Abstract
Mutations in the QUARTET loci in Arabidopsis result in failure of microspore separation during pollen development due to a defect in degradation of the pollen mother cell wall during late stages of pollen development. Mutations in a new locus required for microspore separation, QRT3, were isolated, and the corresponding gene was cloned by T-DNA tagging. QRT3 encodes a protein that is approximately 30% similar to an endopolygalacturonase from peach (Prunus persica). The QRT3 protein was expressed in yeast (Saccharomyces cerevisiae) and found to exhibit polygalacturonase activity. In situ hybridization experiments showed that QRT3 is specifically and transiently expressed in the tapetum during the phase when microspores separate from their meiotic siblings. Immunohistochemical localization of QRT3 indicated that the protein is secreted from tapetal cells during the early microspore stage. Thus, QRT3 plays a direct role in degrading the pollen mother cell wall during microspore development.
Pollen development in species with trinucleate pollen involves a complex series of characteristic changes in cell walls that have been carefully documented cytologically (Heslop-Harrison, 1971a, 1971b; Blackmore and Barnes, 1990; Owen and Makaroff, 1995). Immediately before meiosis, the pollen mother cells produce callose within their primary walls, and plasmodesmatal connections between adjacent pollen mother cells and between pollen mother cells and the tapetum disappear. The walls between adjacent pollen mother cells separate, and after cytokinesis, the microspores become surrounded by callose and synthesize a wall called the primexine. The tetrad wall is then degraded, and the microspores are released into the locule. At about this stage, the tapetal cell walls degrade (Owen and Makaroff, 1995). The microspores expand, whereas the outer layer of exine is deposited on their surface, apparently mediated by secretions from the tapetum. Another wall layer termed the intine is deposited between the exine and the plasma membrane by the microspores during a period of expansion. The microspores then undergo an asymmetric mitosis, yielding a generative cell and a vegetative cell. During this stage, the lumina of the exine wall is filled with a lipophilic material termed tryphine, which is secreted by the tapetum. The generative cell assumes a round morphology and becomes encased in the vegetative cell. The generative cell divides mitotically, giving rise to two sperm nuclei. After a second mitotic division, the pollen grain undergoes dehydration and is released upon anther dehiscence, a process involving changes in the walls of septum tissue (Keijzer, 1987).
Relatively little is known from genetic studies about the gene products that specifically mediate the changes in cell wall structure during pollen formation. A mutation in the dex1 gene of Arabidopsis exhibits a defect in primexine formation, resulting in male sterility (Paxon-Sowders et al., 2001). The DEX1 gene encodes a large, putative membrane-localized, plant-specific protein of unknown function with limited similarity to a hemolysin-like protein from Vibrio cholerae. Another male-sterile mutant, ms33, exhibits defects in the formation of the intine and tryphine deposition, but the molecular basis for the defect is not known (Fei and Sawhney, 2001). Although few mutants are known, there have been many reports of pollen or anther-specific expression of genes encoding cell wall-modifying enzymes (e.g. Brown and Crouch, 1990; Albani et al., 1991; Mu et al., 1994; Tebbutt et al., 1994; Ariizumi et al., 2002). Recent studies using DNA macroarrays resulted in identification of a large number of genes encoding enzymes involved in cell wall modification in anthers (Amagai et al., 2003). More than 20% of the genes identified in this way had sequence identity to enzymes known to be involved in synthesis or modification of pectin. This is consistent with many studies implicating pectin in the elaboration of the pollen wall (Southworth, 1990; Bedinger, 1992; van Aelst and van Went, 1992).
In the qrt (quartet) mutants of Arabidopsis, microspores fail to separate and remain attached in a characteristic tetrahedral cluster of four pollen grains (Preuss et al., 1994). This failure of separation is associated with the persistence of pectic components of the pollen mother cell wall around the microspores after callose degradation (Rhee and Somerville, 1998). The qrt1 and qrt2 genes have not been cloned. However, we previously speculated that persistence of the pectic components in the pollen mother cell wall could be due to a defect in the structural gene for a hydrolytic enzyme required to remove the pollen mother cell wall or to a defect in a mechanism that regulates the release of such hydrolytic enzymes. According to this hypothesis, the persistent pollen mother cell wall is envisioned as holding the microspores in the tetrahedral conformation, whereas exine deposition envelops the four microspores as they expand. This results in the fusion of the exine layer at the proximal ends of the microspores. Alternatively, it is possible that within the pollen mother cell wall, the pectic components of the microspore primexine wall are not modified or degraded to accommodate separation of the expanding microspores from each other.
We describe here the characterization of a novel class of qrt mutations, defined by the QRT3 locus. The QRT3 gene encodes a divergent class of polygalacturonase that is transiently expressed in tapetal cells during pollen formation.
RESULTS
Isolation of QRT3 Mutations
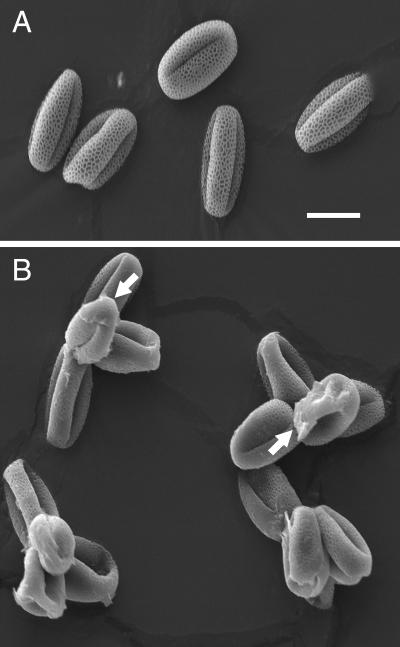
A collection of T-DNA insertional lines deposited in the Arabidopsis Biological Research Center by Ken Feldman were screened for the qrt phenotype as described (Preuss et al., 1994). Screening of approximately 30,000 plants from a population derived from an estimated 3,000 independent T-DNA lines yielded one novel qrt mutation, designated qrt3-1. In addition, a second allele, designated qrt3-2, was found by screening a small population of transgenic plants produced locally. Pollen grains of lines carrying the qrt3 mutations were released as tetrads (Fig. 1) as described previously for the qrt1 and qrt2 mutants (Preuss et al., 1994; Rhee and Somerville, 1998). In contrast to the qrt1 and qrt2 pollen, which were similar to wild type except for the fusion phenotype, the qrt3 pollen frequently had a layer of material deposited on the surface of the distal region of the pollen grains (indicated by arrows, Fig. 1). We hypothesize that this material is residue resulting from rupture of the pollen mother cell wall during the late stages of pollen expansion. The qrt3 mutants had no other noticeable phenotypes.
Figure 1.
Scanning electron micrograph of pollen from the wild type (A) and the qrt3-3 mutant (B). The arrows indicate material deposited on the pollen that appears to derive from the pollen mother cell wall. Bar = 10 μm.
