Abstract
Urea is the major nitrogen (N) form supplied as fertilizer in agricultural plant production and also an important N metabolite in plants. Because urea transport in plants is not well understood, the aim of the present study was to isolate urea transporter genes from the model plant Arabidopsis. Using heterologous complementation of a urea uptake-defective yeast (Saccharomyces cerevisiae) mutant allowed to isolate AtTIP1;1, AtTIP1;2, AtTIP2;1, and AtTIP4;1 from a cDNA library of Arabidopsis. These cDNAs encode channel-like tonoplast intrinsic proteins (TIPs) that belong to the superfamily of major intrinsic proteins (or aquaporins). All four genes conferred growth of a urea uptake-defective yeast mutant on 2 mm urea in a phloretin-sensitive and pH-independent manner. Uptake studies using 14C-labeled urea into AtTIP2;1-expressing Xenopus laevis oocytes demonstrated that AtTIP2;1 facilitated urea transport also in a pH-independent manner and with linear concentration dependency. Expression studies showed that AtTIP1;2, AtTIP2;1, and AtTIP4;1 genes were up-regulated during early germination and under N deficiency in roots but constitutively expressed in shoots. Subcellular localization of green fluorescent protein-fused AtTIPs indicated that AtTIP1;2, AtTIP2;1, and AtTIP4;1 were targeted mainly to the tonoplast and other endomembranes. Thus, in addition to their role as water channels, TIP transporters may play a role in equilibrating urea concentrations between different cellular compartments.
A large variety of organisms can use urea efficiently as a nitrogen (N) source from soils. Because urea is uncharged and permeates slowly through artificial lipid bilayers, it has been suggested that urea can easily cross biological membranes without requiring protein-mediated transport (Galluci et al., 1971). For a non-transpiring organism, however, transport solely along the concentration gradient would not allow significant internal enrichment of urea in view of the rather low urea concentrations usually found in soils and water bodies (Gaudin et al., 1987). Therefore, prokaryotes have evolved energy-dependent transport systems for the uptake of urea across membranes (ElBerry et al., 1993; Valladares et al., 2002). Likewise, higher plants also transport urea actively, as by the H+-urea cotransporter AtDUR3, which is preferentially expressed in roots under N deficiency (Liu et al., 2003). Although urea uptake by plant roots has not yet been properly described at a physiological level, urea uptake in unicellular organisms, including prokaryotes and algae, has been investigated more carefully (Siewe et al., 1998; Wilson and Walker, 1988). Transport assays in Chara australis showed that urea uptake is driven by an active, energy-dependent pathway, whereas at higher external concentrations, urea uptake followed a linear concentration dependency, indicating a second passive or diffusion-controlled transport pathway (Wilson and Walker, 1988; Wilson et al., 1988). This kinetic behavior is in agreement with early investigations on urea transport in other plant and animal systems, where urea transport was best described by facilitated diffusion and was found to be inhibited by mercury, phloretin, or urea analogs (Dainty and Ginzburg, 1964; Stadelman, 1969; Macey, 1984).
Investigations on the molecular nature of urea transport in human or animal cells have shown that urea is mainly transported through facilitative urea transporters of the UT-A and UT-B families (Smith and Rousselet, 2001; Sands, 2003). UT transporters allow a faster equilibration of urea gradients across cell membranes than would be achieved by passive diffusion alone. In addition, four members of the aquaporin (AQP) family of aquaporins were shown by oocyte expression to mediate, in addition to water, the transport of urea also (Ishibashi et al., 1994, 1997, 1998, 2002). These findings led to a thorough reinvestigation of plant aquaporins and their functionality in urea transport. Two plant aquaporins, NtPIP1 and NtTIPa, also mediated 14C-urea fluxes when heterologously expressed in Xenopus laevis oocytes (Eckert et al., 1999; Gerbeau et al., 1999). However, urea transport does not seem to be a general property of aquaporins (Echevarria and Ilundain, 1996), and a possible physiological role of aquaporin-mediated urea transport in plants has not been investigated yet.
In plants, N acquisition could benefit from urea transport systems in roots and in leaves. As indicated by urea recovery from the xylem sap, urea is at least in part taken up by roots as an intact molecule (Hine and Sprent, 1988). Depletion studies suggested that urea uptake is affected by the presence of nitrate and ammonium (Bradley et al., 1989) and by the availability of nickel, which is required as a cofactor for urea-degrading urease (Brown et al., 1987; Gerendás et al., 1998). When supplied to leaves, urea uptake was proportional to the concentration of externally applied urea (Klein and Weinbaum, 1985) or followed the transpiration rate of the leaves, which is indicative for a process driven by the mass flow of water (Palta et al., 1991). Thus, root uptake of urea showed signs of control by N and nickel, whereas leaf uptake reflected more a diffusion-driven process.
To better understand the molecular basis for urea transport in plants, a yeast (Saccharomyces cerevisiae) complementation approach was undertaken to isolate genes encoding urea transport proteins in the model plant Arabidopsis. Here, we report that this approach led exclusively to the isolation of tonoplast intrinsic protein (TIP)-related genes. This novel property of aquaporins was characterized in yeast and X. laevis oocytes and found to fundamentally differ from the recently characterized secondary active urea transport mediated by AtDUR3. Membrane localization provided evidence that TIP-mediated urea transport might occur at different cellular membranes, and transcriptional regulation of TIP genes showed a dependence on the N nutritional status in Arabidopsis.
RESULTS
Isolation of Arabidopsis Genes Mediating Urea Uptake in Yeast
For the isolation of genes involved in urea transport, a yeast complementation assay was established. The yeast mutant YNVW1 (Δdur3 and ura3), which is unable to grow on medium containing <5 mm urea as the sole N source (Liu et al., 2003), was transformed with a cDNA library from Arabidopsis seedlings (Minet et al., 1992), and transformants were screened on 2 mm urea as the sole N source. Of approximately 200,000 independent transformants, 17 colonies were isolated that conferred growth complementation repeatedly after retransformation of YNVW1 and that were predicted to encode integral membrane proteins. All 17 cDNAs represented four genes that belong to the TIPs (Johanson et al., 2001): AtTIP1;1 (γ-TIP1; At2g36830), AtTIP1;2 (γ-TIP2; At3g26520), AtTIP2;1 (δ-TIP1; At3g16240), and AtTIP4;1 (ε-TIP; At2g25810). Except AtTIP2;1, which encodes a δ-TIP originally described as a tonoplastlocated water channel (Daniels et al., 1996), the other three TIPs have not been functionally characterized so far. All four isolated genes complemented the growth of the yeast strain YNVW1 on 2 mm urea as a sole N source; however, their growth rate was still weaker compared with the wild type (23346c), in which the ScDUR3 gene is functional (Fig. 1A). Even on 1 mm urea, yeast transformants expressing AtTIP4;1 still conferred growth complementation, whereas the other three TIPs did not (data not shown). Because yeast complementation by AtTIP1;1 was weakest, this gene was not further investigated.
