Abstract
Wild species tomato (Lycopersicon pennellii) plants bearing a genetic lesion in the gene encoding aconitase (Aco-1; aconitate hydratase EC 4.2.1.3) were characterized at molecular and biochemical levels. The genetic basis of this lesion was revealed by cloning the wild-type and mutant alleles. The mutation resulted in lowered expression of the Aco-1 transcript and lowered levels of both cytosolic and mitochondrial aconitase protein and activity. After in silico analysis, we concluded that in the absence of a recognizable target sequence, the best explanation for the dual location of this protein is inefficient targeting. Biochemical analysis of leaves of the Aco-1 accession suggested that they exhibited a restricted flux through the Krebs cycle and reduced levels of Krebs cycle intermediates but were characterized by elevated adenylate levels and an enhanced rate of CO2 assimilation. Furthermore, the analysis of both steady-state metabolite levels and metabolic fluxes revealed that this accession also exhibited elevated rates of photosynthetic Suc synthesis and a corresponding increase in fruit yield. Therefore, we conclude that the Krebs cycle normally competes with the Suc synthetic pathway for carbon but is not essential for the supply of energy to fuel the operation of this pathway.
The mitochondrial metabolism of plants, like that of all eukaryotes, is dominated by the synthesis of ATP, a fundamental component of this process being the Krebs cycle, which links the pathway of glycolysis to that of the electron transport chain. Despite the fact that the operation and location of the complete Krebs cycle was demonstrated in plant cells decades ago (Beevers, 1961), the function of this important pathway in plants is still far from clear (Hill, 1997; Siedow and Day, 2000). Even fundamental questions such as whether the Krebs cycle operates in illuminated photosynthetic tissue and if it contributes to the energy requirements of photosynthetic Suc synthesis remain controversial (Graham, 1980; Krömer, 1995; Padmasree et al., 2002). In addition to its role in energy production, a second essential feature of the Krebs cycle in plants is meeting the demand for carbon skeletons that is imposed by anabolic processes such as porphyrin and amino acid synthesis (Douce and Neuberger, 1989; Mackenzie and McIntosh, 1999).
The concerted action of citrate synthase, aconitase, and isocitrate dehydrogenase transform acetyl CoA into α-ketoglutarate, which, depending on relative demand, can either be further reduced to succinyl CoA or be utilized as a precursor for Glu synthesis (Hodges, 2002). The operation of the Krebs cycle in the light has been shown to be modified to that in the dark by a combination of at least two factors: the reversible inactivation of the mitochondrial pyruvate dehydrogenase complex in the light (Budde and Randall, 1990) and the rapid export of Krebs cycle intermediates out of the mitochondria (Hanning and Heldt, 1993; Aitkin et al., 2000a). However, the mitochondrial oxidative electron transport continues to be active, despite the limitation that the above modifications must impose on the Krebs cycle, irrespective of illumination (Aitkin et al., 2000b; Padmasree et al., 2002). Although both plant citrate synthase (Landschütze et al., 1995; Koyama et al., 2000) and isocitrate dehydrogenase (Kruse et al., 1998) have been the subject of molecular genetic approaches, aimed at elucidating their in vivo function, these studies were not focused on photosynthetic metabolism, and no such approach has been reported for aconitase.
Here, we describe the molecular and genetic analysis of Aco-1, a tomato (Lycopersicon pennellii) accession that was isolated as deficient in aconitase activity—a trait that was subsequently mapped to chromosome 12 (Bernatzky and Tanksley, 1986; Tanksley et al., 1992). We identified the mutant allele of aconitase and confirmed that the accession it was isolated from was deficient in aconitase transcript, protein, and activity. Having established that this accession was deficient in aconitase activity, we performed detailed physiological and biochemical analyses to assess the role of this enzyme in photosynthetic metabolism.
The results of these analyses will be discussed in the context of current models of the role of the Krebs cycle in photosynthetic metabolism.
