Abstract
Although brassinosteroids (BRs) are known to regulate shoot growth, their role in the regulation of root growth is less clear. We show that low concentrations of BRs such as 24-epicastasterone and 24-epibrassinolide promote root elongation in Arabidopsis wild-type plants up to 50% and in BR-deficient mutants such as dwf1-6 (cbb1) and cbb3 (which is allelic to cpd) up to 150%. The growth-stimulating effect of exogenous BRs is not reduced by the auxin transport inhibitor 2,3,5-triidobenzoic acid. BR-deficient mutants show normal gravitropism, and 2,3,5-triidobenzoic acid or higher concentrations of 2,4-dichlorophenoxyacetic acid and naphtaleneacetic acid inhibit root growth in the mutants to the same extent as in wild-type plants. Simultaneous administration of 24-epibrassinolide and 2,4-dichlorophenoxyacetic acid results in largely additive effects. Exogenous gibberellins do not promote root elongation in the BR-deficient mutants, and the sensitivity to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid is not altered. Thus, the root growth-stimulating effect of BRs appears to be largely independent of auxin and gibberellin action. Furthermore, we analyzed BR interactions with other phytohormones on the gene expression level. Only a limited set of auxin- and ethylene-related genes showed altered expression levels. Genes related to other phytohormones barely showed changes, providing further evidence for an autonomous stimulatory effect of BR on root growth.
Recently, the presence of brassinosteroids (BRs) was demonstrated in Arabidopsis, maize (Zea mays), pea (Pisum sativum), and tomato (Lycopersicon esculentum) roots (Kim et al., 2000; Yokota et al., 2001; Bancos et al., 2002; Shimada et al., 2003). In general, the levels of BRs such as castasterone in roots are clearly below the concentrations in shoots, and roots appear to require active BRs to a much lesser extent than shoots. In agreement with the detection of BRs in roots, genes involved in BR biosynthesis (Bancos et al., 2002) and genes involved in BR signaling (Friedrichsen et al., 2000) are expressed in roots. These findings suggest that BRs are important regulatory substances in roots.
Previous studies merely analyzed the effects of exogenously supplied BRs on root growth. To this end, different systems such as shoot cuttings (analysis of adventitious root formation), cultured excised roots, root segments, and seedling roots (Roddick and Guan, 1991) were used. In general, BRs inhibit adventitious root formation even though root induction has also been reported occasionally (e.g. Sathiyamoorthy and Nakamura, 1990). Root cuttings were used widely to analyze BR effects on main root elongation and lateral root formation. This system can provide valuable hints in determining whether roots respond directly to BRs or indirectly via primary effects on the shoot. Both inhibitory effects (e.g. Guan and Roddick, 1988; Roddick et al., 1993) and stimulating effects (e.g. Romani et al., 1983) were observed. However, potential cross talk with other phytohormones (which may be provided by the shoot and transported into the root) and the nutrient flux are interrupted in this situation. Excised root cultures, which were derived from root cuttings, are afflicted with similar drawbacks. Therefore, the analysis of seedlings and intact plants corresponds to the most natural situation. 24-Epibrassinolide (EBL) showed inhibitory effects on root formation in mung bean (Vigna radiata), wheat (Triticum aestivum), maize (Roddick and Ikekawa, 1992), tomato (Guan and Roddick, 1988), Arabidopsis (Clouse et al., 1993), and soybean (Glycine max) seedlings (Hunter, 2001).
In contrast, a weak increase of root length was observed in dark-grown cress (Lepidium sativum) roots (Yopp et al., 1981) and maize primary roots (Kim et al., 2000). The conflicting results may be explained by culture conditions (e.g. lightor darkgrown plants) and in particular by the mode, level, and time of exposure to BRs. The exogenous application of steroids bears limited resemblance to the in vivo situation where roots would respond to endogenous BRs either synthesized within the root or imported from the shoot. Exogenous phytohormones override endogenous signaling networks, and an impaired balance of pathways may be detrimental. Concordantly, auxin (e.g. Leyser et al., 1996), cytokinin (CK; e.g. Bertell and Eliasson, 1992; Cary et al., 1995), ethylene (e.g. Cary et al., 1995), abscisic acid (for review, see Pilet, 1998), and jasmonate (Staswick et al., 1992) all have the potential to inhibit root elongation, although at the same time, other processes such as auxin-induced lateral root formation (Zolman et al., 2000) and auxin- or ethylene-induced root hair elongation (Pitts et al., 1998) can occur.
The BR-deficient dwf1-6 mutant (Kauschmann et al., 1996), the BR-deficient cbb3 mutant (which is allelic to the cpd mutant; Szekeres et al., 1996), and CPD-antisense plants (Schlüter et al., 2002) were used in the present study. In comparison with mutants affected in reactions specific to BR biosynthesis (such as dwf4 and cbb3/cpd), the dwf1-6 mutant has a rather mild phenotype. The phenotypic changes of CPD-antisense plants were intermediate between BR-deficient mutants such as dwf1-6 and cbb3/cpd and the wild type. The combination of different BR-deficient genotypes provides a means to exclude changes, which are restricted to a specific genotype.
In this article, we show: (a) Positive or negative BR effects on root growth occur according to the applied BR concentration. Low concentrations of exogenous BRs stimulate root growth in wild-type plants and normalize the root length deficit of BR-deficient mutants. Higher concentrations are inhibitory. (b) Exogenous auxins stimulate root growth of BR-deficient plants but do not normalize root length. (c) The auxin transport inhibitor 2,3,5-triidobenzoic acid (TIBA) does not interfere with BR-induced root growth. (d) BRs and auxin stimulate root growth largely additively. (e) A limited number of phytohormone-related genes display altered transcript levels in roots of the BR-deficient dwf1-6 mutant. In summary, we provide evidence for an autonomous growth-stimulating effect of BRs in roots largely independent from other phytohormones.