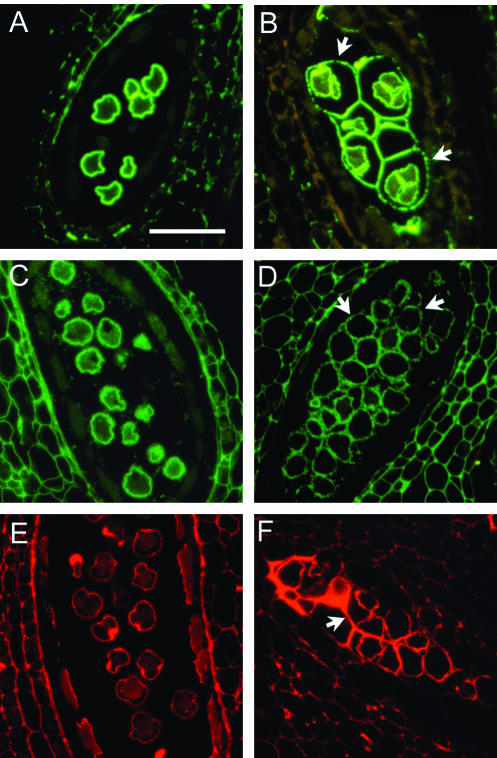
Antibodies that react with unesterified pectin, esterified pectin, and rhamnogalacturonan II (Knox et al., 1990; McCann et al., 1992; Williams et al., 1996) were used to detect pectins associated with developing pollen by immunofluorescence microscopy of tissue sections (Fig. 2). An antibody against unesterified pectin stained the primexine during the early microspore stage of pollen development in both the wild type and the qrt3-1 mutant. At this stage, the pollen mother cell wall in the wild type had been degraded, whereas the pollen mother cell wall remained intact in the mutant and was strongly stained with the antibody (Fig. 2). A similar observation was described for the qrt1 and qrt2 mutants (Rhee and Somerville, 1998). The residual pollen mother cell wall in the qrt3-1 also mutant stained with antibodies against esterified pectin and rhamnogalacturonan II (Fig. 2). In this respect, the qrt3 mutant was distinguishable from the qrt1 mutant, in which the pollen mother cell wall that remains around the microspores did not stain for unesterified pectin (Rhee and Somerville, 1998). This suggests a distinct basis for the quartet phenotype in the qrt1 and qrt3 mutants.
Figure 2.
Immunohistochemical staining of pectin associated with microspores of wild type (A, C, and E) and the qrt3-1 mutant (B, D, and F). Sections were stained with antibodies against unesterified pectin (A and B), methyl-esterified pectin (C and D), and RGII (E and F). In the wild type, the developing pollen grains are separated from each other. In the qrt3 mutant, the developing pollen are enclosed by residual pollen mother cell wall (arrows). Scale bar = 50 μm. The wild-type images were previously published by Rhee and Somerville (1998).
Isolation of the QRT3 Gene
Cosegregation of kanr and the qrt phenotype in the F2 population of a cross between qrt3-1 and wild type suggested linkage of the T-DNA and the qrt3-1 allele. Preliminary mapping experiments indicated that the kanr T-DNA was inserted into a single chromosomal locus (kanr:kans::103:35; X2 =0.01, P < 0.01) that was mapped to approximately 62.9 cM on chromosome 4. Analysis of Southern blots of qrt3-1 genomic DNA probed with T-DNA indicated that the T-DNA was inserted in a complex pattern including head-to-head and head-to-tail repeats (data not shown). Approximately 4.8 kb of genomic DNA flanking one end of the T-DNA insertion was isolated by plasmid rescue using SalI-digested qrt3-1 genomic DNA. Several cDNA clones were isolated by screening an Arabidopsis cDNA library made from inflorescence mRNAs (Weigel et al., 1992) using the flanking DNA as a probe. PCR amplification using primers against the cDNA and T-DNA revealed that the T-DNA was inserted within the open reading frame (ORF; Fig. 3).
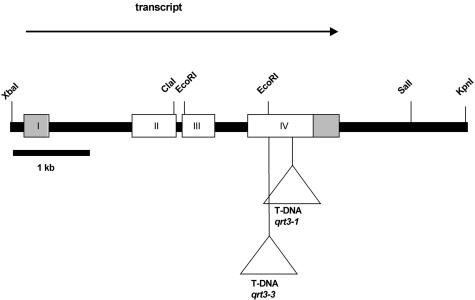
Figure 3.
Structure of the QRT3 locus At4G20050. The regions of the primary transcript present in a putative full-length cDNA are shown as boxes. The 5′ and 3′ non-translated regions are shown as gray boxes. The primary transcript is processed to remove three introns, one of which is in the 5′-non-translated region. The site of insertion of the TDNA inserts corresponding to the qrt3-1 and qrt3-3 mutations are indicated below the figure.
The sequence of the 1,941-bp cDNA clone pSYRC8 was deposited in GenBank as accession number AY268942. The clone contained 343 nucleotides of 5′-untranslated sequence, a 1.4-kb ORF, and 153 nucleotides of 3′-untranslated sequence. To determine if the clone was full length, 5′-RACE was used to amplify poly dC-tailed first strand cDNA from flowers. PCR amplification of tailed cDNAs using a primer corresponding to nucleotides 559 to 589 of the cDNA insert in pSYR8 produced a fragment of approximately 500 nucleotides. Thus, we concluded that pSYR8 contained an essentially full-length cDNA.
Conceptual translation of the pSYRC8 sequence in three frames revealed a single long ORF with a putative translation initiation codon at nucleotide 344. This ATG was flanked by AACCATGGA, which is similar to the most frequent ATG context sequence in plant genes, AACAATGGC (Joshi, 1987; Lütcke et al., 1987; Fütterer and Hohn, 1996). Assuming this ATG to be the initiation codon, the ORF is 481 amino acids in length, and the predicted molecular mass of the protein is 51 kD with a pI of 5.75.
Sequencing of approximately 8 kb of genomic DNA spanning the QRT3 locus resulted in complete agreement with the sequence deposited by the Arabidopsis Genome Initiative as gene At4G20050. Comparison of the genome sequence with the cDNA sequence indicated that the QRT3 primary transcript is interrupted by three introns (Fig. 3). The first intron is 922 nucleotides in length and separates the leader sequence from the coding region, which starts eight nucleotides from the 5′ border of the intron.
Confirmation and Complementation of qrt3 Mutations
To verify that the gene identified by the T-DNA insertion in At4G20050 was, in fact, qrt3, we obtained an additional mutant allele from the collection of sequenced T-DNA insertions produced by Joe Ecker and colleagues (http://signal.salk.edu/tabout.html). The seed pool SALK_093266, which was derived from a plant containing a T-DNA insertion in the fourth exon of the gene At4G20050 (Fig. 3), was found to segregate plants with the qrt phenotype. This mutant allele was designated qrt3-3. Similarly, seed pool SALK_052045, which has an insert at or near the location of the insert in SALK_093266, was also found to segregate a qrt3 phenotype. These results were consistent with the conclusion that At4G20050 is the qrt3 locus.