RESULTS
Genetic Analysis of Aconitase within Tomato
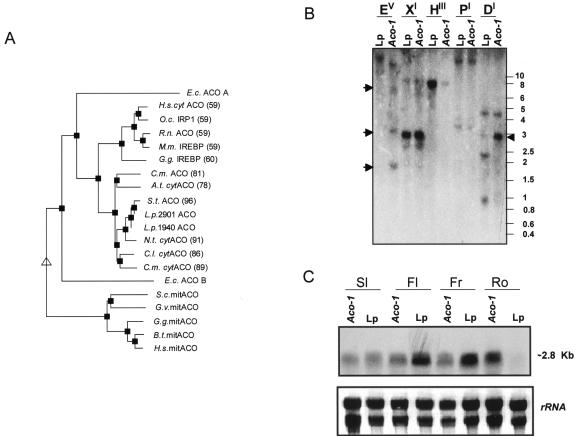
Full-length cDNAs encoding aconitase were isolated from leaves of the tomato accessions LP1940 (Lp) and 2901 (Aco-1) by a PCR-based approach as described in “Materials and Methods.” Using this strategy, we isolated two 2.8-kb clones, the sequences of which have been deposited in GenBank (accession nos. bk524167 and bk524171 for the Lp and Aco-1 genotypes, respectively). Sequence analysis of the clones from both genotypes indicated that the whole coding region had been amplified and revealed open reading frames encoding proteins of 898 amino acids in both instances. The predicted proteins of the two accessions differ in 12 amino acid residues with only four of these producing changes in polarity of the protein (data not shown). Comparison at the nucleotide level revealed that both alleles show between 79 and 97% identity to all plant aconitases in the databases (potato [Solanum tuberosum], X97012; tobacco [Nicotiana tabacum], AF194945; pumpkin [Cucurbita maxima], D29629; Arabidopsis, AY136414; lemon [Citrus limon], AF073507; and melon [Cucumis melo], X82840). In addition to the high identity with other plant aconitases, the two alleles share high identity (50%–60%) with mammalian cytosolic aconitases but not with mammalian mitochondrial isoforms or the bacterial aconitase A (Fig. 1A).
Figure 1.
Genetic and molecular characterization of the tomato genotypes. A, Dendogram of aconitase sequences. Alignments were produced using MULTALIN software. Percentage of protein identity is showed in parentheses. Those accession with <40% are not indicated. Sequences align into two clear clusters representing mitochondrial and cytosolic sequences. E.c., Escherichia coli; H.s., human (Homo sapiens); O.c., Oryctolagus cuniculus; R.n., rat (Rattus norvegicus); M.m., mouse (Mus musculus); G.g., Gallus gallus; C.m., melon; C.m. cyt, pumpkin; A.t., Arabidopsis; S.t., potato; L.p., tomato; N.t., tobacco; C.l., lemon; S.c., yeast (Saccharomyces cerevisiae); G.v., Gracilaria verrucosa; B.t., Bos taurus. B, Southern blot of genomic DNA from tomato lines digested with the restriction enzymes indicated (EV, EcoRV; XI, XbaI; HIII, HindIII; PI, PvuI; and DI, DraI) and probed with the entire aconitase cDNA cloned from the Lp accession. Arrows indicate polymorphisms between both genotypes. Number on the right indicates molecular masses in kilobase pairs. C, Northern-blot containing total RNA extracted from different tissues of 6- to 8-week-old greenhouse-grown plants. Sl, Source leaf; Fl, flower; Fr, fruit; Ro, root. Lp, tomato 1940; Aco-1, tomato 2901.
To assess the complexity of the gene encoding the aconitase enzyme of tomato, Southern hybridization of genomic DNA from both accessions was performed using the full open reading frame of the cloned aconitase cDNA from tomato (Fig. 1B). Using four different tomato expressed sequence tags (ESTs) with high homology with other aconitase genes as probes (for details, see “Materials and Methods”), identical bands were revealed (data not shown).
On comparison of the digestion of the genomic DNA with EcoRV and DraI allelic differences between the genotypes became apparent (Fig. 1B). Despite the fact that the above experiments hinted to the presence of only a single gene when the chromosomal map location of the cloned cDNA was determined by using a set of tomato introgression lines (Eshed and Zamir, 1994), two distinct loci were revealed. Aconitase mapped both to the southern arm of chromosome 7 (ILs 7-2, 7-3, and 7-4) and to the northern telomeric region of chromosome 12.