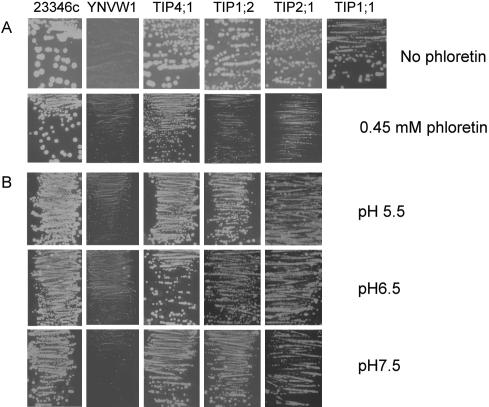
Figure 1.
Functional complementation of the yeast mutant YNVW1 by TIP genes. Growth complementation of urea uptake-defective yeast (YNVW1) by pFL61 harboring AtTIP1;1, AtTIP1;2, AtTIP2;1, or AtTIP4;1 on medium containing 2 mm urea as a sole N source in the absence or presence of 0.45 μm phloretin (A) or at different pH (B).
In transport studies in heterologous systems, phloretin [3-(4 hydroxyphenyl)-1-(2,4,6-trihydroxyphenyl)-1-propanone] is commonly used as an inhibitor for Glc and urea transporters (You et al., 1993; Ishibashi et al., 1994; Tsukaguchi et al., 1998). Relative to the wild type, AtTIP4;1-, AtTIP2;1-, and AtTIP1;2-expressing YNVW1 cells showed a reduced growth rate in presence of 0.45 mm phloretin (Fig. 1A), indicating that urea transport is sensitive to phloretin as found for urea transport mediated by mammalian AQPs in oocytes (Ishibashi et al., 1994). As an alternative to phloretin, Hg2+ has been employed frequently to block aquaporin-mediated transport of urea and water (Ishibashi et al., 1994, 1997, 2002; Kammerloher et al., 1994; Daniels et al., 1996). However, our attempt failed to find a suitable Hg2+ concentration that only inhibited growth of TIP-expressing YNVW1 strains but not of the respective wild type (data not shown).
Because YNVW1 growth complementation on urea by the active urea transporter AtDUR3 was strongly dependent on external pH (Liu et al., 2003), the influence of pH on TIP-mediated urea uptake in yeast was tested. However, the growth rate of AtTIP4;1-, AtTIP2;1-, and AtTIP1;2-expressing YNVW1 was hardly affected by raising the medium pH from 5.5 to 7.5 (Fig. 1B), indicating that external pH had no considerable effect on urea transport in yeast when mediated by one of the isolated TIPs.
AtTIP2;1 Mediates Urea Transport in X. laevis Oocytes
To investigate the mechanism of urea transport by the isolated TIP genes in more detail, one of the isolated genes, AtTIP2;1, was heterologously expressed in X. laevis oocytes. In vitro-synthesized AtTIP2;1 mRNA was micro-injected into oocytes, which were further incubated in choline-based standard buffer solution for 2 to 3 d before assaying urea uptake after supply of [14C]-labeled urea. Because uptake of [14C]urea by water-injected oocytes was low and showed no difference to noninjected oocytes, noninjected oocytes served as a control. In contrast, accumulation of [14C]-urea in AtTIP2;1-expressing oocytes was approximately 3- to 4-fold higher when oocytes were exposed to 100 μm urea for 20 min (Fig. 2A). Uptake of urea into oocytes was independent of external pH between pH 5 and 8. In control oocytes, urea accumulation was not significantly altered by increasing pH but showed higher variability at pH 8 (Fig. 2A). The pH independence in oocytes was in agreement with the observation that urea-dependent growth complementation of TIP-transformed yeast was unaffected by external pH (Fig. 1B).
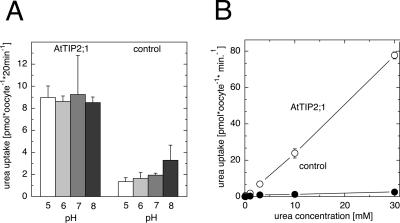
Figure 2.
Characterization of [14C]-labeled urea transport by AtTIP2;1. A, Uptake rates of [14C]urea in AtTIP2;1-expressing and in uninjected control oocytes from X. laevis. Uptake was determined from 100 μm [14C]-labeled urea at pH 5 to 8 after 20 min of incubation. Bars = means ± sd, n = 3, of one representative experiment, except for control oocytes at pH 5 and 8, where n = 7 from two experiments. B, Concentration-dependent uptake of [14C]urea in AtTIP2;1-expressing oocytes was not saturable and displayed a linear concentration dependency up to 30 mm urea. Standard choline solutions at pH 7 were used. Symbols represent means ± sd, n = 3.
Urea accumulation in AtTIP2;1-expressing oocytes increased steeply with urea supply, when external urea concentrations were shifted from 100 μm to 30 mm at pH 7 (Fig. 2B). Similar to the comparatively low increase of urea accumulation in control oocytes, urea accumulation by AtTIP2;1 increased linearly with external concentration. A linear concentration dependence over large substrate concentration ranges is more typical of channel-mediated transport, suggesting that AtTIP2;1 might transport urea by a similar transport mechanism as water.
Subcellular Localization of AtTIP1;2, AtTIP2;1, and AtTIP4;1
To elucidate the potential physiological role of the isolated TIP transporters in urea transport in plants, subcellular localization of AtTIP1;2, AtTIP2;1, and AtTIP4;1 was assayed in PEG-transformed protoplasts derived from an Arabidopsis cell culture precultured in the dark. All TIP proteins were C terminally linked to green fluorescent protein (GFP), and expression of the fusion proteins was driven by a 35S promoter. Protoplasts transformed by the empty plasmid pCF203 carrying GFP alone served as control.
