptional regulator (Martins et al., 1998). It is possible that other Arabidopsis FRO family members function in copper reduction and, in turn, may be regulated by copper.
35S-FRO2 transgenic plants grow better than wild type on low-iron medium. This result is likely due to the fact that these plants display elevated levels of ferric chelate reductase activity under iron starvation; therefore, the transgenic plants may be better able to acquire the low amounts of iron present in the medium. Alternatively, the low-iron tolerance phenotype of the 35S-FRO2 plants may be due to enhanced storage of iron in the seed. In agreement with our data that suggests that expression of FRO2 is controlled posttranscriptionally, 35S-FRO2 plants are not sensitive to growth on high iron. Our results are significant in that they suggest that overexpression of FRO2 may be useful for the construction of plants capable of thriving on low-iron soils. This is particularly exciting because approximately one-third of the world's soils are considered iron deficient.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis wild-type (ecotype Columbia gl-1) and transgenic seeds (35S-FRO2 and FRO2-GUS) were surface sterilized, sown on plates of Gamborg's B5 medium (Sigma-Aldrich, St. Louis) supplemented with 2% (w/v) Suc, 1 mm MES, and 0.6% (w/v) agar, pH 5.8, and placed in the dark for 2 to 4 d at 4°C. Transgenic plants were grown on plates supplemented with either 50 μg mL–1 kanamycin (35S-FRO2) or 25 μg mL–1 hygromycin (FRO2-GUS). Plates were incubated at 22°C under constant illumination for 12 to 14 d until they reached the 4- to 6-true-leaf stage. Yellow filters (acrylic yellow-2208, Cadillac Plastic and Chemical, Pittsburgh) were placed between the lights and plants to prevent the photochemical degradation of Fe(III)-EDTA (Hangarter and Stasinopoulos, 1991).
Seedlings were transferred to either iron-sufficient [50 μm Fe(III)-EDTA] or iron-deficient {300 μm FerroZine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate]; HACH Chemical, Ames, IA} medium. The medium contained 0.6% (w/v) agar, 1 mm MES, pH 6.0, and macronutrients and micronutrients as described previously (Marschner et al., 1982). Plants were incubated for various times after transfer in a growth chamber as described above. For growth on low-iron medium, plants were transferred to plates that contained no added iron but that also did not contain the iron chelator, Ferrozine; thus any iron present in the agar or following purification through a Barnstead Nanopure Diamond water purification system was available to the plants. For growth on zinc supplemented medium, iron-sufficient or -deficient medium was supplemented with either 100 μm or 500 μm ZnSO4.
Construction of the Chimeric FRO2-GUS Gene
An EcoRV/PstI genomic DNA fragment containing 1,204 bp of FRO2 5′ sequence and 63 bp of FRO2 coding sequence was cloned into SmaI/PstI digested pCAMBIA1381Xa (GenBank accession no. AF234303) creating pELC207. Translation is initiated from the FRO2 ATG.
Construction of the Chimeric 35S-FRO2 Gene
The Arabidopsis FRO2 cDNA was cloned into the BamHI site of pCGN18 creating pELC215. pCGN18 was the kind gift of Dr. Thomas Jack (Department of Biological Sciences, Dartmouth College, Hanover, NH; Connolly et al., 2002). The FRO2 cDNA contains 22 bp of sequence 5′ of the FRO2 ATG and 270 bp of sequence downstream of the stop codon.
Plant Transformation
The FRO2-GUS (pELC207) and 35S-FRO2 (pELC215) constructs were used to transform Agrobacterium tumefaciens strains GV3101and ASE, respectively. A. tumefaciens-mediated transformation of Arabidopsis plants (Columbia gl-1) was accomplished using the floral dip protocol (Clough and Bent, 1998). T1 seeds obtained from self-fertilization of the primary transformants were surface sterilized and plated on Gamborg's B5 medium supplemented with either kanamycin (35S-FRO2) or hygromycin (FRO2-GUS). Resistant plants were transferred to soil, and the T2 seeds resulting from self-fertilization were collected. The T2 seeds were surface sterilized and plated on the appropriate medium; transgenic lines that displayed 3:1 segregation for antibiotic resistance to antibiotic sensitivity in the T2 generation and that were 100% antibiotic resistant in the T3 generation were selected for further analysis. All further experiments were performed using T4 or T5 seeds.