To test whether the genomic region containing the QRT3 locus was sufficient for complementation, a cosmid clone (pSYR19) carrying the entire QRT3 coding sequence and approximately 10 kb of 5′ genomic sequence was transformed into plants carrying the qrt3-2 allele. Of 19 independent transgenic plants, four had complete complementation of the qrt phenotype, and three exhibited partial complementation. Additionally, an 11-kb SalI-SstI fragment containing approximately 3.7 kb upstream and 3.2 kb downstream of the QRT3 coding sequence was subcloned into a Ti plasmid, pIW131, to produce plasmid pSYR18. Plasmid pSYR18 was then mutagenized with Tn3, and two plasmids with insertions in the second exon were recovered and named pSYR14 and pSYR15. The plasmids pSYR14, pSYR15, and pSYR18 were used to transform the qrt3-2 mutant. Seven of 12 pSYR18 transgenic lines showed complementation of the qrt phenotype, but none of the 34 transformants obtained with pSYR14 or pSYR15 showed complementation. These results confirmed that the gene At4G20050 is the QRT3 gene.
QRT3 Resembles a Polygalacturonase from Peach (Prunus persica)
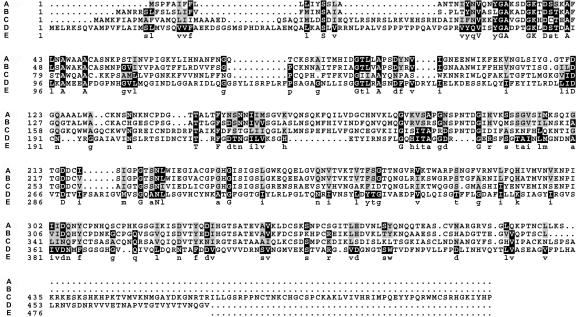
Comparison of the QRT3 amino acid sequence with the nonredundant set of protein sequences in GenBank and Swiss-Prot databases using the BLAST program identified the TAPG2 endopolygalacturonase (PGase) from peach as the most similar non-Arabidopsis protein to QRT3 (BLAST score of 80 and probability of 2.3 × 10–6). A multiple alignment of the most closely related subset of the PGase gene family showed that QRT3 is approximately 30% similar to these PGases (Fig. 4). The sequence similarity of QRT3 to the PGases extends throughout the protein, but conservation is most evident in two regions of the protein between amino acids 75 and 113 and amino acids 362 and 392.
Figure 4.
Alignment of QRT3 with sequences of polygalacturonases. A, Tap2g (GenBank accession no. U70480) was isolated from the abscission zone of tomato (Lycopersicon esculentum). B, PGase (GenBank accession no. X76735) was from peach. C, CRYJ2 was from Japanese cedar (Cryptomeria japonica). D, QRT3. E, Consensus sequence.
Glycosyl hydrolases have been grouped into families based on their primary sequence (Henrissat et al., 2001). The QRT3 protein is sufficiently divergent from known enzymes that it was not listed in the CAZY database of carbohydrate active enzymes (Coutinho and Henrissat, 1999; http://afmb.cnrsmrs.fr/CAZY/). However, the related TAPG2 polygalacturonase from peach is classified as being in family 28 of the glycosyl hydrolases.
Analysis of the QRT3 sequence for potential protein localization site(s) using the PSORT algorithm (http://psort.nibb.ac.jp) indicated that QRT3 has a putative N-terminal signal peptide similar to proteins that are sorted through the vesicular pathway. The proposed site of cleavage in QRT3 is at position 28 of the amino acid sequence. In addition, PSORT assigned a high probability (score 0.77) that QRT3 was a secreted protein.
Comparison of the amino acid sequence of QRT3 with all Arabidopsis proteins indicated that only one other protein in Arabidopsis has significant sequence similarity. That protein, which exhibited 44.7% sequence identity, was encoded by At4G20040, which adjoins QRT3 in the Arabidopsis genome.
Localization of QRT3 Expression
Measurements of the amount of QRT3 gene expression in various tissues was obtained from experiments in which total RNA from leaves, roots, stems, and flowers was hybridized to Affymetrix ATH1 DNA chips (Table I). Analysis of the expression of the floral homeotic gene AGAMOUS was used as a flower-specific control, and the unique GAPC gene for cytosolic glyceraldehydes-3-phosphate dehydrogenase (Shih et al., 1991) was used as measure of ubiquitous gene expression. The results of this experiment indicated that QRT3 mRNA was unambiguously detected above background in extracts from flowers where it was expressed at a similar level to the AGAMOUS gene and at about 10% of the level of the GAPC gene. In the other tissues, both QRT3 and AGAMOUS had signals that were about 1% of the level of the GAPC gene. QRT3 expression in RNA samples from stem tissues was approximately 17% as high as in flowers, raising the possibility that it may be involved in a specialized cell separation function in that tissue.
Table I.
Analysis of QRT3 expression in various tissues
Plants were grown on either agar-solidified medium or in soil, as indicated. The nos. represent the expression score reported by Affymetrix software for each gene. The gene for cytosolic glyceraldehydes-3-phosphate dehydrogenase (gapC; At3G04120) was used as a control for a unique gene that is expressed at a moderately abundant level in all cells (Shih et al., 1991). The expression of agamous (At4G18960) was included as an example of a gene with flower-specific gene expression. The average background score was 56.8.
| Tissue | Replicates | GAPC | AG | QRT3 |
|---|---|---|---|---|
| Leaves (15 d), agar | 3 | 6,860 | 42 | 69 |
| Leaves (14 d), soil | 4 | 8,598 | 129 | 88 |
| Leaves (50 d), soil | 1 | 8,597 | 43 | 32 |
| Flowers (29 d), soil | 4 | 7,907 | 853 | 843 |
| Stems (29 d), soil | 4 | 8,597 | 56 | 191 |
| Roots (15 d), agar | 3 | 10,730 | 153 | 39 |
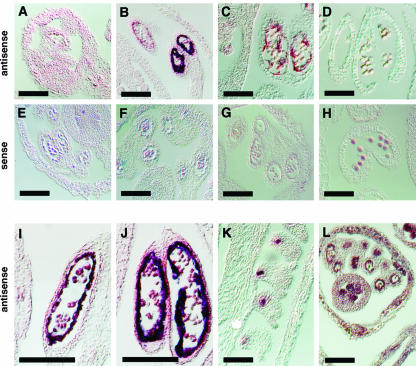
To determine the pattern of QRT3 expression in the inflorescence tissue, in situ hybridization of QRT3 mRNA transcripts in developing inflorescence was carried out using an antisense QRT3 transcript as a probe. As shown in Figure 5, the QRT3 expression pattern is specific both temporally and spatially in the anthers. The gene is expressed during the tetrad stage in the tapetum (Fig. 5, B, I, and J). Expression was not detectable after the microspores were released from the tetrad wall (Fig. 5, C and D). There was also a strong expression in the ovules of open flowers (Fig. 5K). There is no apparent phenotype associated with the qrt3 mutation in the ovules at the histological level or in terms of the ovule function. Sections probed with sense transcripts of QRT3 on similar developmental stages failed to detect a signal (Fig. 5, E–H). An antisense transcript of a translation initiation factor (eIF4A; Metz et al., 1992; Taylor et al., 1993) was used as a positive control and showed expression in all tissues (Fig. 5L).