RESULTS
Exogenous BRs Stimulate Root Growth
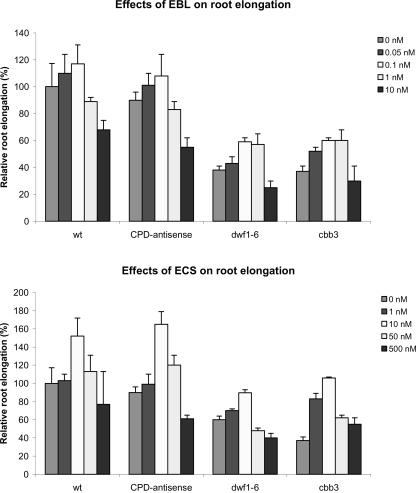
The levels of active BRs such as castasterone in Arabidopsis roots are significantly lower than the levels in shoots (Bancos et al., 2002; Shimada et al., 2003). As a consequence, we applied low concentrations of EBL and 24-epicastasterone (ECS) to consider physiological conditions. Both EBL and ECS stimulate root elongation. The growth-stimulating effect of ECS is most pronounced in the BR-deficient cbb3 mutant: 10 nm ECS raises root length to the level of mock-treated wild-type plants, and higher ECS concentrations (such as 50 and 500 nm) still stimulate root growth in the cbb3 mutant (Fig. 1). Ten micromolar ECS clearly stimulates root growth of wildtype plants, and 0.05 to 0.1 nm EBL consistently showed a tendency to promote root growth in wildtype plants in a series of three independent experiments. Higher EBL concentrations (≥1 nm) inhibit root growth of wild-type plants (Fig. 1). In contrast, 1 nm EBL clearly stimulates root growth of the cbb3 mutant, and higher concentrations such as 10 nm EBL are necessary to inhibit root growth.
Figure 1.
Low levels of exogenous BRs stimulate root growth. Plants were grown on vertically oriented plates in the presence of EBL (top) and ECS (bottom), respectively. Root length was measured 20 ± 1 d after sowing. Relative root lengths are given as the percentage of root length of untreated wild-type plants. Error bars = ±se.
These findings indicate a positive effect of physiological levels of BRs on root growth. BRs act inhibitory if a threshold level is exceeded. The threshold level depends on the biological activity of the applied BR. For instance, the critical concentration is exceeded earlier with EBL in comparison with ECS. The threshold level also depends on the genotype and is reached later in the dwf1-6 and cbb3 mutants, most likely due to low endogenous BR levels.
Auxin Effects on Root Elongation of BR-Deficient Plants
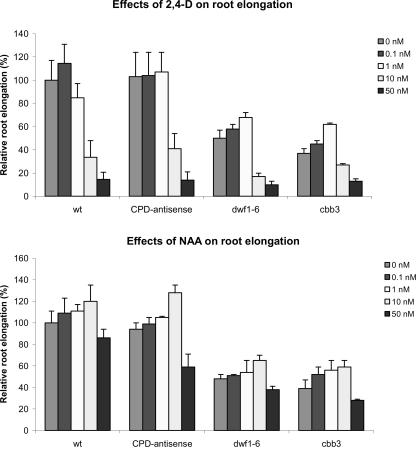
Numerous previous studies pointed to an interaction of BRs and auxins in above ground organs. For example, BR activity was demonstrated in auxin assays (e.g. Grove et al., 1979; Yopp et al., 1981; Takeno and Pharis, 1982; Katsumi, 1985). A positive interaction of BR and auxin in roots could provide the basis for the impaired root growth of BR-mutants. We analyzed the effects of 2,4-dichlorophenoxyacetic acid (2,4-D) and naphtaleneacetic acid (NAA) on root elongation of wild-type plants and BR-deficient mutants. 2,4-D and NAA differ in their transport properties (Delbarre et al., 1996). 2,4-D uptake requires an influx carrier, but 2,4-D is not secreted by an efflux carrier. NAA enters cells by passive diffusion but has its accumulation level controlled by the efflux carrier. Acropetal movement in the root of indole-3-acetic acid (IAA) delivered from the shoot has been implicated in the control of root elongation and lateral root growth, whereas basipetal movement of IAA from the root tip is required for gravity response and has been suggested to affect the initial cell divisions in lateral root initiation.
We found in a series of three independent experiments that low concentrations of both 2,4-D and NAA show a tendency to stimulate root elongation in wild-type plants. The growth-stimulating effects of 2,4-D and NAA are slightly more pronounced in the dwf1-6 and cbb3 mutants (Fig. 2). Higher concentrations of 2,4-D and NAA (≥10 and ≥50 nm, respectively) clearly inhibit root growth of both wild-type and BR-deficient plants. Thus, the BR-deficient mutants have no reduced sensitivity to either 2,4-D or NAA, suggesting that auxin influx and efflux are not impaired and that proper responses to auxin occur. Another evidence for a normal auxin transport in the dwf1-6 and cbb3 mutants is their proper gravitropic response, which is blocked in mutants such as aux1 (Marchant et al., 1999) and pin2 (Müller et al., 1998).
Figure 2.
BR-deficient plants display normal respones to 2,4-D (top) and NAA (bottom). Plants were grown on vertically oriented plates, and root length was measured 20 ± 1 d after sowing. Relative root lengths are given as the percentage of root length of untreated wild-type plants. Error bars = ±se.
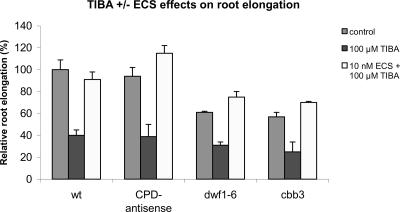
IAA moves out of plant cells through an efflux carrier apparatus that is sensitive to synthetic inhibitors. These inhibitors include N-naphtylphthalamic acid and TIBA, which elicit similar effects (Sabatini et al., 1999). The root growth of CPD-antisense, dwf1-6, and cbb3 mutants is affected by TIBA. Similar to wild-type plants, the mutants show a 50% to 60% reduction of root length in the presence of 100 μm TIBA (Fig. 3). This result indicates that the auxin promoted growth is not impaired by BR deficiency.
Figure 3.
Effects of 100 μm 2,3,5-triiodobenzoic acid (TIBA) in the presence and absence of 10 nm ECS. Plants were grown on vertically oriented plates, and root length was measured 20 ± 1 d after sowing. Relative root lengths are given as the percentage of root length of untreated wild-type plants. Error bars = ±se.