Isolation of RNA, Northern-Blot Analysis, and Real Time RT-PCR
Total RNA was prepared (Verwoerd et al., 1989) from the roots and shoots of 17-d-old plants grown on plates that were either iron sufficient or deficient or from various tissues of soil-grown plants. Northern-blot analysis was performed as described previously (Connolly et al., 2002). An EcoRI fragment containing the 2.47-kb FRO2 cDNA was used as a probe for RNA gel-blot analysis; a NotI fragment containing the 1.4-kb IRT1 cDNA also was used a probe. Previous work demonstrated the specificity of the IRT1 probe (Vert et al., 2002). Arabidopsis contains eight FRO genes; thus, we performed high-stringency Southern-blot hybridization analysis to determine whether the FRO2 probe hybridizes to additional FRO sequences. The analysis revealed that the FRO2 probe hybridizes to a single band in EcoRI- and BamHI-digested genomic DNA and to three bands in HindIII digested genomic DNA, as predicted (data not shown). DNA was radiolabeled according to the random-primer method (Feinberg and Vogelstein, 1984). Quantification of signal resulting from FRO2 hybridization was performed using a Molecular Dynamics Storm phosphoimager. FRO2 values were normalized to rRNA levels; rRNA levels were quantified using the Quantity One gel documentation software package (Bio-Rad Laboratories, Hercules, CA).
For real time RT-PCR, cDNA was prepared from total RNA according to the manufacturer's instructions (SuperScript First-Strand Synthesis System for RT-PCR kit, Invitrogen, Carlsbad, CA). Total RNA was treated with DNase I before cDNA synthesis to remove trace amounts of genomic DNA. Primers were designed to amplify a 103-bp fragment of the FRO2 transcript (FRO2RTFOR, 5′GCCACATCTGCGTATCAAGTT3′; FRO2RTREV, 5′TCCCAAACAAGCTACGACCA3′). The primers amplify a fragment that spans intron 1, allowing the detection of genomic DNA contamination. BLAST searches using the FRO2 primers verified that the primers will anneal only to FRO2 sequences. Primers corresponding to UBQ10 were designed for use as an internal control (UBQ10RTFOR, 5′TGGTGGTATGCAGATTTTCG3′; UBQ10RTREV, 5′GGCTTTCACGTTATCAATGG3′).
Real time RT-PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on the iCycler thermocycler (Bio-Rad Laboratories). To quantify FRO2 and UBQ10 levels in unknown reactions, standards were prepared from serial dilutions of FRO2 and UBQ10 cDNA clones. The standards were run in parallel with the cDNA prepared from the unknown samples and then used to generate a standard curve. Upon completion of the amplification reactions, a threshold level is set; the threshold level is arbitrarily set at a level where the amplification curves are parallel to one another. The threshold values for FRO2 and UBQ10 unknown samples were compared with their corresponding standard curves and assigned an initial starting concentration in femtomoles. To normalize the data, the FRO2 concentrations were divided by the UBQ10 concentrations from each cDNA sample to obtain a relative value for each sample. The lowest FRO2 concentration was set to 1×, and the rest of the samples were assigned relative values based on the 1× sample. Finally, following the real time PCR reaction, melt curves were performed to verify that a single product was amplified; samples were run on agarose gels to confirm the presence of a single band.
Assay of Ferric Chelate and Copper Chelate Reductase Activities
Wild-type (Col gl-1) and 35S-FRO2 transgenic plants were grown for 2 weeks on B5 medium and then transferred to iron-sufficient or -deficient medium for 3 d. Ferric-chelate reductase and copper-chelate reductase activities were determined as described previously, except that copperchelate reductase activity was determined after plants had been grown for 4 d on either iron-sufficient or -deficient medium (Yi and Guerinot, 1996). Two seedlings were pooled and assayed as a group. Each value is the mean of five independent trials. The entire experiment was repeated four times. Statistical analysis of activity data was performed using a Student's t test.
Elemental Analysis
The concentrations of various minerals in the roots and shoots of wild type and three independent 35S-FRO2 transgenic lines was determined using inductively coupled argon plasma spectroscopy in the Nutrient and Element Analysis Lab in the Department of Horticulture at Cornell University (Ithaca, NY). Plants were grown for 2 weeks on B5 medium and then transferred to iron-sufficient or -deficient medium for 6 d. Roots and shoots were separated and dried overnight in a 65°C oven. Approximately 50 to 100 plants were pooled for each sample. Metal ion content of wild type and three independent 35S-FRO2 transgenic lines (13 individual plants each) grown on soil was determined in the lab of Dr. David Salt (Purdue University, West Lafayette, IN).
GUS Histochemical Staining
Wild type and 10 independent, single-insertion, homozygous FRO2-GUS transgenic lines were grown for 2 weeks and then transferred to either iron-sufficient or -deficient medium. After 3 d, plants were subjected to GUS histochemical staining using 5-bromo-4-chloro-3-indolyl β-d-glucoronide as a substrate (Jefferson et al., 1987). GUS histochemical staining was later performed on wild type and six independent FRO2-GUS lines grown either on plates for various times before transfer to iron-sufficient or -deficient medium for 3 d or on soil.