Figure 5.
In situ hybridization of antisense and sense QRT3 transcripts to sections of flowers at various stages of microsporogenesis. A to D, Hybridization of QRT3 antisense probe to sections of developing flowers at various stages. E to H, Hybridization of QRT3 sense probes to sections of flowers at the same stages shown in A to E. I and J, Enlarged images of hybridization of antisense QRT3 probe to developing anthers at the tetrad stage. K, Hybridization of QRT3 antisense to ovules of mature flowers. L, Hybridization of antisense eIF4A transcript to section of developing flower at tetrad stage was used as a positive control. The following developmental stages were characterized: A and E, PMC stage buds; B, F, I, and J, tetrad stage buds; C and G, early microspore stage buds; and D and H, late microspore stage buds. Scale bars = 100 μm.
Detection of QRT3 Protein
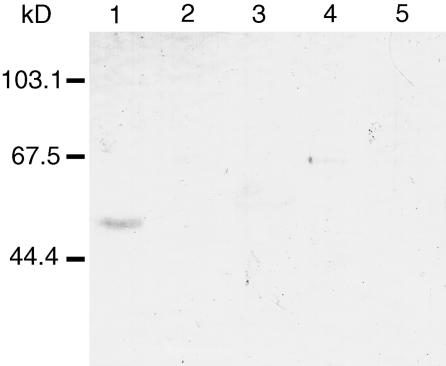
To facilitate characterization of the QRT3 protein, polyclonal antibodies against QRT3 were raised against a his-tagged fusion protein produced in Escherichia coli. To determine the gross expression pattern of QRT3 protein in Arabidopsis, western blots containing protein extracts from wild-type flower buds, open flowers, leaves, and roots were prepared and incubated with antisera. The antibody detected a protein of approximately 51 kD on western blots of protein extracts from unopened flower buds (Fig. 6). QRT3 protein was not detectable in extracts from any other tissue type. The protein was not detectable in extracts from qrt3-1 flower buds (data not shown), indicating that synthesis of QRT3 is abolished in the line homozygous for qrt3-1 (SYR1398).
Figure 6.
Western blot probed with antibody against QRT3. Approximately 20 μg of extracted protein from various tissues of 4-week old plants was loaded in each lane, blotted to nitrocellulose, and detected with an antibody against QRT3 at a dilution of 1:10,000 (v/v). Lane 1, Wild-type unopened flower buds; lane 2, wild type flowers; lane 3, wild-type expanded leaves; lane 4, wild-type roots; lane 5, qrt3-1 unopened flower buds. The positions of Mr standards are indicated on the left.
Immunohistochemical Localization of QRT3
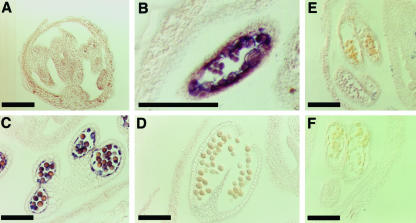
The putative signal peptide of QRT3 and the expression of QRT3 in the tapetum suggested that QRT3 might be secreted into the anther locule. Immunohistochemical localization of QRT3 on sections of developing anthers showed that the protein was detectable in the microspores at the onset of their release from the tetrad wall (Fig. 7B, arrows) and accumulated on the surface of microspores during their expansion. Antibody binding decreased during microspore maturation. It is not known whether the decrease in antibody binding is a result of protein turnover, a specific degradation pathway, or occlusion of the antigen. Pre-immune sera did not bind to the anther tissue of a comparable developmental stage (Fig. 7E). Likewise, omitting the primary antibody abolished staining by the alkaline-phosphatase conjugated secondary antibody (Fig. 7F), indicating specific reaction with the QRT3 antigen.
Figure 7.
Immunohistochemical localization of QRT3 in wild-type flower buds. Ten-micrometer sections of developing anthers were incubated with anti-QRT3 sera at a dilution of 1:1,000 (v/v). A, Tetrad stage; B, early microspore stage; C, late microspore stage; D, mature pollen; E, early microspore stage probed with pre-immune sera; F, early microspore stage probed with secondary antibody. Bars = 100 μm.
The localization of QRT3 to the microspores indicates that the protein is secreted from the tapetum where it is expressed.
Heterologous Expression of QRT3
To test whether QRT3 exhibited pectinase activity, attempts were made to overexpress QRT3 in Arabidopsis by transformation of wild-type plants with a construct in which the cDNA was placed behind the 35S promoter. However, none of the transgenic lines recovered expressed the protein in vegetative tissue (data not shown). It is possible that expression of QRT3 may be deleterious in some cell types.
QRT3 was produced heterologously using E. coli and yeast (Saccharomyces cerevisiae) as hosts. Expression of the full-length QRT3 ORF or a truncated version lacking the first 28 amino acids corresponding to the putative signal peptide in E. coli resulted in accumulation of an insoluble form of the protein (data not shown). No attempt was made to solubilize or assay the protein from E. coli. In contrast, expression of the full-length QRT3 protein and a derivative missing the N-terminal 28 amino acids resulted in accumulation of a soluble protein in yeast (data not shown). The two proteins exhibited similar apparent molecular mass, suggesting that the yeast cells may have removed the putative signal peptide from the full-length protein.
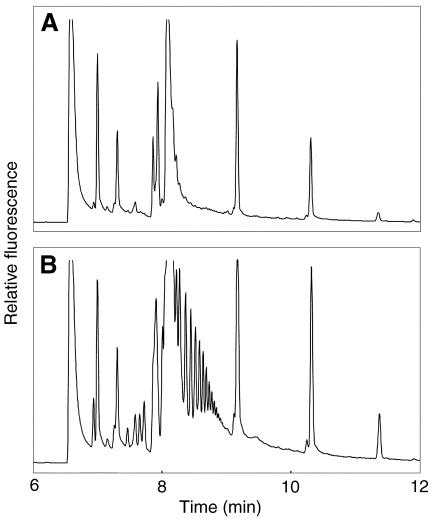
The recombinant QRT3 enzyme was assayed for activity by incubating the yeast extracts with poly-GalUA, then derivatizing the reaction products with a charged flurophore and resolving the various fluorescent species by capillary electrophoresis on an instrument with a laser-induced fluorescence detector (Fig. 8). Extracts from yeast cells expressing QRT3 exhibited polygalacturonase activity as indicated by the appearance of a series of low-molecular mass fragments (Fig. 8) that were absent in reactions containing protein extracts from yeast carrying the vector.
Figure 8.
Electropherograms of reaction products from QRT3-mediated hydrolysis of poly-GalUA. Poly-GalUA was incubated with protein extracts from control yeast cells (A) or cells expressing the QRT3 enzyme (B) for 12 h, and then the reaction products were resolved by capillary electrophoresis. The series of peaks between approximately 8 and 9 min in the QRT3 reaction products correspond to the elution times of oligo-GalUA standards containing two to eight residues. Other peaks correspond to contaminants in the dye and by-products of the derivatization reactions.