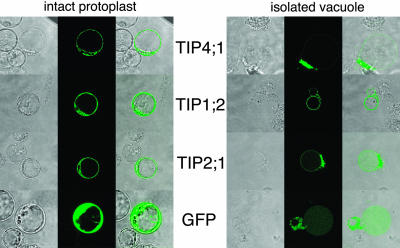
GFP fluorescence of AtTIP4;1-, AtTIP1;2-, and AtTIP2;1-tagged constructs in protoplasts was confined to a ring in the protoplasts and to internal structures but was excluded from the nucleus and the vacuolar lumen (Fig. 3A). This staining pattern was similar to that observed for the vacuolar V-type ATPase subunit C (Allen et al., 2000). After releasing vacuoles from the protoplasts by osmotic shock, the fluorescence signal of AtTIP2;1 and AtTIP1;2 mainly localized to the tonoplast. A strong signal was also observed in the remaining membranes and vesicles of the ruptured protoplast. In contrast, AtTIP4;1-GFP was hardly detectable in the tonoplast after osmotic shock but showed a strong staining of released material. Thus, all three TIPs localized to various internal membranes including the tonoplast, and the image resolution and staining of intact protoplasts did not exclude plasma membrane localization at same time.
Figure 3.
Subcellular localization of GFP-fused TIPs. Transmission image (left), fluorescence image (middle), and fluorescence superimposed over the transmission image (right). A, GFP-derived fluorescence from protoplasts that were transformed with GFP alone or GFP fused C-terminally to AtTIP4;1, AtTIP2;1, or AtTIP1;2. B, Green fluorescence was also assayed after disruption of the protoplast plasma membrane by osmotic shock using 100 mm Na2HPO4 and 10 mm EDTA, pH 6.5.
Transcriptional Regulation of AtTIP1;2, AtTIP2;1, and AtTIP4;1
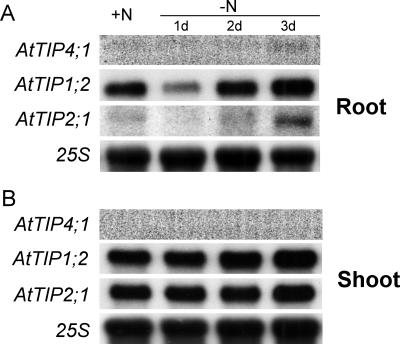
To investigate a potential physiological link of TIP-mediated urea transport to plant N nutrition, expression analysis was performed in 29-d-old Arabidopsis plants grown under axenic conditions. Before harvest, plants were subjected to N deficiency for 1, 2, or 3 d. In roots, expression of AtTIP4;1 was hardly detectable but appeared weakly up-regulated after N starvation for 3 d (Fig. 4A). Transcript levels of AtTIP1;2 and AtTIP2;1 were readily detectable after preculture under high N supply but sharply decreased after 24 h of N starvation. However, prolonging the period of N starvation to 3 d led to an increasing transcript accumulation of both genes (Fig. 4A). In shoots, AtTIP1;2 and AtTIP2;1 seemed to be constitutively expressed and showed no response to the N treatments. AtTIP4;1 could not be detected in shoots by RNA gel-blot analysis (Fig. 4B). Taken together, all three TIP genes clearly exhibited an N-dependent transcriptional regulation in roots, suggesting that the corresponding transporters might be involved in membrane transport processes that are linked to N nutrition or N metabolism.
Figure 4.
N-dependent expression of AtTIPs. RNA gel-blot analysis performed with hydroponically grown plants, which were precultured for 4 weeks in presence of 1 mm NH4NO3 and starved for 1, 2, or 3 d for N. Total RNA from roots (A) or shoots (B) was used for hybridization to the complete open reading frame (ORF) of AtTIP1;2, AtTIP2;1, or AtTIP4;1 and to 25s rRNA.
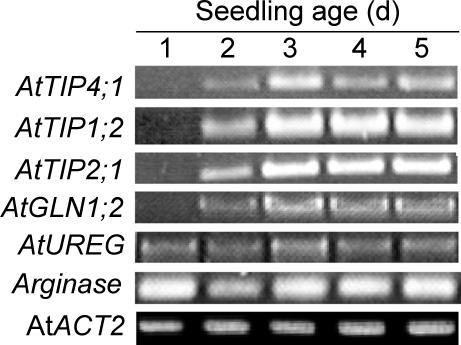
Urea transport might also occur during remobilization of N reserves in the plant. Thus, AtTIP gene expression was investigated in Arabidopsis seedlings, where N is remobilized from cotyledons and seed storage proteins to developing sinks. Because AtTIP transcripts were not well detected by RNA gel-blot analysis, reverse transcriptase (RT)-based PCR was performed using gene-specific primers. To verify that equal amounts of cRNA were used in each PCR reaction, cDNA fragments of the constitutively expressed ACT2 (actin-2) gene were simultaneously amplified by PCR (An et al., 1996). In total RNA isolated from water-germinated seedlings over a 5-d period, a steep increase in transcripts of all three AtTIP genes was observed between the 1st and 2nd d of germination and between the 2nd and 3rd d (Fig. 5). From the 3rd d onwards, expression of AtTIP1;2 and AtTIP2;1 remained constantly high, whereas AtTIP4;1 mRNA levels decreased. A similar expression pattern was only observed for AtGLN1;2, encoding a cytosolic Gln synthetase, whereas AtUREG and arginase showed a more or less constitutive expression over the 5 d of germination (Fig. 5).
Figure 5.
Expression of AtTIPs during germination. Gene-specific RT-PCR performed on total RNA isolated from Arabidopsis seedlings that were germinated in water for 1 to 5 d in a growth chamber (see “Materials and Methods”). AtGLN1;2, Cytosolic Gln synthetase; AtUREG, urease accessory gene; AtACT2, actine gene isoform 2; Arginase, arginase gene.
