Abstract
Peroxiredoxins (Prx) are thiol-dependent antioxidants containing one (1-cysteine [-Cys]) or two (2-Cys) conserved Cys residues that protect lipids, enzymes, and DNA against reactive oxygen species. In plants, the 1-Cys Prxs are highly expressed during late seed development, and the expression pattern is dormancy related in mature seeds. We have expressed the Arabidopsis 1-Cys Prx AtPER1 in Escherichia coli and show that this protein has antioxidant activity in vitro and protects E. coli in vivo against the toxic oxidant cumene hydroperoxide. Although some 1-Cys Prxs are targeted to the nucleus, a green fluorescent protein-AtPER1 fusion protein was also localized to the cytoplasm in an onion epidermis subcellular localization assay. It has been proposed that seed Prxs are involved in maintenance of dormancy and/or protect the embryo and aleurone layer surviving desiccation against damage caused by reactive oxygen species. These hypotheses were tested using transgenic Arabidopsis lines overexpressing the barley (Hordeum vulgare) 1-Cys PER1 protein and lines with reduced levels of AtPER1 due to antisensing or RNA interference. We found no correlation between Prx levels and the duration of the after-ripening period required before germination. Thus, Prxs are unlikely to contribute to maintenance of dormancy. RNA interference lines almost devoid of AtPER1 protein developed and germinated normally under standard growth room conditions. However, seeds from lines overexpressing PER1 were less inclined to germinate than wild-type seeds in the presence of NaCl, mannitol, and methyl viologen, suggesting that Prx can sense harsh environmental surroundings and play a part in the inhibition of germination under unfavorable conditions.
Peroxiredoxin (Prx) antioxidants, first identified in yeast (thiol-specific antioxidant; Kim et al., 1988), can be divided into three classes: typical 2-Cys Prx, atypical 2-Cys Prx, and 1-Cys Prx. Prxs can reduce H2O2, alkyl hydroperoxides, and hydroxyl radicals (Lim et al., 1993; Chae et al., 1994; Netto and Stadtman, 1996; Baier and Dietz, 1997) and have in vitro been shown to have antioxidant activity through protection of lipids, enzymes, and DNA against radical attack. In plants, antioxidant activity has been demonstrated for the 2-Cys BAS1 (Baier and Dietz, 1997) and the 1-Cys PER1 (Stacy et al., 1996) of barley (Hordeum vulgare). Transgenic plants overexpressing the rice (Oryza sativa) 1-Cys Prx (R1C-Prx) exhibit higher resistance to oxidative stress, suggesting antioxidant activity (Lee et al., 2000) in accordance with the function of 1-Cys homologs in other organisms (Shichi and Demar, 1990; Netto and Stadtman, 1996; Peshenko et al., 1996).
The plant 1-Cys Prx genes pBS128 from brome grass (Goldmark et al., 1992), Per1 in barley (also called B15C; Aalen et al., 1994), AtPER1 in Arabidopsis (Haslekås et al., 1998), and FePer1 from buckwheat (Fagopyrum esculentum; Lewis et al., 2000) are expressed in developing seeds. They have high expression levels during the stages of massive water loss at the end of seed development and also in the mature dry seed. pBS128, the first plant 1-Cys Prx identified, was isolated by differential screening based on its dormancy-related expression pattern. AtPER1 and Per1 expression patterns are also dormancy related (Stacy et al., 1996; Haslekås et al., 1998), i.e. during imbibition, the transcripts disappear in germinating seeds, whereas in dormant non-germinating seeds, the transcript levels are maintained or even transiently up-regulated.
Due to this expression pattern, Stacy et al. (1996) suggested 1-Cys Prx involvement in maintenance of dormancy. Some experiments question such a role: High AtPER1 expression level in the non-dormant aba-1 mutant indicated that AtPER1 was not sufficient to induce dormancy (Haslekås et al., 1998), and transgenic tobacco (Nicotiana tabacum) seeds expressing R1C-Prx germinated as efficiently as wild-type (wt) seeds (Lee et al., 2000). The latter experiment was, however, performed with seeds that were exposed to a dormancy-breaking treatment (4°C for 24 h; Lee et al., 2000) and is thus not conclusive.
A function in desiccation tolerance during late stages of seed development has also been suggested (Stacy et al., 1996). One important aspect of desiccation tolerance is the protection against damaging reactive oxygen species (ROS; for review, see Leprince et al., 1993, 1994). Desiccation and resumption of respiration after hydration of dry seeds give rise to ROS, which when not removed or neutralized may have deleterious effects on membranes, proteins, and DNA. Water uptake and resumption of respiration are similar in dormant and non-dormant seeds during early imbibition. In germinating seeds, there is an elevation of germination-specific antioxidants. However, dormant seeds do not enter the last phase of germination and can therefore not be protected by germination-related antioxidants. The level of 1-Cys Prx is high at this stage and may provide the protection needed.
Recently, a new member of the 1-Cys Prx group, XvPer1, was isolated from the resurrection plant Xerophyta viscosa (Mowla et al., 2002). This gene was up-regulated when the plant was dehydrated and during drought and heat stress, which are known to elevate the ROS production as a result of stomatal closing and reduced CO2 availability. In accordance with a putative bipartite nuclear localization signal (NLS) found in plant 1-Cys Prxs, XvPER1 was localized to the nucleus (Mowla et al., 2002). Barley PER1 has also been localized to the nucleus of cells of the developing embryo and aleurone layer, although a weak signal was also detected in the cytoplasm. In imbibed barley embryo and aleurone cells, labeling of similar strength was found in the cytoplasm and in the nucleus (Stacy et al., 1999). A role in protection of nucleic acids against ROS has been suggested (Stacy et al., 1999; Mowla et al., 2002).
In this study, we have investigated the antioxidant activity and subcellular localization of AtPER1. We have also generated transgenic Arabidopsis lines with increased and decreased levels of 1-Cys Prxs to investigate their importance in maintenance of dormancy. Seeds of such lines were also used to study germination frequencies in the presence of osmotic and oxidative stress that inhibit germination.
RESULTS
AtPER1 Has Antioxidant Activity in Vitro and in Vivo
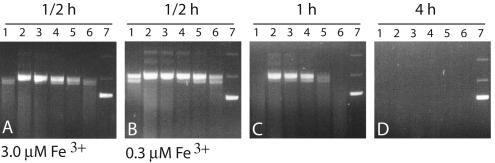
Antioxidant activity of a purified and concentrated AtPER1 fusion protein, produced using the PinPoint system (Promega, Madison, WI), was tested in a DNA protection assay, i.e. DNA was exposed to a mixed function oxidation assay where ROS is generated due to the presence of Fe3+ (Stacy et al., 1996). Protection of plasmid DNA against cleavage by radicals produced in the system increased with increasing concentrations of AtPER1 fusion protein (20–200 μg mL–1; Fig. 1). In a 1-h reaction, a substantial protection against degradation was evident with 50 to 200 μg mL–1 (Fig. 1C, lanes 2–4). In comparison, the DNA was totally degraded without addition of fusion protein or in the presence of 200 μg mL–1 BSA (Fig. 1C, lane 1 and 6). However, the fusion protein did not give full protection against nicking and degradation. Even with 200 μg mL–1 protein, nicking of plasmid DNA was evident after 0.5 h when exposed to 3.0 or 0.3 μm Fe3+ (Fig. 1, A and B) compared with the unnicked, supercoiled plasmid (lane 7). After 4 h, complete DNA degradation was seen also in the presence of AtPER1 (Fig. 1D).