DISCUSSION
During pollen development, many tissue types including the tapetum, pollen mother cells, microspores, and the septum undergo cell wall modification (Stieglitz, 1977; Neelam and Sexton, 1995; Owen and Makaroff, 1995). A number of pectin-degrading enzymes and cDNA clones encoding pectin degrading enzymes have been isolated from anthers of petunia (Petunia hybrida), tobacco (Nicotiana tabacum), Brassica napus, sweet pea (Lathyrus odoratus), and Arabidopsis (Brown and Crouch, 1990; Sexton et al., 1990; Albani et al., 1991; Mu et al., 1994; Tebbutt et al., 1994; Neelam and Sexton, 1995). These enzymes include polygalacturonases, pectic lyases, pectin methylesterases, polymethylgalacturonases, and rhamnogalacturonases. Results presented here indicate that despite the apparent abundance of pectin modifying enzymes expressed during pollen development, the polygalacturonase encoded by the QRT3 gene performs a unique biological function.
Most of the genes encoding pectin degrading enzymes are expressed in mature pollen and in pollen tubes, suggesting a possible role in wall degradation of the pistil tissue during pollen tube penetration and subsequent growth of the tube (Brown and Crouch, 1990). During earlier stages of microspore development, β-1,3-glucanase and endo-cellulase activities are evident (Stieglitz, 1977; Sexton et al., 1990; Neelam and Sexton, 1995). β-1,3-glucanase is required to degrade the callosic wall surrounding the microspores during the tetrad stage (Worrall et al., 1992). The specific role of the cellulase is less defined (Sexton et al., 1990; Neelam and Sexton, 1995). However, degradation of the tapetal and pollen mother cell wall occurs during microspore release, suggesting that cellulase may be involved in degrading these walls (Sexton et al., 1990; Neelam and Sexton, 1995). With the exception of β-1,3-glucanase, the specific roles of cell wall-degrading enzymes and the proteins that resemble cell wall-degrading enzymes expressed during pollen development have not been clearly defined previously.
Immunofluorescence studies of cell wall composition during wild-type Arabidopsis microsporogenesis demonstrated that pectic components of the pollen mother cell wall persist around the microspores until the latter stages of pollen development, when they are apparently degraded (Rhee and Somerville, 1998). The qrt1 and qrt2 mutants fail to degrade the pollen mother cell wall; therefore, the pollen grains do not separate and are held together as a characteristic cluster of four attached pollen grains (Rhee and Somerville, 1998). A similar defect in degradation of the pectic components of the pollen mother cell wall was apparent from immunofluorescence studies of the qrt3 mutants described here. Thus, the QRT3 gene product plays a role in degradation of the pollen mother cell wall.
The QRT3 gene was originally identified by recovery of a TDNA-tagged qrt3 allele. The characterization of three additional qrt3 TDNA alleles, transgenic complementation studies, and the loss of the QRT3 protein in the qrt3-1 mutant support the conclusion that gene At4G20050 corresponds to the QRT3 gene. The QRT3 protein exhibited weak sequence similarity to PGases from peach and tomato and a polymethylgalacturonase from cedar. Polygalacturonases are enzymes that degrade the homogalacturonan region of pectin and are found in saprophytic and pathogenic bacteria, fungi, and higher plants (Brown and Crouch, 1990; Tucker and Seymour, 2002). The activity of polygalacturonases is associated with a number of processes including fruit softening (Kramer et al., 1989; Hadfield and Bennett, 1998) and pollen tube growth (Brown and Crouch, 1990). Polygalacturonases from bacteria, fungi, and plants are diverse in sequence (Stratilová et al., 1993). Fungal PGases may show as little as 36% sequence similarity to each other (Bussink et al., 1992). There are 23 amino acid residues that are strictly conserved among PGases from different organisms, and these residues are considered to be a part of the catalytic site (Rexová-Benková and Mracková, 1978). One of these residues, His-234 in the enzymes from Fusarium moniliforme, has been shown to be essential for activity (Caprari et al., 1996). The region of high similarity between QRT3 and the PGases from tomato and peach and the polymethylgalacturonase from cedar includes the domain that is highly conserved in polygalacturonases. The highly conserved region surrounding the essential His residue lies within one of the conserved domains in QRT3 (amino acids 362–392, Fig. 3).
To test whether QRT3 exhibited pectinase activity, QRT3 protein was produced using E. coli and yeast as hosts. Expression of the full-length QRT3 ORF, or a truncated version lacking the first 28 amino acids corresponding to the putative signal peptide, resulted in accumulation of soluble proteins in yeast. Preliminary results from enzyme assays using QRT3-containing extracts produced from yeast indicated that QRT3 exhibits hydrolytic activity against poly-GalUA. Additional analysis of enzyme function using more sophisticated assays will be necessary to fully characterize the catalytic activity of the QRT3 protein.
Several lines of evidence indicated that expression of the QRT3 gene is restricted to the tapetum during late stages of microsporogenesis. Analysis of gene chip hybridizations to RNA from various tissues showed that the QRT3 transcript is readily detectable in preparations from flowers, is weakly expressed in stems, but is absent from other tissues. Similarly, western-blot analysis using tissue from unopened flower buds, open flowers, roots, and leaves showed that QRT3 accumulates specifically in the unopened flower buds. In situ hybridization of the QRT3 transcript performed on developing anthers showed that the QRT3 mRNA is expressed specifically in the tapetum during the tetrad and early microspore stages of pollen development. Immunohistochemical localization of QRT3 in developing anthers indicated that the protein is secreted into the locule and is located on the microspores during the early microspore stage. The protein accumulates throughout the expansion period of the microspores and disappears as the microspores develop into pollen grains. This localization pattern is consistent with the predicted signal peptide of the QRT3 protein and indicates that the tapetal cells secrete the protein. The localization of QRT3 to the microspores is also consistent with the mutant phenotype of persistent pollen mother cell wall. The tapetal expression pattern is consistent with the genetic evidence that the qrt phenotype is sporophytically controlled (Preuss et al., 1994). In addition, the timing of QRT3 expression is consistent with the cytological data that indicates a defect in wall degradation at the time of microspore release. The apparent transient expression pattern in the anther suggests that QRT3 plays a specific role at a specific stage of microspore development. The disappearance of the protein by the completion of microspore development suggests the presence of a mechanism for removing the potentially damaging QRT3 protein after it has fulfilled its function. In this respect, the QRT3 protein may be a useful experimental tool for investigating the lifecycle of enzymes that modify cell wall polysaccharide structure.
