Abstract
Synthetic biology is an engineering approach that seeks to design and construct new biological parts, devices and systems, as well as to re-design existing components. However, rationally designed synthetic circuits may not work as expected due to the context-dependence of biological parts. Darwinian selection, the main mechanism through which evolution works, is a major force in creating biodiversity and may be a powerful tool for synthetic biology. This article reviews selection-based techniques and proposes strict Darwinian selection as an alternative approach for the identification and characterization of parts. Additionally, a strategy for fine-tuning of relatively complex circuits by coupling them to a master standard circuit is discussed.
Keywords: Darwinian selection, Directed evolution, Synthetic biology
Natural selection is the blind watchmaker, blind because it does not see ahead, does not plan consequences, has no purpose in view. Yet the living results of natural selection overwhelmingly impress us with the appearance of design as if by a master watchmaker, impress us with the illusion of design and planning
Richard Dawkins, The blind watchmaker
Under changing conditions of life, there is no logical impossibility in the acquirement of any conceivable degree of perfection through natural selection
Charles Darwin, On the origin of the species
Synthetic biology: features and tools
Synthetic Biology (SB) is more an approach than a discipline; a framework that includes bio-engineering, systems biology, metabolic engineering and many other disciplines, encompassing the design and construction of new biological parts, devices and systems, as well as the re-design of existing components. SB has also been defined for its potential to embrace the emerging field of designing, synthesizing and evolving new genomes or biomimetic systems. The fundamental principle behind SB is that, analogous to artificial objects, any biological system can be considered as a combination of individual functional elements (de Lorenzo and Danchin 2008). SB approaches are based on three main principles: part-by-part construction of functional elements or biological parts, standardization of these particular parts, and abstraction of the complex underlying information (e.g. the particular DNA sequence).
SB has a very short history; less than a decade in fact. However, the novelty of the discipline is in contrast with the relative antiquity of the tools it uses. In fact, the idea of living organisms as cellular robots might be new, but the techniques used to select, re-design and combine biological parts, in addition to automated sequencing, are standard biotechnological protocols. Unfortunately, experience demonstrates that often rational robot-like, part-by-part approaches simply do not work. For example, Chan et al. (2005) reported that bacteriophages re-designed in order to behave in a more ‘logical’ way, in fact made smaller lysis plaques than their wild-type precursors and might even evolve to get rid of the man-made components. Unlike robots, all living beings tend to perpetuate reproductively over time. Unfortunately, unlike robots, genetically engineered organisms are prone to die. Mutations, changes in the environment and interactions with cellular components thus make synthetic components context-dependent. This means that they might work in one context but not in others. The ultimate factor responsible for this disparity between theory and practice is natural selection.
Creating diversity, selecting fitness
The identification, characterization and optimization of biological parts to be used in SB are often carried out by selection-based approaches. Many screenings aimed at identifying sequences that are suitable as parts for SB combine a method to create a library with high genetic diversity with a Darwinian selection step. Alper et al. (2005) quantitatively characterized a promoter library by muting a constitutive promoter through error-prone PCR and constructing a library with the mutant sequences cloned upstream of a GFP gene. The authors carried out a “pick and test” screening of the resulting library in E. coli on the basis of the fluorescence of the clones, and an additional “dead or alive” confirmation of the constitutive nature of the promoters was performed by cloning them upstream of a chloramphenicol acetyltransferase (cat) gene with chloramphenicol as the selection agent.
In another work, Alper et al. (2006) reported a very interesting approach based on what the authors call global transcription machinery engineering (gTME) on yeast. This is an error-prone PCR-based method in which mutations of a key protein regulating the global transcriptome are produced and a library with the mutants, exhibiting a wide range of diversity at the transcriptional level, is screened. The cited work carried out selection in a medium with high ethanol and glucose concentrations, allowing only tolerant clones to survive. Following this approach, the authors were able to identify several mutants with enhanced ethanol and glucose tolerance, one of which was analysed in detail and found to exhibit differential expression of hundreds of genes compared to the wild type.