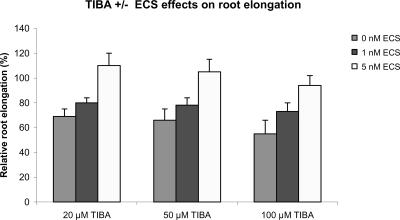
Conversely, TIBA does not interfere with the BR-induced growth. The root length of wild-type plants grown in the presence of 10 nm ECS and 100 μm TIBA is comparable with the root length of mock-treated plants, and 10 nm ECS slightly overcompensates the inhibitory effect of TIBA in the mutants. Thus, BRs clearly stimulate root growth in the presence of TIBA. The degree of root growth stimulation conferred by 10 nm ECS is similar in the presence and the absence of 100 μm TIBA (Figs. 1 and 3). This finding is confirmed by the analysis of ECS-induced root elongation in the presence of different concentrations of TIBA. ECS stimulates root growth in the presence of TIBA in a dose-dependent manner, and the extent of ECS-induced growth is not affected by TIBA (Fig. 4).
Figure 4.
The growth-stimulating effect of ECS is not diminished by 2,3,5-triiodobenzoic acid (TIBA). CPD-antisense plants were grown on vertically oriented plates, and root length was measured 20 ± 1 d after sowing. Relative root lengths are given as the percentage of root length of untreated wild-type plants grown in parallel. Error bars = ±se. Similar results were obtained for wild-type, dwf1-6, and cbb3 plants (data not shown).
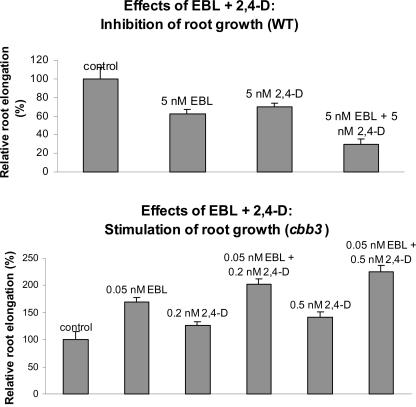
Further evidence for an auxin-independent mode of action of BR comes from the simultaneous administration of EBL and 2,4-D. In case BR and auxin act independently, stimulation and inhibition of root growth conferred by both phytohormones should be additive. In fact, the inhibitory effects of 5 nm 2,4-D and 5 nm EBL were largely additive in wild-type plants (Fig. 5). The growth-promoting effects of both phytohormones were tested in BR-deficient backgrounds because stimulatory effects are weak in wild-type plants. In line with the finding that TIBA does not interfere with BR-induced growth, low concentrations of EBL and 2,4-D additively stimulate root elongation in the cbb3 (Fig. 5) and dwf1-6 mutants (data not shown).
Figure 5.
EBL and 2,4-D effects on root growth are additive. Plants were grown on vertically oriented plates, and root length was measured 20 ± 1 d after sowing. Inhibitory effects (top) and stimulatory effects (bottom) are shown for wild-type and cbb3 plants, respectively. Relative root lengths are given as the percentage of root length of mock-treated wild-type and cbb3 plants, respectively. Error bars = ±se.
Effects of GA and 1-Aminocyclopropane-1-Carboxylic Acid (ACC) on Root Elongation of BR-Deficient Plants
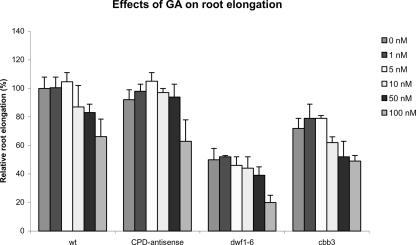
GA-deficient mutants such as ga1-3 show a drastically reduced root system. GA treatments can normalize the root length in this mutant (Fu and Harberd, 2003). To check GA effects on BR-deficient mutants, CPD-antisense, dwf1-6, and cbb3 plants were grown in the presence of different concentrations of GA4/GA7. However, GA did not stimulate root elongation in wild type or in the BR-deficient mutants (Fig. 6). Higher GA concentrations caused inhibition of growth, and the dose response curves of BR-deficient mutants were similar to the dose response curve of wild-type plants.
Figure 6.
GA does not induce root elongation in BR-deficient plants. Plants were grown on vertically oriented plates, and root length was measured 20 ± 1 d after sowing. Relative root lengths are given as the percentage of root length of untreated wild-type plants. Error bars = ±se.
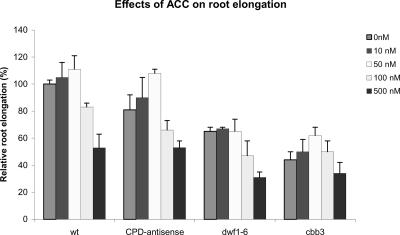
The ethylene sensitivity of BR-deficient plants was checked by means of treatments with the ethylene precursor ACC. The mutants responded in a similar way as the wild-type plants (Fig. 7). This observation indicates that BR deficiency does not alter ethylene responses.
Figure 7.
BR-deficient plants exhibit normal responses to the ethylene precursor ACC. Plants were grown on vertically oriented plates and root length was measured 20 ± 1 d after sowing. Relative root lengths are given as the percentage of root length of untreated wild-type plants. Error bars = ±se.
Analysis of Phytohormone Interactions on the Gene Expression Level
The phytohormone and TIBA treatments shown above suggest a growth-stimulating effect of BRs, which does not depend on either auxin or GA. Another approach to reveal potential interactions with other phytohormones is the analysis of gene expression profiles. We searched for phytohormone-related genes with altered transcript levels in roots of the dwf1-6 mutant. To this end, expression profiles of wild-type and dwf1-6 root material were established by means of Affymetrix ATH1 microarrays. To suppress the effects of biological variability, large pools of plants raised three times independently in a hydroponic system were used for RNA isolation and target synthesis. The targets were checked by means of Test 3 arrays and northern-blot analysis, and only bona fide targets were hybridized to ATH1 arrays. The qualitative and quantitative outcome of the Affymetrix Microarray Suite Version 5.0 software was used to identify genes with altered transcript levels in the dwf1-6 mutant. The information used included the detection P values calculated through single array analyses, the change P values, and signal log ratios determined through comparison analysis, respectively. The detection P value was applied to filter out genes with absent calls (underlying parameters: α 1 = 0.05 and α 2 = 0.065). Simultaneously, the change P value and the signal log ratio algorithms were applied in pair-wise comparisons to identify regulated genes with high reliability. Differences in expression of selected genes were confirmed by means of real-time reverse transcriptase (RT)-PCR experiments. The plant material for real-time RT-PCR was raised two times independently and was also independent from the plant material used for ATH1 array hybridizations.