From these studies, we conclude that the QRT3 protein plays a direct and specific role in degradation of the pectic polysaccharides of the pollen mother cell wall. We speculate that this defect causes the developing pollen grains to remain tightly appressed because of the mechanical constraint imposed by the incompletely decomposed pollen mother cell wall. This leads to inter-digitation of the polysaccharides of the developing cell walls of the pollen grains, resulting in fusion of the exine layers at the point of appression. Intermixing of cell wall polysaccharides leading to abnormal cell fusion has previously been invoked to explain the phenotype of the fiddlehead and other mutants (Pruitt et al., 2000). Additional biochemical studies of the QRT3 enzyme will be required to understand the precise enzymatic function of the protein, which shows only weak homology to any other enzyme. These results provide a partial answer to the question of why plants have a large number of putative pectin-modifying enzymes (Hadfield and Bennett, 1998). Presumably, the other qrt mutants also define genes encoding enzymes involved in degradation of the pollen mother cell wall or the developmental factors that control expression of cell wall-degrading enzymes.
MATERIALS AND METHODS
Plant Material
Arabidopsis plants were grown under natural light in greenhouses at a mean temperature of 23°C under a 16-h photoperiod. For sterile growth of seedlings, seeds were surface sterilized and plated on 0.8% (w/v) agar supplemented with 1× Murashige and Skoog basal culture salts (Sigma, St. Louis). Seedlings were grown under continuous fluorescent illumination at 60 μmol m–2 s–1 at 21°C. The qrt3-1 mutant line SYR1398 was isolated by screening T-DNA insertional lines of the Ws ecotype (Arabidopsis Biological Resource Center, Ohio State University, Columbus) for the quartet phenotype as described (Preuss et al., 1994). The qrt3-2 line SYR1399 was identified as a mutation in a local population of T-DNA transformants of the RLD ecotype. The T-DNA in SYR1399 was lost by segregation so that the line is kanamycin sensitive. The qrt3-3 mutant line SYR1460 was obtained from the Columbia ecotype line SALK_093266, generously donated by Joe Ecker (Salk Institute, La Jolla, CA) to the Arabidopsis Biological Resource Center. The qrt3-4 line SYR1461 was identified in pool SALK_052045. The sequence of DNA flanking the T-DNA inserts in the SALK lines was published online by Ecker and colleagues and is available through The Arabidopsis Information Resource (http://www.arabidopsis.org).
The presence of the kanamycin resistance selectable marker of the T-DNA mutants was scored on 0.8% (w/v) agar containing Murashige and Skoog salts (Sigma) and 100 μg mL–1 kanamycin.
Scanning Electron Microscopy of Pollen
For electron microscopy of developing pollen, anthers were spread on stubs coated with sticky tape to secure the specimen onto the stubs and coated with gold/palladium to 8 nm using a Polaron E5400 sputter coater (Quorum Diagnostics Corporation, New Haven, East Sussex, UK). Observations were made using an SEM525 microscope (Philips, Eindhoven, The Netherlands) at 20 kV.
Immunohistochemical Localization of Pectic Polysaccharides
Pectic polysaccharides were visualized by indirect immunofluorescence as described by Rhee and Somerville (1998). The JIM5 and JIM7 antibodies, which recognize unesterified pectin and methyl-esterified pectin, respectively (Van den Bosch et al., 1989; Knox et al., 1990) were gifts from Paul Knox (Leeds Institute for Plant Biotechnology, Leeds) and Maureen McCann (Purdue University, West Lafayette, IN). The CCRC R1 antibody, which recognizes rhamnogalacturonan II (Williams et al., 1996), was a gift from Michael Hahn (Complex Carbohydrate Research Center, Athens, GA).
Cloning the QRT3-1 Locus
Plasmid rescue of the DNA flanking the T-DNA insertion was carried out as described (Meyer et al., 1996). In brief, genomic DNA was prepared using a CsCl method (Ausubel et al., 1989). Approximately 5 μg of genomic DNA was digested with SalI, extracted with phenol: chloroform (1:1 [w/v]), precipitated with ethanol, resuspended in a volume of 1 mL containing 100 units of T4 ligase (Roche Diagnostics Corporation, Indianapolis), and incubated overnight at 14°C. The DNA was precipitated, resuspended in 20 μL of water, and 3 μL was electroporated into 100 μL of Escherichia coli DH5a cells at 2.5 kV. A clone containing approximately 4.8 kb of the flanking DNA was designated pBRqrt.
A partial cDNA clone of QRT3, pSYRC2, was obtained by using pBRqrt as a probe to screen an inflorescence-specific cDNA library (Weigel et al., 1992) obtained from the Arabidopsis Biological Resource Center. Identification of a full-length cDNA clone, pSYRC8, was accomplished by the 5′-RACE method (Fox et al., 1996). To obtain genomic DNA clones, pBRqrt was used as a probe to screen a kGEM11 Columbia genomic library (J. Mulligan and R. Davis, unpublished data) as described (Ausubel et al., 1989).
Alignment of DNA and protein sequences with those of previously identified genes was carried out using the GCG analysis program (Genetics Computer Group, Madison, WI). The sequence of the full-length cDNA and the genomic sequence of QRT3 were deposited in GenBank as accession numbers AY268942 and AY268941, respectively
Complementation of the qrt3-2 Mutation
Cosmid clone pSYR19, carrying the QRT3 gene and approximately 10 kb of 5′-flanking DNA, was identified by screening a library made from the Columbia ecotype in the vector pBIC20 (Meyer et al., 1994). The clone was introduced into Agrobacterium tumefaciens GV3101 by electroporation, and plants were transformed by the in floral dip method (Clough and Bent, 1998).
To determine whether the At4G20050 gene present on pSYR19 was responsible for the complementation of the qrt phenotype, the gene was disrupted by a Tn3 insertion, and the resulting cosmid was transformed into the qrt3-2 mutant. The pBIC20 vector that was used to build the cosmid library is a derivative of pLAFR3 (Staskawicz et al., 1987) and contains a mobilization gene that enables triparental mating. Tn3 mutagenesis was carried out by using the pHoHoGUS/pSShe Tn3 delivery system (Stachel et al., 1985; Barnett and Long, 1997). Colonies were screened by PCR using a primers against the GUS gene in TN3 and primers against At4G20050. Two cosmid clones that contained a TN3 insertion into the gene, designated pSYR14 and pSYR15, were used to transform qrt3-2 mutant plants.
A second complementing construct was produced by subcloning a 10-kb SstI-PstI fragment containing QRT3 and approximately 3.7 kb of 5′-flanking DNAand 3 kb of 3′-flanking DNA into pBS SK+, yielding pSYR13. An 11-kb SalI-SstI fragment of pSYR13 was subcloned into binary Ti plasmid pIW131 to produce pSYR18, which was then transformed into plants homozygous for the qrt3-2 mutation as described above. Plasmid pIW131, obtained from John Vogel (U.S. Department of Agriculture, Albany, CA) is a derivative of pBI121 in which the BAR gene is used for selection in plants, and the polylinker contains sites for SalI, SmaI, SpeI, and XhoI.
In Situ Hybridization
To make RNA probes from QRT3, a 1.0-kb HindIII fragment of pSYRC2 was subcloned into pBS KS+ vector (Stratagene, San Diego), yielding pSYR3. Plasmid p117S-34, which contains 300 nucleotides of eIF4A, translation initiation factor 4A from Arabidopsis (Metz et al., 1992; Taylor et al., 1993), was used as a control. In situ hybridization was carried out as described (Jackson, 1991).