Figure 1.
In vitro antioxidant activity of AtPER1. Protection of DNA against cleavage by free radical attack by an AtPER1 fusion protein. A, pUC19 plasmid incubated with 3.0 μm Fe3+ for 0.5 h; B through D, pUC19 plasmid incubated with 0.3 μm Fe3+ for 0.5, 1, and 4 h, respectively. Lane 1, Without protein; lane 2, 200 μg mL–1 AtPER1; lane 3, 100 μg mL–1 AtPER1; lane 4, 50 μg mL–1 AtPER1; lane 5, 20 μg mL–1 AtPER1; lane 6, 200 μg mL–1 bovine serum albumin (BSA); and lane 7, untreated pUC19 plasmid DNA. The major pUC19 band in lane 7 is the supercoiled plasmid that upon nicking by radical attack first will convert to a slower-moving open circular plasmid and will later be degraded to a DNA smear.
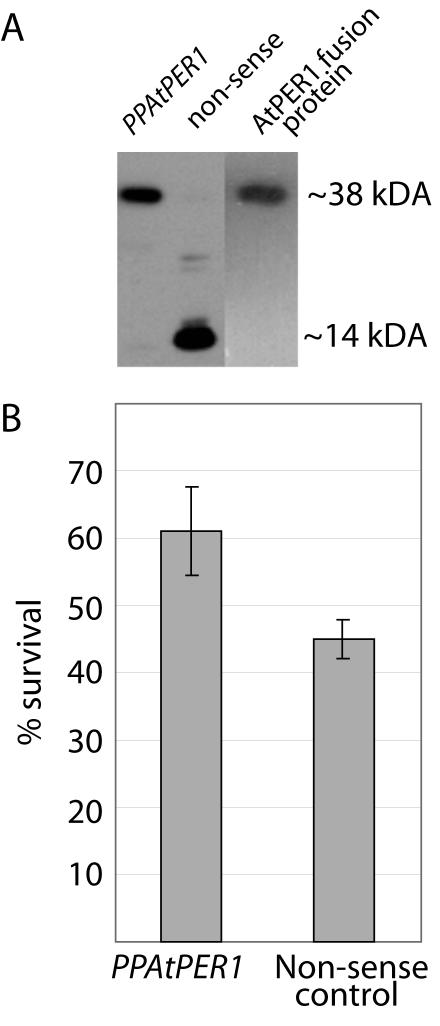
Antioxidant activity of AtPER1 was also tested in vivo. The AtPER1 PinPoint fusion protein was used to produce polyclonal antibodies against AtPER1 (see “Materials and Methods”; Fig. 2A, lane 3). The inframe AtPER1 cDNA construct (PPAtPER1) and the cDNA cloned out of frame in the PinPoint vector system (nonsense construct) were introduced into Escherichia coli JM109 cells. Protein isolated from bacteria harboring the nonsense construct confirmed that this construct did not express a full-length AtPER1 protein. The polyclonal antiserum does, however, recognize the PinPoint tag (Fig. 2A, 14 kD). In bacteria with the PPAtPER1 construct, AtPER1 antiserum detected a protein of the expected size (Fig. 2A, 38 kD), corresponding to the AtPER1 fusion protein purified by means of the N-terminal PinPoint tag (Fig. 2A, lane 3).
Figure 2.
In vivo antioxidant activity of AtPER1. Protection of E. coli cells against cumene hydroperoxide. A, Western blotting using a polyclonal antiserum raised against the AtPER1 PinPoint fusion protein, used on protein isolated from E. coli harboring the PPAtPER1 fusion protein construct, a nonsense construct, or against purified AtPER1 fusion protein. B, Survival of E. coli cells expressing the PPAtPER1 fusion protein or the nonsense protein after exposure to 1.0 mm cumene hydroperoxide. The bars show mean values from five experiments ± sd.
The E. coli cells were tested for survival in the presence of the toxic oxidant cumene hydroperoxide in an assay previously used to show in vivo antioxidant activity of the 2-Cys Prx of barley, BAS1 (Baier and Dietz, 1997). Expression of AtPER1 increased the tolerance of the bacteria to cumene hydroperoxide (Fig. 2B). Less than one-half (45.0% ± 2.8%) of the bacteria harboring the nonsense construct survived 20 min of exposure to 1.0 mm cumene hydroperoxide, compared with 61.0% ± 6.6% of the bacteria expressing AtPER1.
A Green Fluorescent Protein (GFP)-AtPER1 Fusion Protein Is Localized to the Cytoplasm and to the Nucleus
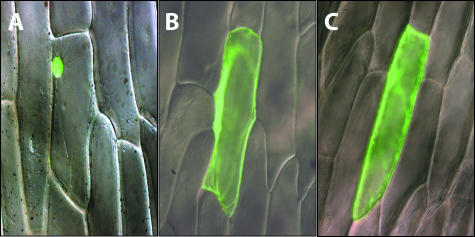
Plant 1-Cys Prxs have been suggested to contain a bipartite NLS at the 3′ end (Stacy et al., 1999; Mowla et al., 2002). Analysis of the AtPER1 sequence using PSORT (http://psort.nibb.ac.jp/) and PredictNLS (http://cubic.bioc.columbia.edu/predictNLS/) predicted cytoplasmic localization to be most likely for AtPER1. To investigate this aspect further, intracellular localization of AtPER1 in a C-terminal fusion with the GFP was examined in a transient expression assay in onion epidermal cells (see “Materials and Methods”). The 35S::GFP-AtPER1 construct as well as 35S::GFP and a positive control for nuclear localization, the cDNA of Heterochromatin Protein1 (HP1) of fruitfly (Drosophila melanogaster) cloned in-frame with GFP were introduced into the inner epidermis of onion using particle bombardment. GFP-HP1 showed a strong signal restricted to the nucleus (Fig. 3A). In contrast, the GFP on its own is present in the cytoplasm and in the nucleus (Fig. 3B). The nuclear localization is thought to result from diffusion, because the GFP protein is smaller than 40 to 60 kD (von Arnim et al., 1998). The signal distribution of the approximately 50-kD GFP-AtPER1 fusion protein was comparable with that of the approximately 25-kD GFP (Fig. 2C).
Figure 3.
Intracellular localization of GFP-AtPER1 protein fusion. Merged differential interference contrast light micrographs and fluorescence micrographs of GFP fusion constructs transiently expressed in onion epidermal cells after particle bombardment. A, 35S::HP1-GFP. The GFP fluorescence signal is only found in the nucleus. B, 35S::GFP. GFP fluorescence signal is found in the cytoplasm and the nucleus. C, 35S::GFP-ATPER1. GFP fluorescence signal is found in the cytoplasm and in the nucleus.