A continuous increase in transcript levels of AtTIP1;2, AtTIP2;1, AtTIP4;1, and AtGLN1;2 during the first 3 d of N starvation was found repeatedly (Fig. 5). Based on the well-characterized transcriptional regulation of AtGLN1;2, which is strongly derepressed under N deficiency (Oliveira and Coruzzi, 1999), it is most likely that the Arabidopsis seedlings started early to suffer from N deficiency and then upregulated N-dependent genes such as AtGLN1;2. The transcriptional coregulation of the three AtTIPs with other N-dependent genes (Fig. 5) may support a physiological relationship between these TIPs and N nutrition or N metabolism in Arabidopsis. Because all three AtTIP genes were apparently not coregulated with the arginase gene or with UREG, encoding a urease accessory protein (Freyermuth et al., 1999; Goldraij and Polacco, 1999), no evidence has been obtained for an involvement of TIPs in N remobilization processes, when N is recycled from the Orn cycle and Arg is broken down to urea.
DISCUSSION
AtTIP1;2, AtTIP2;1, and AtTIP4;1 Function as Urea Permeases
Although urea is a commonly used N form in agricultural plant production, uptake pathways for urea into plant cells are poorly understood. Therefore, a yeast complementation system was developed aiming to isolate urea transporter genes from Arabidopsis. Complementation of the urea uptake-defective yeast strain YNVW1 with cDNAs from Arabidopsis led to the isolation of four genes, AtTIP1;1, AtTIP1;2, AtTIP2;1, and AtTIP4;1. TIPs form a subfamily within the highly conserved superfamily of major intrinsic proteins (MIPs), also referred to as aquaporins, and have been named after their main location in the tonoplast (Johanson et al., 2001). Like MIPs in general, TIPs have been shown to facilitate water transport in heterologous systems (Höfte et al., 1992; Daniels et al., 1996), and for NtTIPa, a close homolog to AtTIP2;1, expression in oocytes also allowed the demonstration of urea permeability (Gerbeau et al., 1999). Furthermore, urea permeability has also been observed in several aquaporins from the animal kingdom (Ishibashi et al., 1994, 1997, 1998, 2002). In the plant kingdom, urea permeability is apparently not restricted to TIP members within the aquaporin family, but also include plasma membrane intrinsic proteins (PIPs; Eckert et al., 1999). This observation raises the question why the employed yeast complementation system exclusively led to the isolation of TIPs. On the one hand, the isolation of TIPs might have been favored by a relatively high abundance of TIP cDNAs in the library. At least AtTIP1;2 and AtTIP2;1 seem to belong to highly expressed TIPs according to northern analysis (Fig. 4; Daniels et al., 1996), whereas for AtTIP4;1, gene expression was low. In addition, the existence of several ESTs points to a frequent expression of AtTIP1;2 and AtTIP2;1. On the other hand, the exclusive isolation of AtTIPs might reflect a high potential of the isolated AtTIPs for urea transport, implying in general a relatively high urea permeability of tonoplast membranes. Using stopped-flow spectrofluorimetry, Gerbeau et al. (1999) found that tonoplast vesicles from tobacco (Nicotiana tabacum) were approximately 75-fold more permeable for urea compared with plasma membrane vesicles. Likewise, when Tyerman et al. (1999) recorded volume changes in membrane vesicles as a response to external urea, they found a 2- to 3-fold higher urea permeability in tonoplast-enriched vesicles compared with plasma membrane-enriched vesicles. To verify whether PIPs also show a comparable urea permeability, we expressed two abundant PIPs, AtPIP1;2 and AtPIP2;1, which were also present in the cDNA library (Minet et al., 1992), in YNVW1 and assayed for growth complementation on 2 mm urea. However, both genes failed to confer urea transport (data not shown). Taken together, the exclusive isolation of TIPs as urea transporters in the yeast complementation assay is consistent with biochemical studies (Gerbeau et al., 1999; Tyerman et al., 1999) and provides new evidence at a molecular level for a higher urea permeability of tonoplast versus plasma membranes.
Similar to many of the mammalian UT-type proteins, which serve at the same time for urea and water transport in vivo (Yang and Verkman, 2002; Sands, 2003), urea transport by TIPs was also inhibited by phloretin. Phloretin is a chalcon derivative with antioxidant activity that is naturally found in fruits in the form of a Glc conjugate and for which an inhibitory action on various classes of urea transporters has been demonstrated (You et al., 1993; Ishibashi et al., 1994; Tsukaguchi et al., 1998). Using erythrocytes from AQP1/UT-B double knockout mice, it has been shown that UT-B permeated not only urea but also water in a phloretin-sensitive manner (Yang and Verkman, 2002). Moreover, phloretin has also been shown to inhibit the activity of Glc transporters, chloride channels, and fatty acid translocases (Cloherty et al., 2001; Hirayama et al., 2001; Helliwell and Kellett, 2002), pointing to a nonspecific inhibitory mechanism on a certain type of transporters independent of the transported substrate. In TIP-expressing yeast transformants, addition of 0.45 mm phloretin led to a distinctly reduced growth rate that was not detected in the wild-type strain 23346c (Fig. 1A). Thus, regarding phloretin sensitivity, TIP-mediated urea transport behaves in a similar manner to urea transporters of the mammalian UT and AQP families.
Besides complementation of the urea uptake-defective yeast strain, urea transport activity was also shown by functional expression in oocytes. For this purpose, accumulation of 14C-labeled urea was measured in AtTIP2;1-expressing oocytes. At 100 μm external urea, accumulation of urea in AtTIP2;1-injected oocytes was independent of external pH, approximately 4-fold higher than in control oocytes, and further increased in a linear concentration dependency (Fig. 2). These results demonstrated that AtTIP2;1 represents a non-saturable urea transporter or urea channel in oocytes that most likely possesses a high capacity for urea transport. Because AtTIP2;1 also mediated water transport in oocytes (Maurel et al., 1993; Daniels et al., 1996), AtTIP2;1 most likely reflects a urea and water channel similar to NtTIPa from tobacco (Gerbeau et al., 1999) and to various animal counterparts.