Expression of QRT3 in E. coli
To express a full-length and the putative mature protein, the QRT3 transcript (pSYRC2) was amplified by PCR using the following primer sets: EXP4-5′ (gggatcccgggcctctagaaaccatggagctaaggaaatctcaagtggccatgcctg) and EXP4-3′ (ggagctcgtcgacccttacaggaaaaataacaaat) and EXP5-5′ (gggatcccgggcctctagaaaccatggagaaagattcaggttcaatgtcccct) and EXP4-3′, respectively. The EXP4-5′ and EXP5-5′ primers contained restriction sites for BamHI, SmaI, and XbaI. The EXP4-3′ primer contained restriction sites for SstI and SalI. The PCR-amplified fragments were cut with BamHI and SstI and cloned into the BamHI and SstI sites of the pQE31 vector (Qiagen, Valencia, CA), resulting in a translational fusion of the protein with six His residues at the N terminus. The plasmid containing the full-length and the mature forms were designated pEXP4 and pEXP5, respectively.
The QRT3 fusion protein was purified from E. coli by chromatography on nickel chelate columns as described in the pET system manual (catalog no. 69755-1, Novagen, Madison, WI). The purified QRT3 protein was further purified by SDS-PAGE and then used as an antigen to raise polyclonal antibodies. Antibody production was carried out by Cocalico Biologicals, Inc. (Reamstown, PA).
Expression of QRT3 in Yeast (Saccharomyces cerevisiae)
The oligonucleotide primers used to construct E. coli expression plasmids were used to PCR amplify the QRT3 sequence. The PCR product was cut with BamHI and SalI and ligated into BamHI and XhoI sites of pYES2 (Stratagene). The plasmid containing the full-length protein was designated as pYES7, and the plasmid containing the predicted mature protein was designated as pYES8. The constructs were electroporated into a yeast strain INVSC2. To induce expression of QRT3, the cells were grown in URA-complex medium containing 2% (w/v) Gal (Sigma, catalog no. G-6404) grown for 4 d at 30°C.
For protein extraction cells were harvested by centrifugation at 5,000g for 10 min, washed once in sterile water, and chilled to 4°C. The cells were resuspended in an equal volume of 25 mm sodium acetate buffer (pH 5). The cells were lysed by two passages through a French press at 16,000 psi. The cell debris was removed by centrifugation at 15,000 rpm for 15 min. The supernatant was dialyzed against 25 mm sodium acetate buffer (pH 5.0).
Immunohistochemical Localization of QRT3
Inflorescence tissue was fixed, embedded, and sectioned as described for in situ hybridization. Immunohistochemical localization was carried out as described by Smith et al. (1992). The primary antibody was pre-adsorbed with acetone-powdered leaf tissue as described elsewhere (Conlon and Rossant, 1992). For pre-adsorbing the primary antibodies, the acetone-powdered tissue was added to the primary antibody to a final concentration of 2% (w/v) and incubated at 4°C with gentle agitation for 2 h. The mixture was centrifuged briefly, and the supernatant was used for immunohistochemical localization at a dilution of 1:1,000 (v/v). The slides were examined using a DMRB (Leitz, Midland, ON) microscope with Normarski optics.
Enzyme Assay
For the assays, 10 μL of 1% (w/v) poly-GalUA was incubated with 50 μl of dialyzed protein extract (2.5 mg mL–1 protein in 25 mm sodium acetate [pH 5.0]) for 12 h at 25°C. The reactions were dried under vacuum, solubilized in 10 μL of 15% (v/v) acetic acid containing 25 mg mL–1 8-aminopyrene-1,3,6-trisulfonic acid (Molecular Probes, Eugene OR), incubated at 55°C for 90 min, diluted 1:500 (v/v) with water, and resolved by capillary electrophoresis on a 50-μm i.d.× 65-cm N-CHO-coated capillary loaded with eCAP carbohydrate separation gel buffer-N (Beckman Coulter, Fullerton CA) at 25 kV. Labeled products were detected by exciting with a 488-nm argon-ion laser and measuring fluorescence at 520 nm.
DNA Chip Analysis
Col-0 plants were grown at a density of four plants per 5-inch pot. All tissue was harvested in the middle of the photoperiod. One set of plants was grown in growth chambers under 125 μmol m–2 s–1 of fluorescent irradiation with a 16-h photoperiod at 25°C during the day and 20°C during the night. A second set was grown under natural light with a similar temperature regime. Expanding leaves were obtained from plants at 12 to 14 d. Leaves were also harvested on the 1st d of inflorescence formation and at 50 d after planting. Stems and flowers were harvested from plants when the inflorescence was 6 inches high. For stem samples, siliques and pedicels were removed from the lower 5 inches of the inflorescence stem, and the remainder was used for RNA preparation. Flower samples were composed of the flowers in which the petals remained attached plus unopened buds. Roots were from 14-d-old plants grown under continuous fluorescent illumination (approximately 30 μmol m–2 s–1) on the surface of vertically oriented 0.8% (w/v) agar medium containing Murashige and Skoog salts. RNA was extracted and hybridized to AtH1 DNA chips (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions.
Distribution of Materials
Upon request, all novel materials described here will be made available in a timely manner.
Acknowledgments
We gratefully acknowledge the contribution of Joe Ecker and colleagues for providing indexed TDNA insertion lines. Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation. We are also grateful to Paul Knox, Maureen McCann, and Michael Hahn for providing antibodies. We appreciate assistance with various aspects of the work provided by Michelle Facette, Beverly Fang, and Joel Griffitts.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.028266.
This work was supported in part by the U.S. Department of Energy (grant no. DOE–FG02–00ER20133).