The ATH1 chip represents 24,000 genes. Two hundred fifty-seven genes displayed stronger expression in the wild type (present in the wild type, change P value < 0.01, signal log ratio ≥ 0.9), and 66 genes displayed stronger expression in the dwf1-6 mutant (present in the mutant, change P value < 0.01, signal log ratio ≥ 0.9). Most genes with altered transcript levels were not characterized hitherto. A complete set of expression data are downloadable at our Web page (http://www.mpimp-golm.mpg.de/BR_reg_gene_expression/).
Similar to the situation in above ground organs (Bancos et al., 2002; Müssig et al., 2002), the steroid 22-hydroxylase DWF4 is clearly BR down-regulated, and the CPD gene shows a tendency to increased transcript levels in the mutant also. This finding is in agreement with the proposed negative feedback inhibition of BR biosynthesis.
In roots of the dwf1-6 mutant, only few auxin-related genes display significantly altered transcript levels (Tables I and II). The NIT3 gene (encoding an enzyme involved in IAA biosynthesis; Kutz et al., 2002) shows higher transcript levels in the mutant. IAA14 (Fukaki et al., 2002), IAA2, and a putative auxin-responsive GH3-like gene show weaker expression in the mutant. Thus, the expression data indicate a potential weak link between BR and auxin action in the root. A positive interaction with ethylene may be more obvious (Table I). Several genes of the ethylene response (such as ERF2 and ERF5, encoding activators of GCC box-mediated gene expression [Fujimoto et al., 2000]) and ethylene biosynthesis (e.g. ACC oxidases) show weaker expression in the dwf1-6 mutant (Table I; Web page). Genes related to GA, CK, ABA, jasmonate biosynthesis, signaling, and response did not show significantly altered transcript levels (see web page).
Table I.
Phytohormone-related genes: stronger expression in wild-type plants
All transcripts meet the presence criterion in the wild-type expression profile after hybridization of an Affymetrix ATH1 array. The change P values are <0.01 (indicating an increase according to the change algorithm of the MAS 5.0 software). Real-time RT-PCR fold change values were calculated from average threshold cycle (CT) values (see “Materials and Methods”). The table shows average CT values (±se) from three replicates at each experiment for both the gene of interest and eEF1α.
| Affymetrix Identification
|
ATH1 Array: Wild Type vs. dwf1-6
|
Annotation, Description
|
Real-Time RT-PCR: Wild Type vs. dwf1-6
|
|
|---|---|---|---|---|
| Experiment no. 1 | Experiment no. 2 | |||
| Fold change | Fold change, CT values | |||
| Ethylene-related genes | ||||
| 253259_at | 5.3 | At4g34410, putative ethylene-responsive element binding factor | nda | nd |
| 258727_at | 3.5 | At3g11930, putative ethylene-responsive protein | nd | nd |
| 258133_at | 3.0 | At3g24500, putative ethylene-responsive transcriptional coactivator | nd | nd |
| 248799_at | 2.0 | At5g47230, ethylene-responsive element-binding factor 5 (ERF5) | 1.3 | 3.1 |
| WT: CT ERF5 23.19 ± 0.19; CT elF 17.2 ± 0.05 | WT: CT ERF5 21.15 ± 0.36; CT elF 17.33 ± 0.13 | |||
| dwf1-6: CT ERF5 23.38 ± 0.12; CT elF 16.99 ± 0.07 | dwf1-6: CT ERF5 22.68 ± 0.32; CT elF 17.21 ± 0.16 | |||
| 265948_at | 2.0 | At2g19590, putative ACC oxidase | 2.2 | 3.7 |
| WT: CT ACCox 25.19 ± 0.2; CT elF 17.2 ± 0.05 | WT: CT ACCox 25.62 ± 0.12; CT elF 17.33 ± 0.13 | |||
| dwf1-6: CT ACCox 26.1 ± 0.15; CT elF 16.99 ± 0.07 | dwf1-6: CT ACCox 27.4 ± 0.15; CT elF 17.21 ± 0.16 | |||
| 248794_at | 1.7 | At5g47220, ethylene responsive element-binding factor 2 (ERF2) | 2.8 | 7.9 |
| WT: CT ERF2 22.44 ± 0.17; CT elF 17.2 ± 0.05 | WT: CT ERF2 21.49 ± 0.14; CT elF 17.33 ± 0.13 | |||
| dwf1-6: CT ERF2 23.69 ± 0.11; CT elF 16.99 ± 0.07 | dwf1-6: CT ERF2 24.36 ± 0.1; CT elF 17.21 ± 0.16 | |||
| 257922_at | 1.7 | At3g23150, ethylene receptor (ETR2) | nd | nd |
| Auxin-related genes | ||||
| 264929_at | 4.9 | At1g60730, putative auxin-induced protein | nd | nd |
| 262099_s_at | 2.1 | At1g59500, putative auxin-regulated protein GH3 | 6.0 | 2.2 |
| WT: CT GH3 19.96 ± 0.3; CT elF 15.28 ± 0.34 | WT: CT GH3 23.05 ± 0.04; CT elF 17.87 ± 0.05 | |||
| dwf1-6: CT GH3 21.97 ± 0.35; CT elF 14.7 ± 0.07 | dwf1-6: CT GH3 23.3 ± 0.1; CT elF 17.01 ± 0.13 | |||
| 245593_at | 1.9 | At4g14550, Aux/IAA protein family (IAA14) | 2.3 | 1.8 |
| WT: CT IAA14 19.52 ± 0.08; CT elF 15.28 ± 0.34 | WT: CT IAA14 22.8 ± 0.1; CT elF 17.87 ± 0.03 | |||
| dwf1-6: CT IAA14 20.14 ± 0.48; CT elF 14.7 ± 0.07 | dwf1-6: CT IAA14 22.79 ± 0.12; CT elF 17.01 ± 0.13 | |||
| 257766_at | 1.7 | At3g23030, Aux/IAA protein family (IAA2) | 1.7 | 2.6 |
| WT: CT IAA2 17.7 ± 0.03; CT elF 15.28 ± 0.34 | WT: CT IAA2 20.8 ± 0.2; CT elF 17.33 ± 0.37 | |||
| dwf1-6: CT IAA2 17.9 ± 0.1; CT elF 14.7 ± 0.07 | dwf1-6: CT IAA2 22.07 ± 0.07; CT elF 17.21 ± 0.6 | |||
a nd, Not determined.