Subcellular Localization Suggests That AtTIPs Might Be Localized at Various Membranes
The identification of four major subfamilies within the MIP gene family (TIPs, PIPs, nodulin-like intrinsic proteins, and small intrinsic proteins) almost implies different physiological roles of the encoded proteins. These proteins are found in at least two different subcellular compartments, the tonoplast (TIPs) and the plasma membrane (PIPs). Whether the prediction of the subcellular localization always holds true has been questioned (Barkla et al., 1999) and requires verification for each protein. Therefore, the subcellular distribution of three of the isolated AtTIPs was assayed by C-terminal fusion of GFP and transient expression in Arabidopsis protoplasts. Relative to the expression of GFP alone, fluorescence of AtTIP fusion proteins suggested localization in several endosomal membrane compartments including the tonoplast (Fig. 3A) but cannot rule out additional localization at the plasma membrane. The fluorescence intensity for the individual AtTIPs, however, differed between membranes. After disruption of the plasma membrane through osmotic shock, only a weak GFP signal remained for AtTIP4;1 in the tonoplast, whereas AtTIP1;2- and AtTIP2;1-derived fluorescence in the tonoplast remained stronger (Fig. 3B), suggesting a relatively higher targeting of these AtTIPs to the tonoplast.
The localization of all investigated AtTIPs in the plasma membrane of heterologous expression systems may require more sensitive assays for subcellular localization (Fig. 1). Using a specific antibody, AtTIP2;1 has been immunolocalized to the tonoplast fraction of membrane proteins (Daniels et al., 1996). In Mesembryanthemum crystallium, analysis of MIP-A yielded localization in the tonoplast fraction, whereas MIP-B and MIP-C were found in the plasma membrane and the tonoplast fraction. Therefore, the authors suggested that putative PIP-like aquaporins can show a more complex localization by being located at different subcellular membranes at the same time (Barkla et al., 1999). However, this issue certainly requires further investigation, e.g. by the use of transgenic plants expressing tagged AtTIP proteins under the control of their native promoters.
All four isolated Arabidopsis TIPs transported urea in heterologous systems, but coincident urea and water transport may be without physiological relevance. Our study suggests that urea transport by TIPs may be a general feature and warrants a closer examination of other substrates for aquaporins besides water (Daniels et al., 1996; Maurel, 1997; Weig et al., 1997). Water transport by PIP aquaporins is of physiological relevance because cellular and wholeplant water relation are affected in mutant plants (Siefritz et al., 2002). From a physiological viewpoint, the identification of TIPs, or MIPs in general, as putative urea transporters is not surprising. When applied to the roots, urea accumulated preferentially in shoots (Gerendás et al., 1998), and when applied to leaves, transport closely followed transpiration rates (Palta et al., 1991). Thus, urea is most likely transported with the mass flow of water in the transpiration stream, which is in favor of a common membrane transport pathway for both substrates. In this case, plasma membrane-localized MIPs could easily explain the physiological link between water and urea uptake by roots, whereas tonoplast-localized MIPs could mediate urea loading and unloading of the vacuole.
N-Dependent Expression of AtTIPs Supports a Role in Plant N Metabolism
If urea transport by AtTIPs is physiologically significant, it is expected that TIPs, as other transporters for nitrogenous compounds, should be regulated by the plant N status. Transcript levels of AtTIP4;1, AtTIP1;2, and AtTIP2;1 increased under N deficiency in Arabidopsis roots (Fig. 4A), similar to the expression pattern of other N-regulated genes, such as AtAMT1;1 or AtDUR3 (Gazzarrini et al., 1999; Liu et al., 2003). Furthermore, in water-cultured Arabidopsis seedlings, transcripts of the same TIP genes clearly increased during the first 3 d of germination (Fig. 5), again going along with a simultaneous upregulation of genes that are derepressed under N deficiency (Oliveira and Coruzzi, 1999; Liu et al., 2003). So far, expression studies of aquaporin genes have shown altered transcriptional regulation during cell enlargement (Smart et al., 1998) after dehydration or after salt stress (Yamada et al., 1995; Mariaux et al., 1998; Smart et al., 2001), clearly supporting their putative role in water movement. In addition, TIP homologs are up-regulated in response to nitrate (Wang et al., 2001) and, therefore, probably responsible for an increased water flux into roots (Hoarau et al., 1996). Thus, enhanced TIP gene expression upon nitrate supply might reflect a root response to altered availability of a signal or an important solute rather than of an N source. The transcriptional upregulation of AtTIP4;1, AtTIP1;2, and AtTIP2;1 in response to N deficiency, however, is more reminiscent of a role of these TIPs in N, i.e. urea, rather than water transport.
Furthermore, the increase in gene expression of all three AtTIP genes was confined to the roots, where acquisition of external urea is possible, whereas AtTIP expression in shoots was more or less constitutive (Fig. 4). In shoots, AtTIPs could play a role in rapid uptake of xylem-derived or even foliar-applied urea and/or in the compartmentation of urea from the cytosol into the vacuole, which might prevent its toxicity under high fertilization rates (Krogmeier et al., 1989). However, to further elucidate a physiological role of urea-transporting AtTIPs in shoots, the determination of urea concentrations in different subcellular compartments must be considered as an essential component.
In conclusion, this study showed that the isolated AtTIPs should be considered as membrane proteins with highest potentials for urea transport and, therefore, will most likely take in a role in urea transport whenever required. In contrast to the high-affinity H+/urea symporter AtDUR3, these AtTIPs provide a less concentration- and pH-dependent transport pathway for urea. Transcriptional up-regulation of the isolated AtTIPs under N deficiency in roots further supports a role in urea transport. With respect to their localization in endosomal membranes and possibly also in the plasma membrane, the identified AtTIPs could potentially facilitate urea transport either from the external growth medium into the cytosol or from the cytosol into the vacuole, e.g. for the storage or detoxification of excessive urea. Thus, although Arabidopsis is not a crop plant, it disposes of several different types of urea transporters for passive and secondary active transport of urea into different cellular compartments. Furthermore, Arabidopsis was shown to dispose of transporters for ammonium, which can be taken up as a urea degradation product and for various N-heterocycles (Desimone et al., 2002). Thus, plants in general have a large number of different transport systems to optimize N uptake independent of the N forms available in the soil. A deeper investigation at a physiological level of urea transport processes in planta and their regulation by N seems required not only to better understand the importance of urea for plant N nutrition but also to improve its utilization as a leaf-applied N fertilizer in agricultural crop production.