Germination Frequencies of Freshly Harvested Seeds Do Not Correlate to Levels of 1-Cys Prx
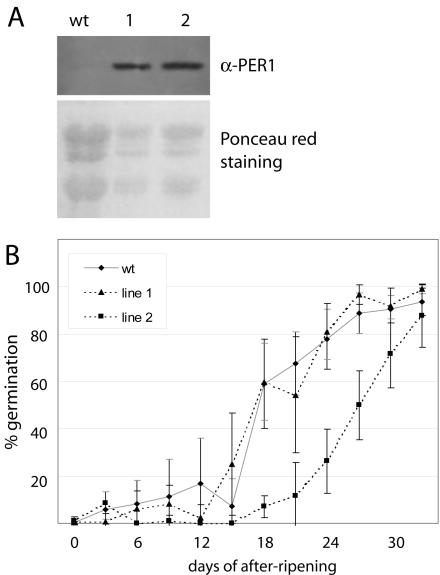
To investigate the relationship between 1-Cys Prx level and maintenance of dormancy, we generated transgenic Arabidopsis plants that overexpressed the barley 1-Cys Prx PER1 (pPCV002 35S::Per1) and plants with down-regulation of the AtPER1 level using an AtPER1-antisense construct (pKOH110 35S::AtPER1; Fig. 4). The expression levels of PER1 in independent 35S::Per1 transformants were tested by immunodetection with PER1 antiserum (Stacy et al., 1999) on western blots with protein isolated from mature seeds. The PER1 antiserum does not crossreact with the Arabidopsis AtPER1 protein (Fig. 5A, wt lane). Two transgenic lines with equally high PER1 levels (Fig. 5A) were used in a germination test. The AtPER1 transcript level in seeds of these lines did not differ significantly from wt seeds (data not shown).
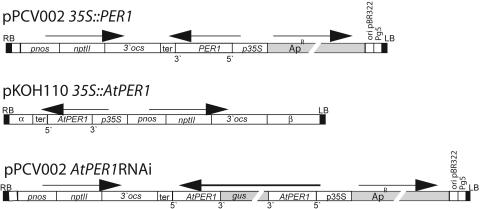
Figure 4.
Schematic drawings of the T-DNA regions from the pPCV002 35S::Per1, pKOH110 35S::AtPER1, and pPCV002 AtPER1RNAi constructs used for Arabidopsis transformation. RB, Right border; LB, left border; p35S, 35S promoter; pnos, nos promoter; ter and 3′ocs, polyadenylation signals; nptII, neomysin phosphotransferase gene; gus, β-glucuronidase stuffer fragment; ApR, ampicillin resistance gene; Pg5, truncated promoter of TL-DNA gene 5; Ori pBR322, ori region from pBR322; β, natural T-DNA sequence; AtPER1 and Per1, cDNA sequences. Horizontal arrows indicate direction of transcription. The orientations of the AtPER1 and Per1 cDNA sequences (i.e. 5′ and 3′ ends) are indicated.
Figure 5.
Germination frequencies of freshly harvested seeds overexpressing PER1 and wt seeds. A, Immunoblot showing PER1 levels in protein isolated from seeds of wt Arabidopsis and of two pPCV002 35S::Per1 transgenic lines using PER1 antiserum. Ponceau red staining of the membrane was used as a loading control. B, Germination frequencies of seeds from wt and the two pPCV002 35S::Per1 transgenic lines scored 8 d after sowing. Seeds from brown, dried, and just opened siliques were sown at the day of harvest and thereafter every 3rd d until 33 d of after-ripening. Percentages are means of five to eight plants ± sd (in gray for wt and in black for transgenic lines).
Siliques were harvested at the same stage of maturity, i.e. the first three brown, dried, and just opened siliques were collected. At the point of harvest, these seeds are completely dormant. Seeds from such siliques were sown every 3rd d after harvest, and germination frequencies were scored 8 d after sowing. In the wt C24 ecotype, germination frequencies are slowly increasing with the number of days of after-ripening. After 12 to 15 d, germination frequencies begin to increase more rapidly, and 60% germination is reached at 18 d of after-ripening (Fig. 5B). After 33 d, more than 95% of the seeds germinate. The pPCV002 35S::Per1 line 1 showed a similar germination pattern as wt seeds, whereas pPCV002 35S::Per1 line 2 needed longer time of after-ripening to reach a similar germination frequency (Fig. 5B). However, after 33 d, 88% of the line 2 seeds also germinated. The PER1 levels are not significantly different in the two lines (Fig. 5A). Thus, differences in germination profiles cannot be attributed to barley 1-Cys Prx but can possibly be attributed to different genomic insertion positions of the T-DNAs in the two 35S::Per1 lines. The variation in germination frequency between seeds of individual wt plants or transgenic plants of the same line, reflected by the sd, can be attributed to environmental variance.
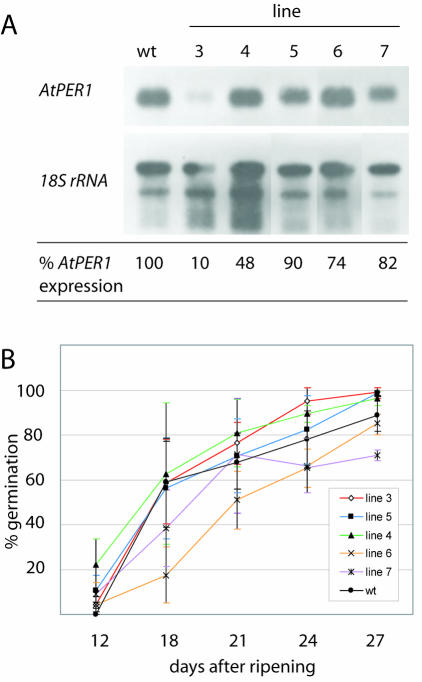
To test whether a reduction in AtPER1 level reduced dormancy, lines transformed with the antisense AtPER1 construct was used. mRNA was isolated from mature seeds of different lines and the level of mRNA investigated by northern hybridization. The lines 3 to 7 showed from 10% to 90% reduction in mRNA level compared with wt seeds (Fig. 6A). Germination tests were performed in the same manner as for the pPCV002 35S::Per1 lines. Average germination frequencies after 8 d of imbibition of seeds sown 18 d after harvest (Fig. 6B) was lower than that of wt seeds for two lines with transcript levels of 74% and 82%, however, only significant for one line. For three lines, among them line 3 with a transcript reduction of 90%, germination frequencies were comparable with wt (about 60%). After 24 d of after-ripening, there was a variation in germination frequencies from 65% to 95% (Fig. 6B). However, the frequencies did not correlate to the level of AtPER1 in the seeds.
Figure 6.