Table II.
Phytohormone-related genes: stronger expression in the dwf1-6 mutant
All transcripts meet the presence criterion in the dwf1-6 expression profile after hybridization of an Affymetrix ATH1 array. The change P values are <0.01 (indicating an increase according to the change algorithm of the MAS 5.0 software). Real-time RT-PCR fold change values were calculated from average CT values (see “Materials and Methods”). The table shows average CT values (±se) from three replicates at each experiment for both the gene of interest and eEF1α.
| Affymetrix Identification
|
ATH1 Array: dwf1-6 vs. Wild Type
|
Annotation, Description
|
Real-Time RT-PCR: dwf1-6 vs. Wild Type
|
|
|---|---|---|---|---|
| Experiment no. 1 | Experiment no. 2 | |||
| Fold change | Fold change, CT values | |||
| 252677_at | 1.9 | Auxin-related genes At3g44320, hydrolysis of indole-3-acetonitrile to indole-3-acetic acid (NIT3) | 2.9 | 3.0 |
| WT: CT NIT3 26.16 ± 0.09; CT eIF 17.2 ± 0.05 dwf1-6: CT NIT3 24.39 ± 0.26; CT eIF 16.99 ± 0.07 | WT: CT NIT3 23.93 ± 0.19; CT eIF 17.33 ± 0.13 dwf1-6: CT NIT3 22.21 ± 0.13; CT eIF 17.21 ± 0.16 | |||
| 252184_at | 2.5 | BR-related genes At3g50660, steroid 22-hydroxylase (DWF4) | 2.2 | 1.9 |
| WT: CT DWF4 23.87 ± 0.19; CT eIF 17.2 ± 0.05 dwf1-6: CT DWF4 22.54 ± 0.06; CT eIF 16.99 ± 0.07 | WT: CT DWF4 23.8 ± 0.04; CT eIF 17.33 ± 0.13 dwf1-6: CT DWF4 22.77 ± 0.08; CT eIF 17.21 ± 0.16 | |||
| 250752_at | 1.3 | At5g05690, steroid 23-hydroxylase (CPD) | nda | 1.9 |
| WT: CT CPD 24.38 ± 0.1; CT eIF 17.8 ± 0.07 dwf1-6: CT CDP 23.76 ± 1.0; CT eIF 18.1 ± 0.32 | ||||
a nd, Not determined.
DISCUSSION
BRs Promote Root Growth
BR-deficient mutants such as dwf1-6 and cbb3 show 40% to 60% shorter roots than wild-type plants. Previous studies predominantly reported inhibitory effects of BRs on root growth (e.g. Guan and Roddick, 1988; Roddick and Ikekawa, 1992), and this inhibitory effect was used to identify or characterize BR-insensitive mutants (Clouse et al., 1996; Koka et al., 2000; Li et al., 2001; Montoya et al., 2002). In this article, we show that the BR effects on root growth are strongly dependent on the BR concentration used. Exogenous BRs stimulate root growth at low concentrations. Inhibition of root growth occurs at higher BR levels, which exceed a certain level. The EBL dose response curve (Fig. 1) corroborates previous results that showed stimulation of root growth of 7-d-old seedlings by 0.1 and 0.5 nm EBL (Clouse et al., 1996). It takes higher concentrations of biologically less active BRs (such as ECS) to exceed this level, and the dwf1-6 and cbb3 mutants tolerate higher concentrations, most likely due to reduced endogenous BR levels.
Interactions with Other Phytohormones
Expression analysis of aerial part of plants revealed a positive BR effect on the expression of a large number of auxin-related genes. Genes such as TCH4 (Xu et al., 1995); SAUR6B (Zurek et al., 1994); and IAA2, IAA3, IAA13, IAA19, IAA22, and SAUR-AC1 (Goda et al., 2002; Müssig et al., 2002) are regulated by BRs and auxin. The expression data indicate a potential weak BR-auxin cross talk in roots also. The IAA2 gene, the IAA14 gene, and a GH3-like gene show weaker expression in the dwf1-6 mutant than in the wild type. This finding could indicate reduced auxin levels, reduced auxin sensitivity, or altered auxin distribution in roots of BR-deficient plants. However, the NIT3 gene, which is involved in auxin biosynthesis, shows stronger expression in the mutant, and root growth of BR-deficient mutants is strongly inhibited by TIBA or higher concentrations of 2,4-D and NAA. These observations argue against impaired auxin responses in the mutants, and BRs may exert a direct regulatory effect on auxin-regulated genes.
The BR-induced root elongation is not diminished by TIBA (Figs. 2, 3, 4), and 100 nm EBL drastically inhibits root growth of the auxin-insensitive axr1 mutant (Clouse et al., 1993). Furthermore, EBL and 2,4-D effects on root growth are largely additive (Fig. 5). These findings provide further evidence for an auxin-independent effect of BRs on root elongation.
Low levels of 2,4-D and NAA slightly promote root growth in wild-type and BR-deficient plants (Fig. 2). Conceivably, the growth-stimulating effect of auxin could be due to a stimulation of BR biosynthesis. To check this, wild-type plants were established in a hydroponic culture system, and 1 nm 2,4-D was added to the liquid medium. CPD and DWF4 mRNA levels were not altered 3 and 24 h after treatments (data not shown), suggesting that auxin does not influence BR biosynthesis.
In contrast to BR-induced root growth, BR-induced gravitropic curvature in maize primary roots is auxin dependent. Kim et al. (2000) have shown that the activation of the gravitropic response by BRs is nullified by application of TIBA. However, the CPD-antisense, dwf1-6, and cbb3 plants show normal gravitropic responses on vertical plates, calling the necessity of BRs for the gravitropic response into question and suggesting functional auxin influx and efflux carriers to be present despite the BR deficiency in these mutants (Müller et al., 1998; Marchant et al., 1999).