MATERIALS AND METHODS
Generation of a Yeast Mutant and Screening of a cDNA Library
The DUR3 gene in the yeast (Saccharomyces cerevisiae) strain ∑ 23346c (Matα, ura3) was deleted by homologous integration of a disruption cassette as described by Liu et al. (2003), resulting in the urea uptake-defective yeast strain YNVW1. For the isolation of urea transporter genes, YNVW1 was transformed with a cDNA library from Arabidopsis seedlings (Minet et al., 1992). Transformants were directly plated on urea-selective medium (yeast N base with 2 mm urea, 2% [w/v] Glc, and 2% [w/v] agarose).
The isolated genes are consistent with the following annotations in the database: AtTIP1;1 (γ-TIP1; At2g36830), AtTIP1;2 (γ-TIP2; At3g26520), At-TIP2;1 (δ-TIP1; At3g16240), and AtTIP4;1 (ε-TIP; At2g25810).
Radiotracer Uptake Studies in Xenopus laevis Oocytes
The ORF of AtTIP2;1 was amplified from a cDNA library (Minet et al., 1992) by Pfu polymerase (Stratagene, La Jolla, CA) using PCR and AtTIP2; 1-specific primers containing a restriction site for BglII: 5′-AACACAAAAACAAAAAGTTTTTTTAATTTTAATCAAAAAAGATCTATGGCTGGAGTTGCCTTTGGTTC-3′ and 5′-GAATGTAAGCGTGACATAACTAATT ACATGACTCGAGAGATCAGATCTTTAGAAATCAGCAGAAGCAAGAGG-3′. The resulting PCR product was first in vivo cloned into the yeast expression vector p426HXT (Wieczorke et al., 1999), sequenced, and the sequence was verified by database comparison. Then, the ORF was ligated into the oocyte expression vector pOO2 (Ludewig et al., 2002) after linearization with BamHI using the BglII overhang. Capped cRNA was transcribed from pOO2-TIP2;1 in vitro using the mMessage mMachine kit (Ambion, Austin, TX) after linearization of the plasmid with XmnI. X. laevis oocytes were removed from adult female frogs by surgery and manually dissected. Oocytes (Dumont stage V or VI) were defolliculated using 10 mg mL–1 collagenase (Boehringer Mannheim, Basel) and trypsin inhibitor (Sigma, St. Louis) for 1 h and injected with 5 to 50 nL of cRNA (15–50 ng per oocyte). Each co-injection experiment was repeated multiple times. Oocytes were kept after injection for 2 to 5 d at 16°C in ND96 solution with the following composition: 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES (pH 7.4), and 20 μg mL–1 gentamycin.
Standard bath solutions for oocytes were also used for uptake experiments. Oocytes were pooled to groups of three and incubated for 20 min at room temperature in 500 μL of the respective buffer containing 10% (v/v) 14C-labeled urea (specific activity 57 mCi mmol–1, Amersham, Buckinghamshire, UK). Then, oocytes were carefully washed five times in 1 mL of ice-cold buffer with 100-fold excess urea and solubilized in 5% (w/v) SDS. After addition of 5 mL of scintillation cocktail (Ultima Gold, Zinsser, Frankfurt, Germany), washed oocytes were measured in a scintillation counter (Wallac, Turku, Finland).
Generation of AtTIP-GFP Constructs and Transformation of Arabidopsis Protoplasts
The ORFs of AtTIP1;2, AtTIP2;1, and AtTIP4;1 were amplified by Pfu polymerase (Stratagene) using the following primers with KpnI cloning sites: TIP4;1, 5′-AGGGTACCATGAAGAAGATCGAGTTAGGGCA-3′ and 5′-AGGGTACCAATTCAACAATGGTTGCTCGTCGTC-3′; TIP1;2, 5′-AGGGTACCATGCCGACCAGAAACATCGC-3′ and 5′-AGGGTACCAGTAATCGGTGGTAGGCAAT-3′; and TIP2;1, 5′-AGGGTACCATGGCTGGAGTTGCCTTTGGTT-3′ and 5′-AGGGTACCAGAAATCAGCAGAAGCAAGAGGA-3′.
PCR products were verified by sequencing and compared with the sequences in the Arabidopsis database. Finally, the ORFs were cloned into the pCF203 vector (kindly provided by C. Fankhauser, ETH Zürich) in frame to a gene encoding GFP driven by a 35S promoter.
Five milliliters of 1-week-old Arabidopsis ecotype Landsberg suspension cell culture (gift of Axel Vögel, ZMBP, Tübingen, Germany) were transferred to 45 mL of growth medium as described by Liu et al. (2003) and subcultured for 3 d under dark conditions (26°C and rotation at 120 rpm). The cell culture was centrifuged at 400g for 5 min and washed with 25 mL of cell wall digestion buffer solution without digestion enzyme. Protoplasts were prepared and transformed according to the protocols of Merkle et al. (1996) and Negrutiu et al. (1987). After transformation, the protoplasts were visualized using a confocal microscope. Plasma membranes of protoplasts were disrupted by osmotic shock, adding 5 to 10 μL of a 10 mm EDTA and 100 mm KH2PO4 (pH 6) solution to protoplasts, and after incubation for 2 to 3 min, the fluorescence was investigated using the confocal microscope.
Plant Culture and Gene Expression Analyses
Arabidopsis seeds (ecotype Columbia 0) were germinated and precultured axenically in magenta boxes containing 50 mL of nutrient solution as described by Liu et al. (2003). Except for the control treatment with 1 mm NH4NO3, plants were subjected to N deficiency for 1, 2, and 3 d before harvest. Plant roots and shoots were harvested separately for total RNA extraction. To obtain plant material from germinating seedlings, surface-sterilized seeds were placed in petri dishes and germinated in autoclaved distilled water and harvested every day during a 5-d period.
Isolation of total RNA and RNA gel-blot analysis were conducted as performed in Gazzarrini et al. (1999). The full-length cDNAs of the three TIPs were used as a probe for hybridization to total RNA. A 25S rRNA probe was used as a RNA loading control for quantification achieved by a PhosphorImager (Storm, Molecular Dynamics, Sunnyvale, CA). For RT-mediated PCR analysis, mRNAs were converted to cDNAs by reverse transcriptase according to the manufacturer's protocol (MBI, St. Leon-Roth, Germany), and cDNA fragments of AtTIP1;2, AtTIP2;1, AtTIP4;1, arginase gene, UREG, and AtGLN1;2 were amplified using specific primers for the corresponding genes. To ensure that equal amounts of cRNA were used in each PCR reaction, a cDNA fragment of the constitutively expressed ACT2 gene was simultaneously amplified by PCR (An et al., 1996). The verification of the amplicons was made by sequencing.