These works focused on the selection of particular genetic variants for useful purposes. However, it has to be noted that differences in biological fitness on a particular trait depend, particularly in microorganisms, not only on genetic variations in terms of homology but also on the copy number. Thus, copy number manipulation has been proposed as a potentially powerful strategy to engineer microorganisms displaying new phenotypes: Christ and Chin (2008) reported that evgA gene amplification allowed E. coli to survive at extreme and otherwise lethal temperatures.
Besides error-prone PCR, there are also non-recombinant alternatives to the rapid production of genetic variants. For example, DNA shuffling is known to mimic evolutionary processes. Whole genome shuffling, a process combining multi-parental crossing obtained by DNA shuffling with standard breeding has been successfully implemented in bacteria (Zhang et al. 2002). This method has been found to be faster than sequential random mutagenesis and screening for the production of improved organisms.
Protein engineering and directed evolution
The linkage between genotype and phenotype is the basis of selection-based evolution, and it is also the basis of protein engineering approaches for identification and characterization of gene products. Persson et al. (2008) combined random mutagenesis and phage display selection strategies of various stringencies, which gave a considerable increase in apparent affinities for several of the selected populations. Phage display has also been combined with proteolysis selection in order to generate novel proteins with stable folds (Riechmann and Winter 2000). Ribosome display (Hanes and Plückthun 1997) and in vitro compartmentalization (Tawfik and Griffiths 1998) can be also used to evolve proteins for their binding interactions. For example, an approach has been developed aimed at mimicking natural selection (fitter genes having more “offspring”) by coupling the amplification of a gene to the formation of product by the enzyme it encodes (Kelly and Griffiths 2007). All these works suggest that directed evolution strategies could successfully complement in vitro selection.
Improving the fittest: adaptive evolution
Adaptation is the evolutionary process whereby an organism becomes better able to live in its habitat. An experimental strategy based on adaptive evolution has been developed as a tool for improving genetic circuits. The experimental procedure basically involves maintaining exponential growth by daily passage of cultures into fresh medium under the selection pressure. It has been reported that this simple strategy can be used to achieve in silico predicted biological functions. For example, Ibarra et al. (2002) showed that placing E. coli under growth selection pressure by using glycerol as the sole carbon source led to an increase in the growth rate from a sub-optimal to the optimal rate predicted from a whole-cell in silico model. The compatibility of selection-based approaches with in silico design was also reported by Fong et al. (2005a), who used an integrative in silico + adaptive evolution approach in order to select for lactic acid production in E. coli. The authors succeeded in constructing highly productive strains based on the computationally predicted designs, and the production of lactic acid was further increased after adaptive evolution was carried out, mainly because growth rate was coupled to lactate secretion rate.
Adaptive evolution approaches such as those cited above often use adaptation as a final improvement step, with the main source of genetic variants being achieved by muting an in silico design and further selection implemented to perfect the fitness of the design. However, adaptive evolution might be used as a simple and rapid tool for the selection of desired phenotypes (Fong et al. 2005b). Although the mechanism underlying this fast adaptation is not fully understood, it is known that compensatory gene expression changes occur as part of the initial adaptive response followed by further positive selection of beneficial gene expression changes.
Learning from Darwin: further simple selection approaches
It is well known that Darwin compiled a huge amount of data on biological diversity and adaptations during the Beagle’s voyage, and also during his fieldwork in the United Kingdom. However, the strength of Darwin’s argument lies mainly in artificial, rather than natural, selection. The origin of the species by means of natural selection (Darwin 1859) is, in fact, full of dozens of examples of artificial selection of cattle and crops, and the power of artificial selection to produce biological diversity was described in detail 9 years later in a book dedicated to the topic (Darwin 1868). The mechanism behind both natural and artificial selection is the same: the fittest (with respect to the environment in natural selection and with respect to human requirements for artificial selection) survive. Domestication confirms the tremendous power of selection in order to fix genetic variants and produce phenotypic traits that are mainly (but not only) quantitative and often astoundingly distant from those of the original natural population they originated from. A classic example is the dairy cow, many breeds of which can produce 10,000 l of milk or more per cow every year.
The simple mechanism of natural and artificial selection has only been partially mimicked in SB, which often uses a directed evolution approach. By this approach, natural intraspecific variation is substituted by random mutations and selection is usually performed on the basis of screenings for a desired trait (e.g. enzymatic activity). Often, hundreds of bacterial colonies are individually picked from the pool of mutants, the desired trait analysed and clones are selected or discarded depending on the existence and/or intensity of the desired trait.