Germination frequencies of freshly harvested seeds with reduced level of AtPER1 and wt seeds. A, Northern blot with mRNA isolated from seeds of wt and five independent pKOH110 35S::AtPER1 transgenic lines hybridized with an AtPER1 probe and as a loading control an rRNA probe. AtPER1 expression level relative to wt is indicated. B, Germination frequency of seeds from wt and the five pKOH110 35S::AtPER1 transgenic lines scored 8 d after sowing. Seeds from brown, dried, and just opened siliques were harvested and sown at the indicated days of after ripening. Percentages are means of five to eight plants ± sd.
Arabidopsis Seeds with Significantly Reduced Levels of AtPER1 Display No Obvious Phenotype
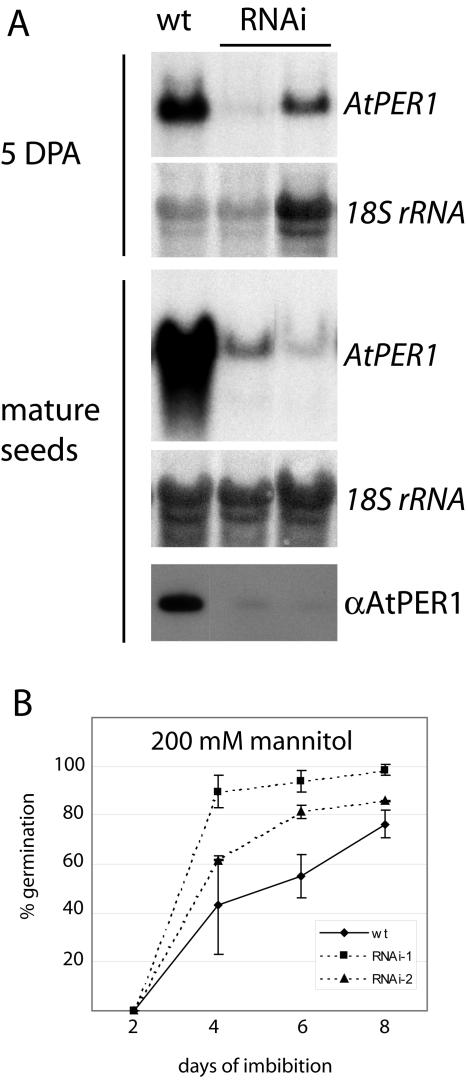
Because the antisense construct only to a limited extent lowered AtPER1 expression levels, an RNA interference (RNAi) construct (pPCV002 AtPER1RNAi) was made. Two copies of the AtPER1 cDNA were cloned in inverse orientations on each side of a stuffer fragment (Fig. 4). Transcription from the 35S promoter would make possible the generation of a double-stranded AtPER1 cDNA intended for down-regulation of the endogenous AtPER1 expression by RNAi (Chuang and Meyerowitz, 2000). Arabidopsis plants transformed with this construct were tested for mRNA and protein expression levels. Two independent lines with significantly lowered mRNA levels at 5 DPA and in mature seeds (Fig. 7A) were selected for further experiments. As anticipated, the protein levels detectable by the antiserum specific for AtPER1 (Fig. 2A) were drastically reduced in these two lines (Fig. 7A).
Figure 7.
Germination behavior of two AtPER1RNAi lines with reduced levels of AtPER1 mRNA and protein. A, Northern blots of total RNA isolated from 5 DPA and mature seeds were hybridized to an AtPER1 probe and the rRNA loading control probe. The AtPER1 level in protein isolated from mature seeds was detected by western blotting using AtPER1 antiserum. B, Germination on 200 mm mannitol of seeds from the two AtPER1RNAi lines of A compared with wt seeds. Germination was scored at d 2, 4, 6, and 8 of imbibition and is given as the average of three parallel tests ± sd. An average of 265 seeds was tested for each sample in each parallel.
No obvious phenotype, e.g. embryo lethality or morphological changes, was detected in the primary RNAi transformants or their progeny plants that also had drastically reduced AtPER1 levels (data not shown). The seeds developed normally, and the dormancy level correlated closely to that of seeds from wt plants grown and harvested at the same time as the RNAi lines (data not shown). In after-ripened seeds, the germination frequencies were similar, i.e. between 95% and 98%. The number of abnormal germinating seedlings was not significantly different either between AtPER1RNAi lines and the wt or between the RNAi lines and 35S::Per1-overexpressing lines.
Only one of our tests indicated a role for AtPER1. Mature, after-ripened, thus non-dormant, AtPER1RNAi seeds were germinated under ionic osmotic stress (10–175 mm NaCl) and nonionic osmotic stress (10–500 mm mannitol), which both are known to inhibit germination (see Espelund et al., 1995). On average, a slightly higher (although in most cases not significant) germination frequency was observed for the RNAi lines compared with wt seeds grown and harvested at the same point of time, both on NaCl and on mannitol (data not shown). At a concentration of 200 mm mannitol, the RNAi lines germinated significantly faster than wt: Four days after sowing 72% and 92% of the total number of germinating RNAi seeds had already germinated, compared with only 57% of the total germinating wt seeds (Fig. 7B). Total germination frequencies after 8 d were 86% and 98% for the AtPER1 RNAi lines, while 76% for the wt seeds. This suggested that AtPER1 normally contributes to inhibition of germination under harsh conditions.
Seeds with an Elevated Level of 1-Cys Prx Have a Lower Germination Frequency Than wt on NaCl, Mannitol, and Methyl Viologen (MV)
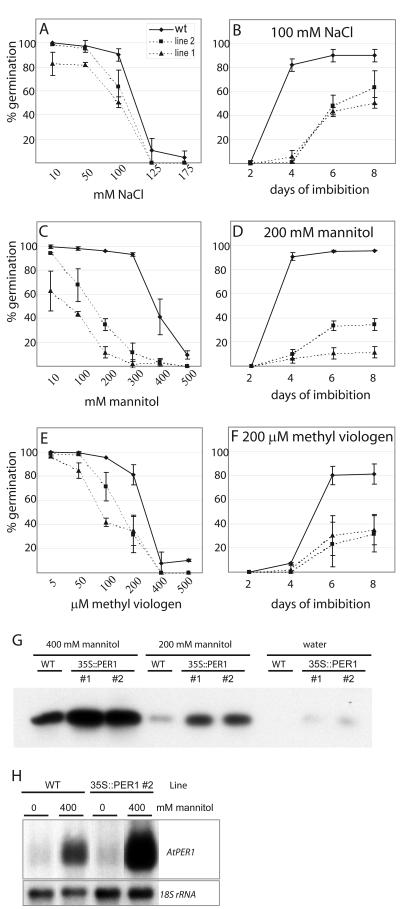
To further investigate the influence of seed 1-Cys Prx levels under conditions shown to inhibit germination, we compared germination frequencies between seeds from wt and 35S::Per1 lines (Fig. 5A). Germination frequencies were scored on 10 to 175 mm NaCl, 10 to 500 mm mannitol, and 5 to 500 μm MV, a compound known to create superoxide anions and H2O2.