Analysis of mRNA northern blots using the Lp aconitase cDNA as a probe suggests constitutive expression of this gene, the transcript being present in leaves, flowers, fruits, and roots (Fig. 1C). Comparison of the relative mRNA levels between the genotypes suggests a considerably lower expression level of aconitase mRNA in leaves, flowers, and fruits of the Aco-1 genotype but an elevated expression in the roots.
Determination of Aconitase Protein Levels and Activity in the Tomato Genotypes
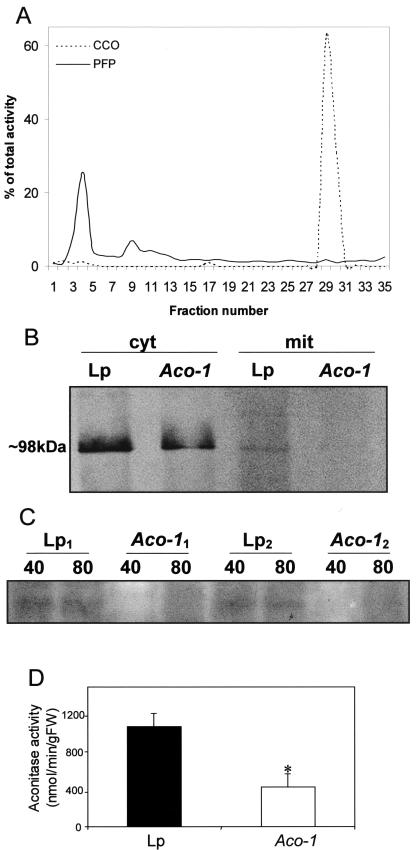
Having demonstrated that leaves of the Aco-1 plants exhibited lower expression of the aconitase gene, we next turned our attention to determining the effect this had on the protein amount and activity. Given the considerable difficulty of measuring aconitase activity in crude extracts, we attempted activity elution using the method of Slaughter et al. (1977). This zymogram analysis revealed that the Aco-1 plants have much lower total aconitase activity than the control genotype (Fig. 2C). This fact was confirmed by measuring the activity in desalted total cell extracts; however, such measurements reveal little about the subcellular location of the aconitase efficiency. To test this, we isolated mitochondria from leaves of both Aco-1 and Lp plants using a Percoll gradient purification method (Millar et al., 2001). Figure 2A documents a typical enzyme profile obtained after fractionation of tomato leaf extracts on a Percoll gradient with the profile of pyrophosphate-dependent PFP demarcating the cytoplasm and that of CCO the mitochondria. From this figure, it can be seen that the cytosolic contamination of the mitochondrial fractions is minor (below 2.4% in each accession).
Figure 2.
Analysis of the aconitase protein of the tomato genotypes. A, Activity of marker enzymes along a Percoll density gradient used for mitochondrial preparation. Each fraction corresponds to 1 mL of the gradient (from top to base). Each fraction was desalted and assayed for Ppi-dependent phosphofructokinase (PFP) and cytochrome c oxidase (CCO) activities as described in “Materials and Methods”. Fractions 25 to 33 were pooled to form the “mitochondrial fraction.” B, Western-blot analysis of aconitase in cytosolic and mitochondrial fractions. Total protein from the first supernatant and from the pooled mitochondrial fractions (25–33) were analyzed by western blotting, using antiserum raised against pumpkin aconitase that recognizes a protein of approximately 98 kD. C, Zymogram of aconitase activity in leaves of 6-week-old plants from Lp and Aco-1 genotypes. Numbers above the gel indicate the total amount of protein loaded in micrograms. 1 and2, Independent plants. D, Mitochondrial aconitase activity. Aconitase activity was determined in the mitochondrial fractions of Percoll gradients. Data represent the mean ± se of measurements from four plants per genotype. An asterisk indicates those that were determined by the Student's t test to be significantly different between genotypes.