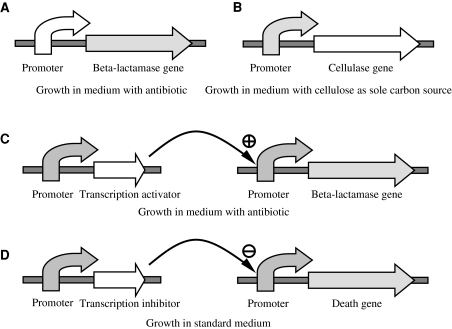
It is tempting to envisage variations of this strategy applying strict Darwinian selection, in which a large pool of natural genetic variants would be selected on the biological fitness of the variants in a given environment. The use of naturally occurring DNA sequences would imply a first round of selection, since wild-type coding sequences have already been shaped by natural selection. In fact, natural variants that are selected are a priori superior to randomly produced mutants, as demonstrated by the success of pharmaceutical screenings on natural compounds to be used as antibiotics, anti-tumorals or for many other therapeutic applications (Li and Vederas 2009). Ideally, the initial genetic pool might be genomic, meta-genomic or even a combination of several meta-genomic DNA libraries, although experimental handling limitations would define the size of the starting pool. Figure 1 shows four examples of a strategy based on Darwinian selection applied to the identification of biological parts to be used in SB, such as strong promoters (Fig. 1a), protein coding sequences (1B) and sequences coding for transcription regulators (1C and 1D).
Fig. 1.
Strategies based on the proposed Darwinian selection approach aimed at identifying biological components suitable for SB, such as strong promoters (a), protein coding sequences such as cellulases (b) and sequences coding for transcription regulators: activators (c) and inhibitors (d). Natural (i.e. meta-genomic) DNA variants are cloned as the components subjected to selection (white arrows). Dark and light gray arrows correspond to other components. Suitable media allowing selection of the fittest components are indicated. Notice than solid, rather than liquid media should be used, since B-lactamase, as well as some cellulases are secreted and bystander cells might survive if liquid media was used
Interestingly, there are many simple biotechnological screenings that are very similar to the Darwinian strategy proposed here for SB. In classical biotechnology, screening for enzymes and, less frequently, promoters or other regulatory sequences often follows procedures utilizing the deleterious effect of a selective medium on the vast majority of the screened clones. For example, screening for cellulases is usually carried out on CMC (carboxymethylcellulose)-based media, which results in the selection of cellulolytic isolates (Yan et al. 2001). A particularly interesting selection-based strategy is that reported by Kubota et al. (1991), where E. coli promoters were screened by cloning hundreds of DNA fragments from a genomic library upstream of the ampC gene into a promoter-probe plasmid. By selecting with antibiotic so that only clones with sequences promoting ampC expression survived, the authors were able to identify and characterize several naturally occurring strong promoters. This strategy and the aforementioned screening for cellulases are in fact two examples of Darwinian selection approaches for SB, which are shown in Fig. 1a, b, respectively.
Complexity problems and evolutionary solutions
Genetic networks able to integrate multiple inputs in the generation of cellular responses have been constructed. Sayut et al. (2009) developed an AND logic gate displaying clear, logical responses that could be described using a mathematical model. However, the analysis of even relatively simple synthetic networks often reveals that there is a surprisingly large diversity of complex behaviors (Guet et al. 2002). A common problem when multiple genes are used in a synthetic circuit is that the expression level of each gene must be controlled independently. In practice, certain promoters are known to suffer from cross talk (an inducer of one of the promoters affects the expression of the other). This has been reported for the IPTG-inducible Plac and the arabinose-inducible PBAD promoters, with IPTG being in fact an inhibitor of PBAD. This problem has been overcome by applying a directed evolution approach to screen mutants of the arabinose-binding regulatory protein AraC in order to construct an arabinose-inducible system compatible with IPTG (Lee et al. 2007).
The sensitivity of complex circuits to a range of parameters, from protein and RNA stabilities to culture temperature, might result in synthetic circuits working imperfectly. Again, directed evolution can be used to complement rationally designed circuits, which can be optimized by screening randomly mutated circuits (Yokobayashi et al. 2002).