Primers used for RT-PCR were: TIP1;2, 5′-ATGCCGACCAGAAACATCGC-3′ and 5′-TCAGTAATCGGTGGTAGGCAA-3′; TIP2;1, 5′-ATGGCTGGAGTTGCCTTTGGTT-3′ and 5′-TTAGAAATCAGCAGAAGCAAG AGG-3′; TIP4;1, 5′-ATGAAGAAGATCGAGTTAGGGC-3′ and 5′-TTAATTCAACAATGGTTGCT CGT-3′; Arginase, 5′-GATATGTCGAGGATTATTGGTA-3′ and 5′-GTTTATCGATCTGATCCC AAC-3′; UREG, 5′-GAAGGCGTCGTGGGTGGG-3′ and 5′-AAGTATTGAAAGAGTTCCATTCA-3′; GLN1;2, 5′-ATGAGTCTTCTTGCAGATCTT-3′ and 5′-TCAAGGGTTCCAGAGGAGT-3′; and ACT2, 5′-TCCAAGCTGTTCTCTCCTTG-3′ and 5′-GAGGGCTGGAACAAGACTTC-3′.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Dr. Karin Schumacher (ZMBP Tübingen, Germany) and Dr. Christian Fankhauser (ETH Zürich) for help with the GFP constructs, Catherina Brancato (ZMBP Tübingen) for excellent technical assistance with the protoplast transformation, and Dr. Pia Walch-Liu (Lancaster University, UK) for help with the oocyte uptake studies.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027409.
This work was supported by the Deutsche Forschungsgemeinschaft (Bonn; grant no. Wi 1728/2 to N.v.W.).
References
- Allen G, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF et al. (2000) Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science 289: 2338–2342 [DOI] [PubMed] [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Pantoja O, Kirch H-H, Bohnert HJ (1999) Aquaporin localization: how valid are the TIP and PIP labels? Trends Plant Sci 4: 86–88 [DOI] [PubMed] [Google Scholar]
- Bradley DP, Morgan MA, O'Toole P (1989) Uptake and apparent utilization of urea and ammonium nitrate in wheat seedlings. Fertil Res 20: 41–49 [Google Scholar]
- Brown PH, Welch RM, Cary EE (1987) Nickel: a micronutrient essential for higher plants. Plant Physiol 85: 801–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloherty EK, Levine KB, Carruthers A (2001) The red blood cell glucose transporter presents multiple, nucleotide-sensitive sugar exit sites. Biochemistry 40: 15549–15561 [DOI] [PubMed] [Google Scholar]
- Dainty J, Ginzburg BZ (1964) Permeation of the cell membranes of Nitella translucens to urea, and the effect of high concentrations of sucrose on this permeability. Biochim Biophys Acta 79: 112–121 [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ (1996) Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell 8: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimone M, Catoni E, Ludewig U, Hilpert M, Schneider A, Kunze R, Tegeder M, Frommer WB, Schumacher K (2002) A novel superfamily of transporters for allantoin and other oxo derivatives of nitrogen heterocyclic compounds in Arabidopsis. Plant Cell 14: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarria M, Ilundain AA (1996) Aquaporins. J Physiol Biochem 54: 107–118, 1996 [PubMed] [Google Scholar]
- Eckert M, Biela A, Siefritz F, Kaldenhoff R (1999) New aspects of plant aquaporin regulation and specificity. J Exp Bot 50: 1541–1545 [Google Scholar]
- ElBerry HM, Majumdar ML, Cunningham TS, Sumrada RA, Cooper TG (1993) Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J Bacteriol 175: 4688–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyermuth SK, Forde BG, Polacco JC (1999) Nucleotide sequence of cDNA encoding an Arabidopsis urease accessory protein (accession no. AF109374) (PGR99-012). Plant Physiol 119: 364 [Google Scholar]
- Galluci E, Micelli C, Lippe C (1971) Non-electrolyte permeability across thin lipid membranes. Arch Int Physiol Biochem 79: 881–887 [DOI] [PubMed] [Google Scholar]
- Gaudin R, Dupuy J, Bournat P (1987) Suivi du contenue en azote de la solution du sol d'une rizière après placement d'urée. Agron Trop 42: 13–19 [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeau P, Guclu J, Ripoche P, Maurel C (1999) Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. Plant J 18: 577–587 [DOI] [PubMed] [Google Scholar]
- Gerendás J, Zhu Z, Sattelmacher B (1998) Influence of N and Ni supply on urease activity in rice. J Exp Bot 49: 1545–1554 [Google Scholar]
- Goldraij A, Polacco JC (1999) Arginase is inoperative in developing soybean embryos. Plant Physiol 119: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell PA, Kellett GL (2002) The active and passive components of glucose absorption in rat jejunum under low and high perfusion stress. J Physiol 544: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine JC, Sprent JI (1988) Growth of Phaseolus vulgaris on various nitrogen sources: the importance of urease. J Exp Bot 39: 1505–1512 [Google Scholar]
- Hirayama BA, Diez-Sampedro A, Wright EM (2001) Common mechanisms of inhibition for the Na+/glucose (hSGLT1) and Na+/Cl-/GABA (hGAT1) cotransporters. Br J Pharmacol 134: 484–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau J, Barthes L, Bousser A, Deléens E, Prioul J-L (1996) Effect of nitrate on water transfer across roots of nitrogen pre-starved maize seedlings. Planta 200: 405–415 [Google Scholar]
- Höfte H, Hubbard L, Reizer J, Ludevid D, Herman EM, Chrispeels MJ (1992) Vegetative and seed-specific forms of tonoplast intrinsic protein in the vacuolar membrane of Arabidopsis thaliana. Plant Physiol 99: 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Ishibashi K, Yamauchi K, Kageyama Y, Saito-Ohara F, Ikeuchi T, Marumo F, Sasaki S (1998) Molecular characterization of human Aquaporin-7 gene and its chromosomal mapping. Biochim Biophys Acta 1399: 62–66 [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Kuwahara M, Gu Y, Kageyama Y, Tohsaka A, Suzuki F, Marumo F, Sasaki S (1997) Cloning and functional expression of a new water channel abundantly expressed in the testis permeable to water glycerol, and urea. J Biol Chem 272: 20782–20786 [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Morinaga T, Kuwahara M, Sasaki S, Imai M (2002) Cloning and identification of a new member of water channel (AQP10) as an aquaglyceroporin. Biochim Biophys Acta 19: 335–340 [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Sasaki S, Fushimi K, Uchida S, Kuwahara M, Saito H, Furukawa T, Nakajima K, Yamaguchi Y, Gojobori T (1994) Molecular cloning and expression of a member of the aquaporin family with permeability to glycerol and urea in addition to water expressed at the basolateral membrane of kidney collecting duct cells. Proc Natl Acad Sci USA 91: 6269–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, Karlson M, Johnsson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126: 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerloher W, Fischer U, Piechottka GP, Schäffner AR (1994) Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J 6: 187–199 [DOI] [PubMed] [Google Scholar]