References
- Albani D, Altosaar I, Arnison PG, Fabijanski SF (1991) A gene showing sequence similarity to pectin esterase is specifically expressed in developing pollen of Brassica napus: sequences in its 5′ flanking region are conserved in other pollen-specific promoters. Plant Mol Biol 16: 501–513 [DOI] [PubMed] [Google Scholar]
- Amagai M, Ariizumi T, Endo M, Hatakeyama K, Kuwata C, Shibata D, Toriyama K, Watanabe M (2003) Identification of anther-specific genes in a cruciferous model plant, Arabidopsis thaliana, by using a combination of Arabidopsis macroarray and mRNA derived from Brassica oleracea. Sex Plant Reprod 15: 213–220 [Google Scholar]
- Ariizumi T, Amagai M, Shibata D, Hatakeyama K, Watanabe M, Toriyama K (2002) Comparative study of promoter activity of three anther-specific genes encoding lipid transfer protein, xyloglucan endotransglucosylase/hydrolase and polygalacturonase in transgenic Arabidopsis thaliana. Plant Cell Rep 21: 90–96 [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1989) Preparation of genomic DNA from plant tissue. In FM Ausubel, R Bent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology, Vol. 1. Wiley, New York, pp 2.3.1–2.3.3 [Google Scholar]
- Barnett MJ, Long SR (1997) Identification and characterization of a gene on Rhizobium meliloti pSyma, syrB, that negatively affects syrM expression. Mol Plant-Microbe Interact 10: 550–559 [DOI] [PubMed] [Google Scholar]
- Bedinger P (1992) The remarkable biology of pollen. Plant Cell 4: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmore S, Barnes SH (1990) Pollen wall development in angiosperms. In S Blackmore, R B Knox, eds, Microspores Evolution and Ontogeny. Academic Press, San Diego, pp 173–192
- Brown SM, Crouch ML (1990) Characterization of a gene family abundantly expressed in Oenothera organensis pollen that shows sequence similarity to polygalacturonase. Plant Cell 2: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink HJD, Buxton FP, Fraaye BA, Graaff LHD, Visser J (1992) The polygalacturonases of Aspergillus niger are encoded by a family of diverged genes. Eur J Biochem 208: 83–90 [DOI] [PubMed] [Google Scholar]
- Caprari C, Mattei B, Basile ML, Salvi G, Crescenzi V, Lorenzo GD, Cervone F (1996) Mutagenesis of endopolygalacturonase from Fusarium moniliforme: histidine residue 234 is critical for enzymatic and macerating activities and not for binding to polygalacturonase-inhibiting protein (PGIP). Mol Plant-Microbe Interact 9: 617–624 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conlon RA, Rossant J (1992) Endogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development 116: 357–368 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B (1999) Carbohydrate-active enzymes: an integrated database approach. In HJ Gilbert, G Davies, B Henrissat, B Svensson, eds, Recent Advances in Carbohydrate Bioengineering. The Royal Society of Chemistry, Cambridge, UK, pp 3–12
- Fei HM, Sawhney VK (2001) Ultrastructural characterization of male sterile 33 (ms33) mutant in Arabidopsis affected in pollen desiccation and maturation. Can J Bot 79: 118–129 [Google Scholar]
- Fox DK, Westfall B, Nathan M, Hughes AJ, Rashtchian A, Schuster DM (1996) Striding new distances with 5′ RACE: long 5′ RACE of human APC and TSC-Z cDNA. Focus 18: 33–37 [Google Scholar]
- Fütterer J, Hohn T (1996) Translation in plants: rules and exceptions. Plant Mol Biol 32: 159–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KA, Bennett AB (1998) Polygalacturonases: many genes in search of a function. Plant Physiol 117: 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PM, Davies G (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55–72 [PubMed] [Google Scholar]
- Heslop-Harrison J (1971a) The pollen wall: structure and development. In J Heslop-Harrison, ed, Pollen: Development and Physiology. Butterworth, London, pp 75–98
- Heslop-Harrison J (1971b) Wall pattern formation in angiosperm microsporogenesis. Symp Soc Exp Biol 25: 277–300 [PubMed] [Google Scholar]
- Jackson D (1991) In situ hybridisation in plants. In DJ Bowles, SJ Gurr, M McPhereson, eds, Molecular Plant Pathology, A Practical Approach. Oxford University Press, Oxford, pp 163–174
- Joshi CP (1987) An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res 15: 6643–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer CJ (1987) The processes of anther dehiscence and pollen dispersal: I. New Phytol 105: 487–498 [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K (1990) Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Kramer M, Sheehy RE, Hiatt WR (1989) Progress towards the genetic engineering of tomato fruit softening. Trends Biotechnol 7: 191–194 [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Wells B, Roberts K (1992) Complexity in the spatial localization and length distribution of plant cell-wall matrix polysaccharides. J Microscopy 166: 123–136 [Google Scholar]
- Metz AM, Timmer RT, Browning KS (1992) Sequences for two cDNAs encoding Arabidopsis thaliana eukaryotic protein synthesis initiation factor 4A. Gene 120: 313–314 [DOI] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93: 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mu J-H, Stains JP, Kao T-h (1994) Characterization of a pollen-expressed gene encoding a putative pectin esterase of Petunia inflata. Plant Mol Biol 25: 539–544 [DOI] [PubMed] [Google Scholar]
- Neelam A, Sexton R (1995) Cellulase (endo β-1, 4 glucanase) and cell wall breakdown during anther development in the sweet pea (Lathyrus odoratus L.): isolation and characterization of partial cDNA clones. J Plant Physiol 146: 622–628 [Google Scholar]
- Owen HA, Makaroff CA (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Paxon-Sowders DM, Dodrill CH, Owen H, Makaroff CA (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexová-Benková L, Mracková M (1978) Active groups of extracellular endo-d-galacturonases of Aspergillus niger derived from pH effect on kinetic data. Biochim Biophys Acta 523: 162–169 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR (1998) Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J 15: 79–88 [DOI] [PubMed] [Google Scholar]
- Sexton R, Campillo ED, Duncan D, Lewis LN (1990) The purification of an anther cellulase (β1:4-glucan hydrolase) from Lathyrus odoratus L. and its relationship to the similar enzyme found in abscission zones. Plant Sci 67: 169–176 [Google Scholar]
- Shih MC, Heinrich P, Goodman HM (1991) Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate-dehydrogenase from Arabidopsis thaliana. Gene 104: 133–138 [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates Development 116: 21–30 [DOI] [PubMed] [Google Scholar]
- Southworth D (1990) Exine biochemistry. In S Blackmore, RB Knox, eds, Microspores: Evolution and Ontogeny. Academic Press, London, pp 193–212
- Stachel SE, Gynheung A, Flores C, Nester EW (1985) A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J 4: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B, Dahlbeck D, Keen N, Napoli C (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea J Bacteriol 169: 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz H (1977) Role of β-1, 3-glucanase in postmeiotic microspore release. Dev Biol 57: 87–97 [DOI] [PubMed] [Google Scholar]
- Stratilová E, Markovic O, Skrovinová D, Rexová-Benková L, Jörnvall H (1993) Aspergillus sp. polygalacturonase: multiplicity, divergence, and structural patterns linking fungal, bacterial, and plant polygalacturonases. J Protein Chem 12: 15–22 [DOI] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ (1993) RNS2: A senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA 90: 5118–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbutt SJ, Rogers HJ, Lonsdale DM (1994) Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol Biol 25: 283–297 [DOI] [PubMed] [Google Scholar]
- Tucker GA, Seymour GB (2002) Modification and degradation of pectins. In GB Seymour, JP Knox, eds, Pectins and their Manipulation. Blackwell, Oxford pp 150–173
- van Aelst AC, van Went JL (1992) Ultrastructural immunolocalization of pectin and glycoproteins in Arabidopsis thaliana pollen grains. Protoplasma 168: 14–19 [Google Scholar]
- Van den Bosch KA, Bradley DJ, Knox JP, Perotto S, Butcher GW, Brewin JJ (1989) Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J 8: 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Williams MNV, Freshour G, Darvill AG, Albersheim P, Hahn MG (1996) An antibody Fab selected from a recombinant phage display library detects de-esterified pectic polysaccharide rhamnogalacturonan II in plant cells. Plant Cell 8: 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D, Hird DL, Hodge R, Paul W, Draper J, Scott R (1992) Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]