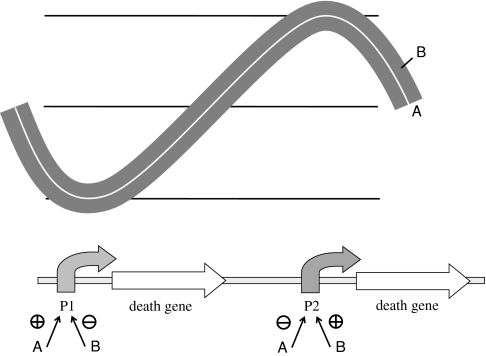
Theoretically, any pool of living organisms with genetic variation and vigorous reproduction is suitable for Darwinian selection. However, fully selection-based shaping of sophisticated synthetic systems such as oscillators or computational biological devices (i.e. counters) is difficult to implement because of their complexity. Genetic circuits that yield simple ON/OFF outputs can be tuned by specifically designed selection modules (Yokobayashi and Arnold 2005). However, selection of oscillatory circuits would need oscillatory levels of selection pressure, which might be difficult to implement in liquid cultures. For this particular case, a possible selection approach could be implemented whereby (1) individual components of the circuit are first screened on the basis of their fitness (Fig. 1), and (2) the complete network is subjected to further selection through specifically designed selection modules yielding an oscillatory selection pressure. This could be achieved by combining the circuit to be tested with a master circuit exhibiting a standard behavior. The output of the master circuit would be the selection pressure aimed at maintaining the behavior of the tested circuit within certain levels. Figure 2 shows an example of this proposed approach in which the fitness of an oscillator is selected by a master circuit through transcriptional activation/inhibition of a death gene on a third coupling circuit. Using previously defined robust synthetic circuits as master, fine tuning of developing circuits might be achieved. With this strategy, fluctuations in the selection pressure might be more easily achieved than with chemical modification of the growth medium.
Fig. 2.
Strategy for directing the evolution of an oscillatory circuit though a well calibrated master circuit. An oscillatory circuit A (white line) is subjected to fluctuating selection by a master B oscillator (thick grey line) through the action of a coupler circuit (shown below). P1 and P2, promoter sequences. The behavior of circuit A must be within certain limits marked by the output of B by transcriptional activation/inhibition of a death gene on the coupling circuit. When the output of master circuit B decreases, so too should the output of circuit A, in order to keep the death gene under the control of P1 inhibited. When the output of master circuit B rises, high inhibitory levels of A must balance activation of P2 by B
Directed evolution on a higher level of biological complexity, i.e. multicellular systems, is a particularly challenging field. Microchemical interface technology has been proposed as a powerful tool to interface with developing biological systems thus achieving unprecedented levels of spatial and temporal control of chemical environments (Ismagilov and Maharbiz 2007). Theoretically, selection pressure could be administered using this technique and multicellular systems tuned by directed evolution. However, true Darwinian selection of multicellular systems would require a vast pool of genetically diverse multicellular systems (each composed of genetically identical cells) which would need to be exposed to a range of selection factors in order to discard imperfectly working systems. Such a strategy is technically unapproachable. Additionally, it should be noted that group (multicellular system) rather than individual (cell) selection should occur. Selection needs genetic variations to shape, and although a multicellular system can mute, mutations will certainly originate and operate at an individual (cell) level, and the whole system might not be affected. Darwinian selection of multicellular systems would need beneficial mutations (in terms of the desired behavior of the circuit) to somehow extent from individual cells to the whole system, in order for the new genetic trait to produce a particularly behavior (phenotype) of the system that would be sensitive to selection pressure. Today, there are no strategies for Darwinian selection of multicellular systems: in SB, the individual cell seems to be the threshold of complexity that Darwinian selection can cope with.