On the lowest concentrations of NaCl used (10–50 mm), germination frequencies of wt and 35S::Per1 lines were almost similar, but on 100 mm NaCl the 35S::Per1 lines germinated to a lower extent than wt seeds (Fig. 8A), i.e. after 8 d, 90% of wt seeds but only 50% to 60% of the 35S::Per1 seeds germinated. Germination of the latter was also slower, because after 4 d, almost all of the wt seeds that germinated had germinated, whereas hardly any of the 35S::Per1 seeds had started to germinate (Fig. 8B). One hundred and twenty-five and 175 mm NaCl drastically impaired germination of wt seeds, and transgenic seeds did not germinate at all (Fig. 8A).
Figure 8.
Germination on NaCl, mannitol, and MV of seeds from two independent pPCV002 35S::Per1 transgenic lines with elevated level of 1-Cys Prx compared with wt seeds. Germination was scored after 8 d of imbibition on 10 to 175 mm NaCl (A), 10 to 500 mm mannitol (C), and 5 to 500 μm MV (E). Germination was scored at d 2, 4, 6, and 8 of imbibition on 100 mm NaCl (B), 200 mm mannitol (D), and 200 μm MV (F). Germination is given as the average of three parallel tests ± sd. An average of 115 seeds was tested for each sample in each parallel. G, Immunoblot showing AtPER1 levels in 50 μg of protein isolated from seeds imbibed for 8 d on 400 mm mannitol, on 200 mm mannitol, or on water of wt Arabidopsis and of the two pPCV002 35S::Per1 transgenic lines using AtPER1 antiserum. H, Northern blots of mRNA isolated from wt and 35S::Per1 seeds imbibed for 8 d on water or 400 mm mannitol were hybridized to an AtPer1 probe and the rRNA loading control probe.
Concentrations of 10 to 300 mm mannitol had only minor effect on germination of wt, whereas germination in 35S::Per1 lines showed lower total germination frequencies after 8 d with increasing concentrations (Fig. 8C). Germination was also slower (Fig. 8D shows this for 200 mm mannitol). At concentrations of 400 and 500 mm mannitol, wt germination was also critically reduced (Fig. 8C).
Germination on the lowest concentration (5 μm) of MV did not result in any difference between wt and 35S::Per1 seeds (Fig. 8E). At 50 to 200 μm MV, germination of wt and transgenic seeds were gradually delayed and also much lower after 8 d. Wt seeds could, however, withstand higher concentrations than 35S::Per1 seeds (Fig. 8, E and F). Four hundred and 500 μm MV dramatically impaired germination of wt and 35S::Per1 seeds (Fig. 8E). In summary, the 35S::Per1 lines were slower and less inclined to germinate when exposed to osmotic and oxidative stress.
To investigate the fate of the AtPER1 protein during imbibition with and without stress, we determined protein levels of wt and 35S::Per1 seeds imbibed on 400 and 200 mm mannitol or on water, using western blotting (Fig. 8G). In wt seeds and 35S::Per1 seeds, AtPER1 levels correlate inversely to germination frequency, i.e. the level is highest in seeds imbibed on 400 mm mannitol and lowest in seeds germinated on water for 8 d. On water, with a germination frequency of near to 100%, only traces of AtPER1 protein was seen in the wt—with double the amount of total protein and 10 times longer exposure time that in Figure 8G, a faint AtPER1 protein band could be detected (not shown). It should also be noted that compared with wt, the AtPER1 level is higher in both 35S::Per1 lines (Fig. 8G), which have significantly lower germination frequencies than wt on mannitol (Fig. 8C).
To investigate whether the AtPER1 protein level was reflected in the AtPER1 transcript level, mRNA from seeds imbibed on water and on 400 mm mannitol was isolated and subjected to northern hybridization with an AtPER1 probe (Fig. 8H). Although the AtPER1 transcript is hardly visible in the mRNA from seeds germinated on water, the AtPER1 transcript level is high in seeds imbibed on mannitol, and considerably higher in 35S::Per1 seeds than in wt, consistent with the lower germination frequencies of the 35S::Per1 seeds.
DISCUSSION
It has been proposed that one function of seed 1-Cys Prxs is protection against ROS (Stacy et al., 1996), because this is an important aspect of desiccation tolerance (Leprince et al., 1994). Protection against ROS is also needed during imbibition of mature dry seeds when respiration is resumed. Previously, antioxidant activity of the barley PER1 has been shown in vitro in a mixed function oxidation assay (Stacy et al., 1996). Using the same assay, we here show in vitro activity of the AtPER1 fusion protein (Fig. 1). However, the protection level of AtPER1 appears lower than previously shown for PER1 (Stacy et al., 1996). On the other hand, the in vivo protection demonstrated by enhanced survival of E. coli bacteria subjected to cumene hydroperoxide (Fig. 2) is similar to that shown for the 2-Cys barley BAS1 protein in the same assay (Baier and Dietz, 1997).
In support of a protective function, Lee et al. (2000) have shown that transgenic tobacco plants overexpressing the rice R1C-Prx exhibited higher resistance to oxidative stress. Recently, Mowla et al. (2002) have shown that expression of XvPer1 from the resurrection plant X. viscosa was induced upon dehydration of vegetative tissue, a condition known to induce ROS production. Tr155, a homolog from Tortula ruralis is expressed in the gametophyte in response to desiccation and rehydration (Wood et al., 2000).
In the promoters of Per1 and AtPER1, a putative antioxidant-responsive element has been identified (Haslekås et al., 1998; Aalen, 1999). Consistent with a protective function of AtPER1 via ROS induction, oxidative stress has been demonstrated to induce expression of AtPER1 (C. Haslekås, P.E. Grini, S. Nordgard, T. Thorstensen, M.K. Viken, V. Nygaard, and R. B. Aalen, unpublished data). However, our AtPER1 RNAi lines almost devoid of AtPER1 protein (Fig. 7A) documented that low AtPER1 level is not deleterious to the developing and germinating seed, at least under favorable growth room conditions. It is reasonable to assume that other antioxidant systems in the seed can substitute for the absence of AtPER1.
Where in the cells does AtPER1 exert its function? In mammals, Prx isoforms have been found localized to the cytosol, to the nucleus, and to mitochondria and peroxisomes (Wood et al., 2003b). In plants, the 2-Cys BAS1 protein is found in the chloroplast (Baier and Dietz, 1997). In the C-terminal end of plant 1-Cys Prxs, a putative bipartite NLS has been identified (Stacy et al., 1999; Mowla et al., 2002), and XvPER1 from X. viscosa has been found localized to the nucleus (Mowla et al., 2002). PER1 was mainly found localized to the nucleus of developing embryo and aleurone cells, but in imbibed embryo and aleurone cells, the labeling was of similar strength in the cytoplasm and the nucleus (Stacy et al., 1999). Despite the putative NLS, the human 1-Cys protein was found localized to the cytosol (Kang et al., 1998). Analysis of AtPER1 in PSORT and PredictNLS Software predicted the most likely location to be the cytoplasm.