An impaired auxin transport is also indicated by the administration of different auxins, which possess different transport properties. For instance, root growth of the aux1 mutant is less sensitive to auxins requiring carrier-mediated uptake (2,4-D and IAA), but the dose response curves for wild type and aux1 root elongation in the presence of varying concentrations of 1-NAA are identical (Marchant et al., 1999). However, roots of BR-deficient plants do not show reduced sensitivity to either 2,4-D or 1-NAA in comparison with wild-type plants. For instance, 10 nm 2,4-D and 50 nm NAA clearly inhibit root elongation in all genotypes (Fig. 2). We also did not find significantly altered transcript levels of genes involved in the auxin transport (see Web page).
Another root growth-promoting phytohormone is GA. Roots of GA-deficient mutants (e.g. ga1-3 from Arabidopsis [Fu and Harberd, 2003] and na from pea [Yaxley et al., 2001]) are shorter than those of the wild type. GA treatment normalizes the root length of ga1-3 mutant plants to wild-type levels, showing the role of GA in the regulation of root growth (Fu and Harberd, 2003). Auxin is necessary for GA-mediated control of root growth because attenuation of polar auxin transport by means of N-naphtylphthalamic acid or decapitation of plants restrains GA-induced root growth (Fu and Harberd, 2003). GA treatments do not normalize root length of the dwf1-6 and cbb3 mutants. The dose response curves of BR-deficient mutants are identical to the dose response curve of wild-type plants (Fig. 6).
First hints to GA and BR signaling cross talk were derived from the finding that bri1-201 seedlings have drastically reduced transcript levels of the GA-repressed GA5 gene and elevated transcript levels of the GA-inducible GASA1 gene. BR treatment of cpd seedlings resulted in decreased GASA1 and increased GA5 expression, showing that BRs affect the expression of the GA5 and GASA1 genes antagonistically to GA (Bouquin et al., 2001). However, no GA-related genes (including the GA5 and GASA1 genes) show significantly altered transcript levels in the dwf1-6 mutant. Thus, the BR effect on root elongation most likely is independent from GA.
Putative BR Effects on Ethylene Production and Responses
The elongation of roots is inhibited by ACC and ethylene (Cary et al., 1995). This also holds true for BR-deficient plants (Fig. 7). Higher concentrations of auxin inhibit root growth as well, suggesting a common mechanism (for review, see Casson and Lindsey, 2003). In fact, auxin stimulates ethylene biosynthesis, and elevated ethylene levels inhibit root elongation potentially via the inhibition of polar auxin transport. This mechanism constitutes an auxin-ethylene feedback loop.
Likewise, BRs may stimulate ethylene biosynthesis and trigger ethylene responses in roots. BRs are known to stimulate the production of ethylene in shoots and roots (e.g. Schlagnhaufer et al., 1984; Arteca et al., 1985; Woeste et al., 1999; Yi et al., 1999; Arteca and Arteca, 2001). In line with these observations, our expression data point to a positive BR effect on genes involved in ethylene biosynthesis and ethylene response (Table I; Web page). The positive BR effect on ethylene production could have an impact on the auxin-ethylene feedback loop. However, this cannot account completely for the BR-induced growth inhibition because EBL clearly inhibits root growth in the axr1 mutant (Clouse et al., 1993). A direct BR-ethylene feedback loop may exist, which specifically interferes with BR transport, BR biosynthesis, or BR responses.
A close ethylene interaction in roots was shown for CKs also. Synthetic CKs inhibit root growth, and this effect is coupled to ethylene (e.g. Bertell and Eliasson, 1992; Cary et al., 1995; for review, see Casson and Lindsey, 2003). Transgenic plants with reduced CK content display enhanced root growth (Werner et al., 2001). However, there is little evidence for a BR-CK interaction, and dwf1-6 roots did not show significantly altered transcript levels of CK-related genes. Furthermore, the dwf1-6 mutant shows decreased rather than increased transcript levels of genes involved in the ethylene response or ethylene biosynthesis. Therefore, the reduced root growth of BR-deficient plants most likely is not related to CKs.
Conclusion and Outlook
Our experiments have shown that BRs promote root elongation. This effect appears to be largely independent from auxin and GA. In future experiments, we intend to analyze the inhibitory effect of higher concentrations of BRs, which may depend on ethylene. The simultaneous application of high concentrations of BRs and ethylene biosynthesis inhibitors (such as aminoethoxyvinyl-Gly) or ethylene signaling inhibitors (such as silver thiosulfate) and the analysis of BR effects on ethylene mutants allows verification of the proposed BR-ethylene interaction.
The Affymetrix expression profiles and real-time RT-PCR experiments (Table I; Web page; data not shown) indicate altered expression levels of the SCARECROW transcription factor (Di Laurenzio et al., 1996; stronger expression in dwf1-6), the ROOT HAIRLESS 1 gene (Schneider et al., 1998; stronger expression in dwf1-6), and SOLITARY-ROOT/IAA14 gene (weaker expression in dwf1-6; Fukaki et al., 2002). These genes are required for the radial organization of the Arabidopsis root, formation of root hairs, and involved in lateral root formation, respectively. Thus, BRs may play a role in root differentiation. Although BR-deficient plants produce lateral roots and root hairs, it may be worthwhile to carry out a detailed morphological analysis of the root system (in particular, of the root meristem).
MATERIALS AND METHODS
Hydroponic Growth Conditions
Arabidopsis cv 24 (wild type) and the BR-deficient mutant dwf1-6 (cbb1; Kauschmann et al., 1996) were grown hydroponically in modified 0.25× Hoagland medium. Two different conditions were applied to establish plants. Black 0.5-mL reaction tubes (Treff AG, Degersheim Switzerland) were filled with agar-solidified medium (0.8% [w/v] agar in 0.25× modified Hoagland medium), and the bottom of the tube was cut after the agar became solid. The medium composition was 1.5 mm Ca(NO3)2, 0.28 mm KH2PO4, 0.75 mm MgSO4, 1.25 mm KNO3, 0.5 μm CuSO4, 1.0 μm ZnSO4, 5 μm MnSO4, 25 μm H3BO3, 0.1 μm Na2MoO4, 50 μm KCl, 3 mm MES-KOH (pH 5.7), and 20 μm FeEDTA. Single seeds were placed on top of the agar-filled tubes, and the tubes were placed into holes in the cover of boxes filled with medium. Thus, light entry into the medium was largely prohibited. Boxes were transferred into a greenhouse, and the medium was changed weekly. Plants were harvested 24 d after sowing. This method was used for raising the material for microarray hybridizations.