Concluding remarks and future prospects
By comparing the complexity and diversity of natural organisms with those of man-made biological constructions, it can be concluded that adaptation is the key factor behind the superiority of the evolutionary process on rational design. Thus, it seems logical that artificial construction of living circuits relies, at least partially, on selection processes. Selection-based approaches have been successfully used in SB, mainly as a complement of in silico designs. Identification, characterization and improvement of biological parts can be partially achieved through selection-based approaches, and directed evolution has proven a successful strategy to adapt rationally designed simple circuits to a context-dependent environment. Fully Darwinian selection strategies (applied on natural rather than artificially mutated parts and with “dead or alive” screenings) might also be implemented.
The difficulty of applying true selection to complex circuits might be partially overcome by using specifically designed selection modules (Yokobayashi and Arnold 2005) as well as ad hoc master circuits, like the one proposed in this work. However, highly sophisticated biological networks as well as multicellular systems are still recalcitrant to Darwinian selection mainly because of the difficulties of producing and selecting among a sufficiently vast pool of genetically diverse systems.
As a final remark, it seems that the growing complexity of synthetic circuits is linked to a need for powerful evolutionary approaches in order to adapt the circuits and to improve their performance in a context-dependent environment. Such strategies based on selection, that brilliant ‘blind engineer’, may prove key to the construction of complex living systems in the near future.
Acknowledgments
I am indebted to Fabiola Barraclough for the correction of the English text and to two anonymous referees for their constructive comments and suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA. 2005;102:12678–126283. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1:0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ D, Chin JW. Engineering Escherichia coli heat-resistance by synthetic gene amplification. Protein Eng Des Sel. 2008;21:121–125. doi: 10.1093/protein/gzm085. [DOI] [PubMed] [Google Scholar]
- Darwin Ch. On the origin of species by means of natural selection or the preservation of favoured races in the struggle for life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Darwin Ch. The variation of animals and plants under domestication. London: John Murray; 1868. [Google Scholar]
- Dawkins R. The blind watchmaker. Londan: Longman scientific & Technical; 1986. [Google Scholar]
- de Lorenzo V, Danchin A. A Synthetic biology: discovering new worlds and new words. EMBO Rep. 2008;9(9):822–827. doi: 10.1038/embor.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BØ. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng. 2005;91:643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- Fong SS, Joyce AR, Palsson BØ. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression rates. Genome Res. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1469. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Ismagilov RF, Maharbiz MM. Can we build synthetic, multicellular systems by controlling developmental signaling in space and time? Curr Opin Chem Biol. 2007;11:604–611. doi: 10.1016/j.cbpa.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT, Griffiths AD. Selective gene amplification. Protein Eng Des Sel. 2007;20:577–581. doi: 10.1093/protein/gzm060. [DOI] [PubMed] [Google Scholar]
- Kubota M, Yamazaki Y, Ishihama A. Random selection of promoters from Escherichia coli and classification based on the promoter strength. Jpn J Genet. 1991;66:399–409. doi: 10.1266/jjg.66.399. [DOI] [PubMed] [Google Scholar]
- Lee SK, Chou HH, Pfleger BF, Newman JD, Yoshikuni Y, Keasling JD. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl Environ Microbiol. 2007;73:5711–5715. doi: 10.1128/AEM.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JW, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- Persson H, Wallmark H, Ljungars A, Hallborn J, Ohlin M. In vitro evolution of an antibody fragment population to find high-affinity hapten binders. Protein Eng Des Sel. 2008;21:485–493. doi: 10.1093/protein/gzn024. [DOI] [PubMed] [Google Scholar]
- Riechmann L, Winter G. Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc Natl Acad Sci USA. 2000;97:10068–10073. doi: 10.1073/pnas.170145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayut DJ, Niu Y, Sun Y. Construction and enhancement of a minimal genetic AND logic gate. Appl Environ Microbiol. 2009;75:637–642. doi: 10.1128/AEM.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nat Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- Yan Y, Smant G, Davis E. Functional screening yields a new beta-1, 4-endoglucanase gene from Heterodera glycines that may be the product of recent gene duplication. Mol Plant Microbe Interact. 2001;14(1):63–71. doi: 10.1094/MPMI.2001.14.1.63. [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y, Arnold FH. A dual selection module for directed evolution of synthetic circuits. Nat Comp. 2005;4:245–254. doi: 10.1007/s11047-004-7442-x. [DOI] [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a synthetic circuit. Proc Natl Acad Sci USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WPC, del Cardayré SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]