In the onion epidermis cell transient expression assay, our GFP-AtPER1 fusion protein was localized to the cytoplasm and to the nucleus (Fig. 3). The signal distribution of GFP-AtPER1 was similar to that of GFP, which does not contain a NLS. The partial nuclear localization of GFP is caused by bidirectional diffusion through the nuclear pore complex (von Arnim et al., 1998). The exclusion limit for this bidirectional diffusion has been estimated to 40 to 60 kD (Gorlich and Mattaj, 1996). The predicted size of the GFP-AtPER1 fusion protein of 52 kD underpasses the size exclusion limit, so we cannot exclude that the partial nuclear localization seen for GFP-AtPER1 is due to diffusion. The difference in localization, when compared with the closely related PER1 and XvPER1, might be caused by a single amino acid change in the AtPER1 putative bipartite NLS (see Mowla et al., 2002). Intracellular localization of AtPER1 could also be dependent on type of tissue, developmental stage, or environmentally conditions/factors that are not met by the onion cells used in the transient expression assay. There is also the possibility that the high level of fusion protein expressed in bombarded cells may result in inefficient targeting to the nucleus. It would therefore be of interest to investigate subcellular localization of AtPER1 or GFP-AtPER1 fusion protein in Arabidopsis plants.
Most seeds possess some degree of dormancy at the completion of seed development, but so far, no genes exclusively controlling dormancy have been isolated (Koornneef et al., 2002). However, 1-Cys Prxs have been proposed a role in maintenance of dormancy under normal growth conditions (Goldmark et al., 1992; Stacy et al., 1996; Haslekås et al., 1998). Later results are conflicting with this hypothesis. Even though the transcript disappears upon germination, the protein is still present, e.g. PER1 protein was stable up to 11 d postimbibition (Stacy et al., 1999), and tobacco transformed with R1C-Prx expressed protein until 15 d postimbibition (Lee et al., 2000).
To thoroughly test the hypothesis on 1-Cys Prx involvement in maintenance of dormancy, we have performed germination trials on Arabidopsis seeds with reduced or increased level of 1-Cys PRXs, i.e. lines with high expression of the barley PER1, elevating the total level of 1-Cys Prx in the seeds, and antisense lines with 10% to 90% reduction in AtPER1 levels. Instead of bulk-harvested seeds, we have used seeds harvested at precisely the same stage of maturity. Neither for the five antisense 35S::AtPER1 lines nor for the 35S::Per1 lines was there any correlation between 1-Cys Prx level and the length of the after-ripening period required for germination (Figs. 5 and 6). Thus, our results do not support 1-Cys Prx involvement in maintenance of dormancy.
Dormancy is thought to be beneficial for the seeds because this quality enhances the possibility of survival when the seeds are allowed to germinate, e.g. the seeds do not germinate during non-favorable conditions as when they fall to the ground in the autumn. Similarly, other non-favorable conditions like oxidative or osmotic stress might inhibit germination. How can the seeds sense such unfavorable conditions? Answers to this question may come from mutant lines with reduced or enhanced sensitivity to NaCl and mannitol during germination and from Quantitative Trait Locis (QTLs) mapping close to genes involved in abscisic acid (ABA) responses, biosynthesis, or modulation (Quesada et al., 2002). ABA concentration increases in the response to salt or osmotic stress. The Catalase1 gene of maize (Zea mays; Cat1), active in seeds, is induced by ABA, but also via H2O2, and H2O2 level is increased upon ABA elevation (Guan et al., 2000). The AtPER1 promoter is also induced by ABA and oxidative stress (C. Haslekås, P.E. Grini, S.H. Nordgard, T. Thorstensen, M.K. Viken, V. Nygaard, and R.B. Aalen, unpublished data). Germination-inhibiting concentrations of mannitol, salt, and ABA have been shown to induce barley Per1 expression (Aalen et al., 1994; Espelund et al., 1995), and likewise imbibition of Arabidopsis seeds on high concentrations of mannitol results in high AtPER1 protein and transcript levels (Fig. 8, G and H).
The AtPER1 promoter contains an ABA-responsive element (Aalen, 1999) likely to be bound by ABI5, suggested to be a sensor of osmotic status in the seed environment (Carles et al., 2002). A slightly lower sensitivity to germination inhibition by mannitol in our AtPER1RNAi lines (Fig. 7B) and a higher sensitivity to NaCl, mannitol, and MV in 35S::Per1 lines (Fig. 8) support the involvement of 1-Cys Prxs in sensing environmental conditions. H2O2 has been implicated to play a role in the triggering of germination in planta (Fontaine et al., 1994). Under stress conditions on the other hand, the H2O2 generated should be kept down. Because Prxs are able to remove H2O2, an elevated level of 1-Cys Prx could possibly reduce H2O2 to a lower level, resulting in a reduced germination frequency. This hypothesis is reminiscent to the floodgate model proposed for 2-Prx action in mammals (Wood et al., 2003a). In this respect, it will be important to investigate whether 1-Cys Prx can be inactivated by overoxidation, as has been shown for mammalian 2-Cys Prx (Wood et al., 2003a).
Is there a threshold level of 1 Cys Prx that is critical? The marginal effect of lowering the AtPER1 level suggests that other genes are also involved in sensing harsh conditions and to a considerable extent can compensate for the absence of AtPER1. On the other hand, the results using the 35S::Per1 lines imply that higher levels of 1-Cys Prx increase the sensitivity of imbibed seeds with regard to unfavorable conditions.
Although a role for AtPER1 in the maintenance of dormancy seems unlikely, we suggest that the antioxidant function of 1-Cys Prx is employed to sense and/or react to seed environmental conditions, thus preventing germination to take place under unfavorable conditions. It would therefore be of interest to compare the germination ability of different Arabidopsis ecotypes under such conditions and to investigate whether putative differences are correlated to AtPER1 levels.
MATERIALS AND METHODS
Purification of the AtPER1 Fusion Protein for Antiserum Production
AtPER1 cDNA was amplified by reverse transcriptase-PCR using the gene-specific primers 5′-CGG GAT CCG GTA AAA TGC CAG GGA TC-3′ and 5′-GGA ATT CGT AGG CTT TTG TTA TTA TCT TTT C-3′, containing BamHI site and EcoRI sites. The AtPER1 cDNA digested with EcoRI, blunted, and then digested by BamHI was ligated into the BamHI and SmaI sites of PinPoint Xa3 vector (Promega), generating the construct PPAtPER1. Sequencing confirmed the cDNA sequence to be in the correct reading frame following the polypeptide tag. AtPER1 fusion protein was purified using the PinPoint batch purification protocol (Promega), with 50 mm HEPES and 5 mm biotin as elution buffer. Purified AtPER1 fusion protein was used for antiserum production in two rabbits (AGRI SERA AB, Vännäs, Sweden).
Antioxidant Activity Assay
Antioxidant activity of purified AtPER1 fusion protein, concentrated in a Centricon-10 concentrator (Millipore, Bedford, MA), was tested in a DNA cleavage assay as by Stacy et al. (1996). Control reactions were performed on flow-through from the Centricon column without AtPER1 protein and by the addition of 200 μg mL–1 BSA.