Alternatively, plants were raised in one-half-concentrated Murashige and Skoog medium supplemented with 1% (w/v) Suc and solidified with 0.7% (w/v) agar under a 16-h day (140 μmol m-2 s-1, 22°C)/8-h night (22°C) night regime. Plants were transferred to boxes filled with 0.25× modified Hoagland medium after 10 d of growth and then treated as described above. This method produces larger plants and was applied for auxin treatments of wild-type plants and for production of wild-type and dwf1-6 plant material for real-time RT-PCR.
Root Growth Assays
For monitoring effects of different phytohormones on root growth of wild type, CPD-antisense (Schlüter et al., 2002), dwf1-6, and cbb3 plants (Kauschmann et al., 1996), plants were grown in one-half-concentrated Murashige and Skoog medium supplemented with 0.8% (w/v) agarose under a 16-h day (140 μmol m-2 s-1, 22°C)/8-h night (22°C) regime. Phytohormones and inhibitors were added as described in the text. BRs (CIDtech Research Inc., Cambridge, ON), GA (61.2% GA4 and 30.1% GA7, Duchefa, Haarlem, The Netherlands), and TIBA (Duchefa) were solved in MeOH, ACC (Sigma, Taufkirchen, Germany) was solved in water, 2,4-D (Duchefa) in ethanol, and NAA (Duchefa) in dimethyl sulfoxide. All plates of a given experiment (e.g. four different EBL concentrations and the appropriate control) contained the same amount of solvent (e.g. in case of EBL treatment, all plates contained 5 × 10-4% [v/v] MeOH) to rule out solvent effects. Approximately 3 cm of medium was removed after the medium became solid, and seeds were placed on top of the cutting area. Only for the experiments shown in Figure 5, no medium was removed, and seeds were placed directly on the agar surface. Plates were placed upright, and root length was measured 20 ± 1 d after sowing. All experiments were performed three times (BR treatments [Fig. 1], auxin treatments [Fig. 2], and TIBA treatments [Figs. 3 and 4]) or two times (auxin and BR treatment [Fig. 5], GA treatment [Fig. 6], and ACC treatments [Fig. 7]). Twenty-five plants per experiment, treatment, and genotype were analyzed.
Hybridization of Affymetrix Genome Arrays
Total RNA was isolated by means of the TRIzol Reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. The protocol was modified with respect to the RNA precipitation step (1/20 volume 3 m NaOAc was added to increase yield). The quality and quantity was checked using the Bioanalyzer 2100 (Agilent Technologies, Böblingen, Germany) and MOPS-formaldehyde agarose gels. Twenty micrograms of total RNA was used for double-stranded cDNA synthesis (SuperScript Choice System, Invitrogen). Biotin-labeled cRNAs were synthesized using the BioAray High Yield RNA Transcript Labeling Kit (Enzo, New York). All cRNA samples were checked for degradation by gel analysis according to the Affymetrix technical manual. In addition, all targets were checked by hybridizations of Test 3 arrays. Only bona fide probes were used for ATH1 array hybridizations (Affymetrix, Santa Clara, CA). Hybridization, washing, staining, and scanning procedures were performed as described in the Affymetrix technical manual. Expression analysis via the Affymetrix Microarray Suite Version 5.0 software was performed with standard parameters. The output of every experiment was multiplied by a scaling factor to adjust its average intensity to a target intensity of 100. Thus, scaling allows comparisons between any two experiments. To consider biological variability, RNA from three independent experiments was pooled for the synthesis of the Affymetrix target. Selected expression data were verified two times by means of real-time RT-PCR with independent plant material.
Real-time RT-PCR
Total RNA was isolated using the Invisorb Spin Plant RNA kit (Invitek, Berlin). One microgram of total RNA was then reverse transcribed with the Superscript II RT (Invitrogen) in a reaction volume of 28.5 μL to generate first strand cDNA. Every cDNA was checked for contamination with genomic DNA by means of PCR using the intron-spanning primers SPY_fw 5′ gga atc tag ctt tcg att gtt ttt ctg a 3′, SPY_rev 5′ tca aag ttt gga gac aca gct aga cat c 3′, CCA1_fw 5′ aac agc aac aac aat gca act act gat t 3′, and CCA1_rev 5′ aca aac aga gac aag aga caa gac atg g3′. Occasionally, genomic PCR products appeared after 35 cycles, and these cDNAs were discarded. Real-time RT-PCR was performed with 1 μL of a 1:3.5 dilution of the first strand cDNA reaction and the SYBR Green reagent (Applied Biosystems, Foster City, CA) in a 25-μL volume on a Perkin Elmer Geneamp 5700 machine (PerkinElmer, Boston) using the primers ERF2_fw 5′acggactcctcaaagatgccttc 3′, ERF2_rev 5′ctcctccatcgccgtaaagttct 3′ (At5g47220), ERF5_fw 5′tgacgttaacggtggagagacg 3′, ERF5_rev 5′tgaggagataacggcgacagaag 3′ (At5g47230), ACC_ox_fw 5′gtcagccattaccctccttgtcc 3′, ACC_ox_rev 5′ctgaaggccatcatattcatcg 3′ (At2g19590), NIT3_fw 5′ atgatcctactgtctccggaggtg 3′, NIT3_rev 5′ ccaagatcaagatcagctgtgacg 3′ (At3g44320), GH3_fw 5′ tgtcaagcttggtcaggaatacg 3′, GH3_rev 5′ cgctttgttcttgaaaccagtca 3′ (At1g59500), IAA2_fw 5′ cccgtaagaacaacaacagtgtga 3′, IAA2_rev 5′ ctctaacgctttgagaagctcgg 3′ (At3g23030), IAA14_fw 5′ tatgtgccaagctacgaggacaa 3′, IAA14_rev 5′ ccaactgcttcagatcccttcat 3′ (At4g14550), DWF4_fw 5′ cagacgatgatcttttgggatgg 3′, DWF4_rev 5′ agaagaagtctcatgtccggcaa 3′ (At3g50660), CPD_fw 5′ cagagcaactcggtaacgacagg 3′, CPD_rev 5′ gcggtgaaggaaaacagagagtg 3′ (At5g05690), eIF1α _fw 5′ ttgacaggcgttctggtaagg 3′, eIF1α _rev 5′ cagcgtcaccattcttcaaaaa 3′ (At5g60390), Ubq10_fw 5′ cacactccacttggtcttgcgt 3′, and Ubq10_rev 5′ tggtctttccggtgagagtcttca 3′ (At4g05320). All primer pairs amplified single products, as shown by the melting temperatures of the amplicons and gel electrophoresis. The use of different control genes (either eEF1α or Ubq10) did not bias the results with respect to the fold change values. Therefore, only the eIF1α-based data are shown in Tables I and II.