- Klein I, Weinbaum SA (1985) Foliar application of urea to almond and olive: leaf retention and kinetics of uptake. J Plant Nutr 8: 117–129 [Google Scholar]
- Krogmeier MJ, McCarty GW, Bremner JM (1989) Phytotoxicity of foliar-applied urea. Proc Natl Acad Sci USA 86: 8189–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Ludewig U, Frommer WB, von Wirén N (2003) AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. Plant Cell 15: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig U, von Wirén N, Frommer WB (2002) Uniport of ammonium by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem 277: 13548–13555 [DOI] [PubMed] [Google Scholar]
- Macey RI (1984) Transport of water and urea in red blood cells. Am J Physiol 246: 195–203 [DOI] [PubMed] [Google Scholar]
- Mariaux JB, Bockel C, Salamini F, Bartels D (1998) Desiccation- and abscisic acid-responsive genes encoding major intrinsic proteins (MIPs) from the resurrection plant Craterostigma plantagineum. Plant Mol Biol 38: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Maurel C (1997) Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol 48: 399–429 [DOI] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ (1993) The vacuolar membrane protein γ-TIP creates water specific channels in Xenopus oocytes. EMBO J 12: 2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T, Leclerc D, Marshallsay C, Nagy F (1996) A plant in vitro system for the nuclear import of proteins. Plant J 10: 1177–1186 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNA. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Negrutiu I, Shillito R, Potrykus I, Biasini G, Sala F (1987) Hybrid genes in the analysis of transformation conditions. Plant Mol Biol 8: 363–373 [DOI] [PubMed] [Google Scholar]
- Oliveira IC, Coruzzi GM (1999) Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol 121: 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palta JA, Fillery IR, Mathews EL, Turner NC (1991) Leaf feeding of (15N)urea for labelling wheat with nitrogen. Aust J Plant Physiol 18: 627–636 [Google Scholar]
- Sands JM (2003) Mammalian urea transporters. Annu Rev Physiol 65: 543–566 [DOI] [PubMed] [Google Scholar]
- Siefritz F, Tyree MT, Lovisolo C, Schubert A, Kaldenhoff R (2002) PIP1 plasma membrane aquaporins in tobacco: from cellular effects to function in plants. Plant Cell 14: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewe RM, Weil B, Burkovski A, Eggeling L, Krämer R, Jahns T (1998) Urea uptake and urease activity in Corynebacterium glutamicum. Arch Microbiol 169: 411–416 [DOI] [PubMed] [Google Scholar]
- Smart LB, Moskal WA, Cameron KD, Bennett AB (2001) MIP genes are down-regulated under drought stress in Nicotiana glauca. Plant Cell Physiol 42: 686–693 [DOI] [PubMed] [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 116: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CP, Rousselet G (2001) Facilitative urea transporters. J Membr Biol 183: 1–14 [DOI] [PubMed] [Google Scholar]
- Stadelman EJ (1969) Permeability of the plant cell. Annu Rev Plant Physiol 20: 585–606 [Google Scholar]
- Tsukaguchi H, Shayakul C, Berger UV, Mackenzie B, Devidas S, Guggino WB, van Hoek AN, Hediger MA (1998) Molecular characterisation of a broad selectivity neutral solute channel. J Biol Chem 273: 24737–24743 [DOI] [PubMed] [Google Scholar]
- Tyerman SD, Bohnert HJ, Maurel C, Steudle E, Smith JAC (1999) Plant aquaporins: their molecular biology and significance for plant water relation. J Exp Bot 50: 1055–1071 [Google Scholar]
- Valladares A, Montesinos ML, Herrero A, Flores E (2002) An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol Microbiol 43: 703–715 [DOI] [PubMed] [Google Scholar]
- Wang YH, Garvin DFL, Kochian LV (2001) Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol 127: 345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig A, Deswarte C, Chrispeels MJ (1997) The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol 114: 1347–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E (1999) Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 464: 123–128 [DOI] [PubMed] [Google Scholar]
- Wilson MR, O'Donoghue SI, Walker NA (1988) The transport and metabolism of urea in Chara australis: III. Two specific transport systems. J Exp Bot 39: 763–774 [Google Scholar]
- Wilson MR, Walker NA (1988) The transport and metabolism of urea in Chara australis: I. Passive diffusion, specific transport and metabolism of urea and methylurea. J Exp Bot 39: 739–751 [Google Scholar]
- Yamada S, Katsuhara M, Kelly WB, Michalowski CB, Bohnert HJ (1995) A family of transcripts encoding water channel proteins: tissue-specific expression in the common ice plant. Plant Cell 7: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BX, Verkman AS (2002) Analysis of double knockout mice lacking aquaporin-1 and urea transporter UT-B: evidence for UT-B-facilitated water transport in erythrocytes. J Biol Chem 277: 36782–36786 [DOI] [PubMed] [Google Scholar]
- You G, Smith CP, Kanai Y, Lee WS, Stelzner M, Hediger MA (1993) Cloning and characterization of the vasopressin-regulated urea transporter. Nature 365: 844–874 [DOI] [PubMed] [Google Scholar]