Expression and in Vivo Activity Test of Recombinant AtPER1 Protein in Escherichia coli
An AtPER1 nonsense construct was made by PCR amplification with the primers 5′-CGG GAT CCG TAA AAT GCC AGG GAT CAC-3′ and 5′-GGA ATT CGT AGG CTT TTG TTA TTA TCT TTT C-3′, which contain BamHI and EcoRI sites in their 5′ ends, respectively. The 5′ end primer contained one additional nucleotide that generated an out-of-frame fusion to the PinPoint tag when cloned between the BamHI and EcoRI sites of PinPoint Xa3 vector. The nonsense construct and the PPAtPER1 construct (previous section) were used in an in vivo activity assay in E. coli JM109 according to (Baier and Dietz, 1997). Survival of bacteria exposed to 1.0 mm cumene hydroperoxide were analyzed and compared.
Protein Isolation and Western Blotting
Pelleted E. coli cells expressing the PPAtPER1 and nonsense constructs, dissolved in loading buffer (62 mm Tris-HCl, pH 6.8, 10% [v/v] glycerol, 2% [w/v] SDS, 0.7 m 2-β-mercaptoethanol, and 0.0012% [w/v] bromphenol blue) were used for protein SDS-PAGE.
Isolation of total protein from plant tissue, SDS-PAGE, western blotting, and immunodetection were performed as described by Stacy et al. (1999). A 1:1,000 (v/v) dilution of the PER1 antiserum and 1:750 (v/v) dilution of the AtPER1 antiserum were used. Equal loading in all lanes was confirmed by staining the membranes with Ponceau red before detection.
Nuclear Localization Assay
For investigation of intracellular localization of AtPER1, the cDNA was amplified by PCR using the primers 5′-GAA GAT CTA TGC CAG GGA TCA CAT A-3′ and 5′-GCT CTA GAT CAA GAC CTC TGT GTA C-3′, which include the restriction sites BglII and XbaI, respectively. The PCR product was cloned behind the GFP reporter gene between the BglII and XbaI sites of pAVA319 (von Arnim et al., 1998). The in-frame fusion was confirmed by DNA sequencing. Control constructs used were pAVA319 containing 35S::GFP (von Arnim et al., 1998), and 35S::HP1-GFP in the pKEx-327 vector (Baumbusch et al., 2001). Transient expression of the GFP fusion constructs was investigated through bombardment of onion epidermal cells as by Baumbusch et al. (2001) using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Hercules, CA; Stacy et al., 1996).
Constructs for Transformation of Arabidopsis Plants
The pPCV002 35S::Per1 construct was generated by digesting the Per1 cDNA (B15C; Aalen et al., 1994) from pBluescript by SalI and XbaI and ligation between the respective sites of pPCV002 35S (Koncz and Schell, 1986; Fig. 4).
The pKOH110 35S::AtPER1 antisense construct was generated by digesting the AtPER1 cDNA in pBluescript with EcoRI, blunting, and subsequent XbaI digestion. This fragment was ligated into the SmaI and XbaI sites between a 35S promoter and terminator cloned in pUC18. The 35S-AtPER1-ter cassette was excised using EcoRI and ligated into the EcoRI site of pKOH110 (Meza et al., 2001; Fig. 4).
The pPCV002 35S::AtPER1 RNAi construct was generated by digesting the AtPER1 cDNA in pBluescript with SmaI and EcoRI, and this fragment was ligated into the respective sites of a pBluescript plasmid harboring a fragment of the GUS (uidA) gene in the EcoRV site of the polylinker. A second AtPER1 cDNA was excised using SmaI and HindIII, and this fragment was ligated between the HindIII and a blunted XhoI site of pBluescript-AtPER1-GUS from the first cloning step. This resulting AtPER1-GUS-AtPER1 construct was digested by ApaI, followed by blunting and XbaI digestion before ligation between the SmaI and XbaI of pPCV002 35S (Fig. 4).
Production of Transgenic Arabidopsis Plants
For transformation of Arabidopsis plants, alternative methods were used; a root transformation protocol from Valvekens et al. (1988), modified by Mandal et al. (1993), a vacuum transformation method modified after Bechtold et al. (1993), or the floral-dip method of Clough and Bent (1998). Transformants were selected on MS-2 plates supplemented by 50 mg L–1 kanamycin.
Plant Material
Arabidopsis ecotype C24 (wt) and transgenic plants were cultivated in growth chambers at 22°C and 8 h of dark/16 h of light (100 μE m–2).
For germination tests, siliques were harvested at a specific stage; the first three brown, dried, and just opened siliques, were collected and after-ripened at room temperature. Seeds were sown on MS-2 plates at harvest and every 3rd d until 27 to 33 d of after-ripening. Germination was scored every 2nd d until 8 d after sowing.
For germination analyses during osmotic and oxidative stress, after-ripened seeds were imbibed on paper discs soaked in concentrations ranging from 10 to 175 mm NaCl, 5 to 500 μm MV, or 10 to 500 mm mannitol. Germination was scored every 2nd d until 8 d after sowing.
Seeds used in the same experiment were harvested from wt plants and transgenic plants grown at the same time and in the same growth chamber.
RNA Isolation and Northern-Blot Analysis
mRNA was extracted using magnetic Genoprep mRNA beads (Geno-Vision, Oslo, Norway). Total RNA isolation was performed after a modified protocol from Downing et al. (1992): Plant tissue was homogenized in lysis buffer (100 mm Tris-HCl, pH 8.0, 500 mm LiCl, 10 mm EDTA, pH 8.0, 1% [w/v] LiDS, and 5 mm dithiothreitol), extracted twice with phenol:chloroform:isoamylalcohol (25:24:1, v/v), and once with pure chloroform. Total RNA was precipitated in one-third volume of 8 m LiCl on ice for 14 to 20 h. After centrifugation (9,000 rpm, 30 min, and 4°C), the RNA pellet was resuspended in 100 μL of Tris-EDTA solution (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA, pH 8.0), precipitated in 150 μL of 5 m KAc, pH 6.5, for 3 to 5 h on ice, and centrifuged as above, followed by a final ethanol precipitation. For northern analyses 0.5 μg of mRNA or 10 μg of total RNA was separated on a denaturing RNA gel, blotted, and hybridized as described (Galau et al., 1986).
Hybridizing probes include AtPER1 (Haslekås et al., 1998) and Per1 (B15C; Aalen et al., 1994). An rRNA probe from Pyropyxis rubra was used as a loading control (Haslekås et al., 1998). Northern blots were scanned using a SnapScan 600. Signals were quantified using Gel-Pro Analyzer 2.0 software (Media Cybernetics, Silver Spring, MD).