Data were normalized to eIF1α and then compared according to the formula (considering as example DWF4):
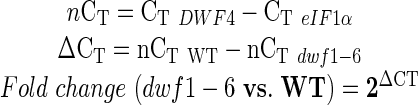 |
Acknowledgments
We thank Peggy Lange for her excellent technical assistance. We are grateful to Michael Udvardi for the possibility to use the Perkin Elmer Geneamp 5700 machine.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.028662.
References
- Arteca JM, Arteca RN (2001) Brassinosteroid-induced exaggerated growth in hydroponically grown Arabidopsis plants. Physiol Plant 112: 104–112 [DOI] [PubMed] [Google Scholar]
- Arteca RN, Bachman JM, Yopp JH, Mandava NB (1985) Relationship of steroidal structure to ethylene production by etiolated mung bean segments. Physiol Plant 64: 13–16 [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertell G, Eliasson L (1992) Cytokinin effects on root growth and possible interactions with ethylene and indole-3-acetic acid. Physiol Plant 84: 255–261 [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127: 450–458 [PMC free article] [PubMed] [Google Scholar]
- Cary AJ, Liu W, Howell SH (1995) Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyls elongation in Arabidopsis thaliana seedlings. Plant Physiol 107: 1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Lindsey K (2003) Genes and signalling in root development. New Phytol 158: 11–38 [Google Scholar]
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker ME (1993) Physiological and molecular effects of brassinosteroids on Arabidopsis thaliana. J Plant Growth Regul 12: 61–66 [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2, 4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CAP, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC (1979) Brassinolide, a plant growth promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Guan M, Roddick JG (1988) Epibrassinolide-inhibition of development of excised, adventitious and intact roots of tomato (Lycopersicon esculentum): comparison with the effects of steroidal estrogens. Physiol Plant 74: 720–726 [Google Scholar]
- Hunter WJ (2001) Influence of root-applied epibrassinolide and carbenoxolone on the nodulation and growth of soybean (Glycine max L.) seedlings. J Agron Crop Sci 186: 217–221 [Google Scholar]
- Katsumi M (1985) Interaction of a brassinosteroid with IAA and GA3 in the elongation of cucumber hypocotyls sections. Plant Cell Physiol 26: 615–625 [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T (1996) Genetic evidence for an essential role of brassinosteroids in plant development. Plant J 9: 701–713 [Google Scholar]
- Kim S-K, Chang SC, Lee EJ, Chung W-S, Kim Y-S, Hwang S, Lee JS (2000) Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol 123: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD (2000) A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol 122: 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz A, Müller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW (2002) A role for nitrilase 3 in the regulation of root morphology in sulphurstarving Arabidopsis thaliana. Plant J 30: 95–106 [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroidinsensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ (2002) Cloning the tomato Curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14: 3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 23: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilet PE (1998) Some cellular and molecular properties of abscisic acid: its particular involvement in growing plant roots. Cell Mol Life Sci 54: 851–865 [Google Scholar]
- Pitts RJ, Cernac A, Estelle M (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Roddick JG, Guan M (1991) Brassinosteroids and root development. In HG Cutler, T Yokota, G Adam, eds, Brassinosteroids; Chemistry, Bioactivity and Applications, American Chemical Society Symposium Series. American Chemical Society, Washington, DC, pp 231–245
- Roddick JG, Ikekawa N (1992) Modification of root and shoot development in monocotyledon and dicotyledon seedlings by 24-epibrassinolide. J Plant Physiol 140: 70–74 [Google Scholar]
- Roddick JG, Rijnenberg AL, Ikekawa N (1993) Developmental effects of 24-epibrassinolide in excised roots of tomato grown in vitro. Physiol Plant 87: 453–458 [Google Scholar]
- Romani G, Marrè MT, Bonetti A, Cerana R, Lado P, Marrè E (1983) Effects of a brassinosteroid on growth and electrogenic proton extrusion in maize root segments. Physiol Plant 59: 528–532 [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sathiyamoorthy P, Nakamura S (1990) In vitro root induction by 24-epibrassinolide on hypocotyls segments of soybean [Glycine max (L.) Merr.]. Plant Growth Regul 9: 73–76 [Google Scholar]
- Schlagnhaufer C, Arteca RN, Yopp JH (1984) A brassinosteroid-cytokinin interaction on ethylene production by etiolated mung bean segments. Physiol Plant 60: 347–350 [Google Scholar]
- Schlüter U, Köpke D, Altmann T, Müssig C (2002) Analysis of carbohydrate metabolism of CPD antisense plants and the brassinosteroid-deficient cbb1 mutant. Plant Cell Environ 25: 783–791 [Google Scholar]
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K (1998) The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev 12: 2013–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálman Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Takeno K, Pharis RP (1982) Brassinosteroid-induced bending of the leaf lamina of dwarf rice seedlings: an auxin-mediated phenomenon. Plant Cell Physiol 23: 1275–1281 [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Vogel JP, Kieber JJ (1999) Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol Plant 105: 478–484 [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB (2001) Gibberellin biosynthesis mutations and root development in pea. Plant Physiol 125: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi HC, Joo S, Nam KH, Lee JS, Kang BG, Kim WT (1999) Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.). Plant Mol Biol 41: 443–454 [DOI] [PubMed] [Google Scholar]
- Yokota T, Sato T, Takeuchi Y, Nomura T, Uno K, Watanabe T, Takatsuto S (2001) Roots and shoots of tomato produce 6-deoxo-28-norcathasterone, 6-deoxo-28-nortyphasterol and 6-deoxo-28-norcastasterone, possible precursors of 28-norcastasterone. Phytochemistry 58: 233–238 [DOI] [PubMed] [Google Scholar]
- Yopp JH, Mandava NB, Sasse JM (1981) Brassinolide, a growth-promoting steroidal lactone: I. Activity in selected auxin bioassays. Physiol Plant 53: 445–452 [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD (1994) Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol 104: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]