In Situ Hybridization
The plasmid pELC214 (FRO2 cDNA in BamH1 site of pBluescript) was used to generate sense and antisense probes for in situ hybridization. Sense and antisense probes were labeled with digoxigenin-11-UTP (Roche Diagnostics, Indianapolis) according to the manufacturer's protocol. For antisense probes, pELC214 was linearized with EcoR1 and transcribed with T3 RNA polymerase. For sense probes, pELC214 was linearized with SacII and transcribed with T7 RNA polymerase. Tissue samples were fixed and embedded as described (Vert et al. 2002), and in situ hybridization was performed according to previous protocols (Long et al., 1996; Long and Barton, 1998).
Distibution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Accession Number
The GenBank accession number for the FRO2 genomic clone is Y09581.
Acknowledgments
We are grateful to David Salt and Brett Lahner for elemental analysis of 35S-FRO2 transgenic plants. We thank Loubna Kerkeb, Beth Krizek, Rob McClung, and the members of our labs for critical reading of this manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025122.
This work was supported by the National Science Foundation (grant no. IBN–9974837 to M.L.G.) and by the U.S. Department of Agriculture National Research Initiatives Competitive Grants Program (grant nos. 9900598 and 0188925 to E.L.C.).
References
- Briat J-F, Lobréaux S (1997) Iron transport and storage in plants. Trends Plant Sci 2: 187–193 [Google Scholar]
- Brüggemann W, Maas-Kantel K, Moog PR (1993) Iron uptake by leaf mesophyll cells: the role of the plasma membrane-bound ferric-chelate reductase. Planta 190: 151–155 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Guardia MD, Alcantara E (1996) Ferric chelate reduction by sunflower (Helianthus annuus L.) leaves: influence of light, oxygen, iron-deficiency and leaf age. J Exp Bot 47: 669–675 [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 137: 266–267 [DOI] [PubMed] [Google Scholar]
- Georgatsou E, Alexandraki D (1994) Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol Cell Biol 14: 3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatsou E, Mavrogiannis LA, Fragiadakis GS, Alexandraki D (1997) The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J Biol Chem 272: 13786–13792 [DOI] [PubMed] [Google Scholar]
- Gitan RS, Eide DJ (2000) Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem J 346: 329–336 [PMC free article] [PubMed] [Google Scholar]
- Gitan RS, Luo H, Rodgers J, Broderius M, Eide D (1998) Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J Biol Chem 273: 28617–28624 [DOI] [PubMed] [Google Scholar]
- Grusak MA (1995) Whole-root iron(III)-reductase activity throughout the life cycle of iron-grown Pisum sativum L. (Fabaceae): relevance to the iron nutrition of developing seeds. Planta 197: 111–117 [Google Scholar]
- Grusak MA, Welch RM, Kochian LV (1990) Does iron deficiency in Pisum sativum enhance the activity of the root plasmalemma iron transport protein? Plant Physiol 94: 1353–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Eide D (1999) Zeroing in on zinc uptake in yeast and plants. Curr Opin Plant Biol 2: 244–249 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1992) Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett 307: 108–112 [DOI] [PubMed] [Google Scholar]
- Hangarter RP, Stasinopoulos TC (1991) Effect of Fe-catalyzed photooxidation of EDTA on root growth in plant culture media. Plant Physiol 96: 843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with broad specificity. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Lescure A-M, Proudhon D, Pesey H, Ragland M, Theil EC, Briat J-F (1991) Ferritin gene transcription is regulated by iron in soybean cell cultures. Proc Natl Acad Sci USA 88: 8222–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H-Q, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Barton MK (1998) The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Marschner H, Römheld V (1994) Strategies of plants for acquisition of iron. Plant Soil 165: 375–388 [Google Scholar]
- Marschner H, Römheld V, Ossenberg-Neuhaus H (1982) Rapid method for measuring changes in pH and reducing processes along roots of intact plants. Z Pflanzenphysiol 105: 407–416 [Google Scholar]
- Martins LJ, Jensen LT, Simons JR, Keller GL, Winge DR (1998) Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J Biol Chem 273: 23716–23721 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Proctor CM, Connolly EL, Guerinot ML (1999) A ferricchelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Sadjuga, Groom QJ (1997) The froh gene family from Arabidopsis thaliana: putative iron-chelate reductases. Plant Soil 196: 245–248 [Google Scholar]
- Susin S, Abadia A, Gonzalez-Reyes JA, Lucena JJ, Abadia J (1996) The pH requirement for in vivo activity of the iron-deficiency-induced “Turbo” ferric chelate reductase (a comparison of the iron-deficiency-induced iron reductase activities of intact plants and isolated plasma membrane fractions in sugar beet). Plant Physiol 110: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin LO (1966) Iron translocation: plant culture, exudate sampling, iron citrate analysis. Plant Physiol 45: 280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat J-F, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Blevins DG, Eide DJ (2002) Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol 129: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Theil EC (2000) Identification and characterization of the iron regulatory element in the ferritin gene of a plant (soybean). J Biol Chem 275: 17488–17493 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, Eide D (1998) Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem 273: 28713–28720 [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide D (1997) Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol 17: 5044–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]