Acknowledgments
We thank Prof. Gunter Reuter (Martin Luther University of Halle, Germany) for the pKEx-35S::HP1-GFP construct; Lars O. Baumbusch (University of Oslo, Norway) for providing the protocol for total RNA isolation from seeds; and Solveig Hauge Engebretsen, Roy Falleth, and Kirsten E. Rakkestad (University of Oslo, Norway) for technical assistance.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025916.
This work was supported by the University of Oslo (grant to C.H.) and by the Research Council of Norway (project no. 140429/130).
References
- Aalen RB (1999) Peroxiredoxin antioxidants in seed physiology. Seed Sci Res 9: 285–295 [Google Scholar]
- Aalen RB, Opsahl-Ferstad HG, Linnestad C, Olsen OA (1994) Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. Plant J 5: 385–396 [DOI] [PubMed] [Google Scholar]
- Baier M, Dietz KJ (1997) The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J 12: 179–190 [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In-planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis-thaliana plants. C R Acad Sci Ser III Sci Vie 316: 1194–1199 [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269: 27670–27678 [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Downing WL, Mauxion F, Fauvarque MO, Reviron MP, Devienne D, Vartanian N, Giraudat J (1992) A Brassica napus transcript encoding a protein related to the Kunitz protease inhibitor family accumulates upon water-stress in leaves, not in seeds. Plant J 2: 685–693 [PubMed] [Google Scholar]
- Espelund M, Debedout JA, Outlaw WH, Jakobsen KS (1995) Environmental and hormonal-regulation of barley late-embryogenesis-abundant (lea) messenger-RNAs is via different signal-transduction pathways. Plant Cell Environ 18: 943–949 [Google Scholar]
- Fontaine O, Huault C, Pavis N, Billard JP (1994) Dormancy breakage of Hordeum vulgare seeds: effects of hydrogen-peroxide and scarification on glutathione level and glutathione-reductase activity. Plant Physiol Biochem 32: 677–683 [Google Scholar]
- Galau GA, Hughes DW, Dure L (1986) Abscisic-acid induction of cloned cotton late embryogenesis-abundant (lea) messenger-RNAs. Plant Mol Biol 7: 155–170 [DOI] [PubMed] [Google Scholar]
- Goldmark PJ, Curry J, Morris CF, Walker-Simmons MK (1992) Cloning and expression of an embryo-specific mRNA up-regulated in hydrated dormant seeds. Plant Mol Biol 19: 433–441 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW (1996) Protein kinesis: nucleocytoplasmic transport. Science 271: 1513–1518 [DOI] [PubMed] [Google Scholar]
- Guan LM, Zhao J, Scandalios JG (2000) Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J 22: 87–95 [DOI] [PubMed] [Google Scholar]
- Haslekås C, Stacy RA, Nygaard V, Culianez-Macia FA, Aalen RB (1998) The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy Plant Mol Biol 36: 833–845 [DOI] [PubMed] [Google Scholar]
- Kang SW, Baines IC, Rhee SG (1998) Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem 273: 6303–6311 [DOI] [PubMed] [Google Scholar]
- Kim K, Kim IH, Lee KY, Rhee SG, Stadtman ER (1988) The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed-function oxidation system. J Biol Chem 263: 4704–4711 [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The Promoter of TL-DNA Gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–36 [DOI] [PubMed] [Google Scholar]
- Lee KO, Jang HH, Jung BG, Chi YH, Lee JY, Choi YO, Lee JR, Lim CO, Cho MJ, Lee SY (2000) Rice 1Cys-peroxiredoxin over-expressed in transgenic tobacco does not maintain dormancy but enhances antioxidant activity. FEBS Lett 486: 103–106 [DOI] [PubMed] [Google Scholar]
- Leprince O, Atherton NM, Deltour R, Hendry GAF (1994) The involvement of respiration in free-radical processes during loss of desiccation tolerance in germinating Zea mays L.: an electron-paramagnetic-resonance study. Plant Physiol 104: 1333–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, Hendry GAF, McKersie BD (1993) The mechanisms of desiccation tolerance in developing seeds. Seed Sci Res 3: 231–246 [Google Scholar]
- Lewis ML, Miki K, Ueda T (2000) FePer 1, a gene encoding an evolutionarily conserved 1-Cys peroxiredoxin in buckwheat (Fagopyrum esculentum Moench), is expressed in a seed-specific manner and induced during seed germination. Gene 246: 81–91 [DOI] [PubMed] [Google Scholar]
- Lim YS, Cha MK, Kim HK, Uhm TB, Park JW, Kim K, Kim IH (1993) Removals of hydrogen peroxide and hydroxyl radical by thiol-specific antioxidant protein as a possible role in vivo. Biochem Biophys Res Commun 192: 273–280 [DOI] [PubMed] [Google Scholar]
- Mandal A, Lang V, Orczyk W, Palva ET (1993) Improved efficiency for T-DNA-mediated transformation and plasmid rescue in Arabidopsis thaliana. Theor Appl Genet 86: 621–628 [DOI] [PubMed] [Google Scholar]
- Meza TJ, Kamfjord D, Hakelien AM, Evans I, Godager LH, Mandal A, Jakobsen KS, Aalen RB (2001) The frequency of silencing in Arabidopsis thaliana varies highly between progeny of siblings and can be influenced by environmental factors. Transgenic Res 10: 53–67 [DOI] [PubMed] [Google Scholar]
- Mowla SB, Thomson JA, Farrant JM, Mundree SG (2002) A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa Baker. Planta 215: 716–726 [DOI] [PubMed] [Google Scholar]
- Netto LE, Stadtman ER (1996) The iron-catalyzed oxidation of dithiothreitol is a biphasic process: hydrogen peroxide is involved in the initiation of a free radical chain of reactions. Arch Biochem Biophys 333: 233–242 [DOI] [PubMed] [Google Scholar]
- Peshenko IV, Novoselov VI, Evdokimov VA, Nikolaev YV, Shuvaeva TM, Lipkin VM, Fesenko EE (1996) Novel 28-kDa secretory protein from rat olfactory epithelium. FEBS Lett 381: 12–14 [DOI] [PubMed] [Google Scholar]
- Quesada V, Garcia-Martinez S, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol 130: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichi H, Demar JC (1990) Nonselenium glutathione-peroxidase without glutathione S-transferase activity from bovine ciliary body. Exp Eye Res 50: 513–520 [DOI] [PubMed] [Google Scholar]
- Stacy RAP, Munthe E, Steinum T, Sharma B, Aalen RB (1996) A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol 31: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Stacy RAP, Nordeng TW, Culianez-Macia FA, Aalen RB (1999) The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J 19: 1–8 [DOI] [PubMed] [Google Scholar]
- Valvekens D, Vanmontagu M, Vanlijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Duff RJ, Zeng Q, Oliver MJ (2000) Molecular architecture of bryophyte genes: putative polyadenylation signals in cDNA 3′-ends of the desiccation-tolerant moss Tortula ruralis. Bryologist 103: 44–51 [Google Scholar]
- Wood ZA, Poole LB, Karplus PA (2003a) Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653 [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schroder E, Harris JR, Poole LB (2003b) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40 [DOI] [PubMed] [Google Scholar]