The aconitase protein level in the pooled fractions was determined by western blot of cytoplasmic and mitochondrial proteins with antiserum raised against pumpkin aconitase (Hayashi et al., 1995). The western blot revealed a minor reduction in the protein located in the cytoplasm but a preferential loss of that located in the mitochondria (down to 30% of that seen in the Lp plants) in the Aco-1 plants (Fig. 2B). The mitochondrial aconitase activity in tomato leaves is comparable with previously reported activity for potato (Jenner et al., 2001), and the Aco-1 plants show a similar quantitative loss in mitochondrial aconitase activity as in the protein itself (Fig. 2D). However, surprisingly, the total aconitase activity of the genotypes (79.95 ± 10.28 and 147.12 ± 10.81 nmol min–1 g fresh weight–1 for Aco-1 and Lp genotypes, respectively; values presented as mean ± se, n = 6) are lower than the mitochondrial activities. This is most probably due to the presence of inhibitory substances that are removed during the mitochondrial preparation. For this reason, we are only able to estimate the quantitative loss of cytoplasmic activity; however, this estimate is in close agreement with the loss in cytoplasmic protein (Fig. 2B).
Characterization of the Tomato Genotypes
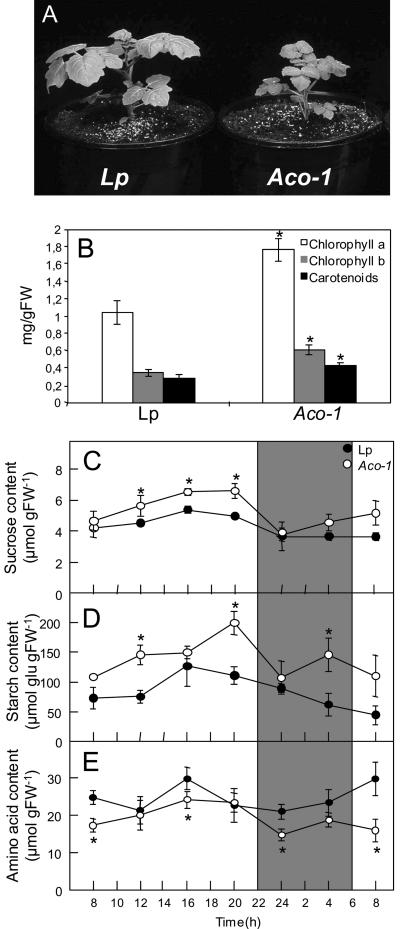
Having confirmed that accession 2901 (Aco-1) that we obtained from the University of California (Davis) Stock Center was deficient in aconitase activity (with respect to the control accession LP1940 [Lp]), we grew both alongside one another in 2.5-L pots in the greenhouse and determined morphological parameters of the genotypes. The genotypes were distinguishable from one another at a very early stage. Up to 3 weeks after transfer to the greenhouse, the Aco-1 plants displayed stunted growth (Fig. 3A). Over the course of development, however, this growth phenotype became less pronounced and by the time of fruit set was no longer visibly apparent. Leaves from the Aco-1 plants were also characterized by a deeper pigmentation color (Fig. 3A). To confirm this, we next determined the chlorophyll and carotenoid content of leaf samples harvested from 6-week-old plants. These determinations revealed significantly higher levels of chlorophyll a and b and carotenoids in leaves from the Aco-1 plants (Fig. 3B). As a further experiment, we recorded the growth, flowering, and biomass of the different organs of the plants. Despite the fact that the Aco-1 plants initially showed a stunted phenotype after 10 weeks of growth, they were marginally taller than the wild-type accession.
Figure 3.
Primary characterization of the tomato genotypes. A, Visual phenotype of the tomato genotypes. Aco-1 is stunted at very early growth stages (up to 3 weeks after transfer to the greenhouse) and has visibly darker leaves; however, neither of these observations are apparent at later stages of growth. B, Photosynthetic pigment content of 6-week-old plants. Source leaves were harvested 6 h into the photoperiod. Values are mean ± se of measurements from six plants per genotype. Diurnal changes in leaf Suc (C), starch (D), and total amino acid (E) content in leaves of 6-week-old plants. At each time point, samples were taken from mature source leaves, and the data represent the mean ± se of measurements from four plants per genotype. An asterisk indicates those that were determined by the Student's t test to be significantly different between genotypes. Gray bars, Dark period; white bars, light period.
After 20 weeks of growth, we determined the biomass of separate organs of the plants. Although there were no significant difference between the mass of stem or leaf across the genotypes, the Aco-1 plants produced slightly less root matter but considerably more fruit matter per plant (Table I). Interestingly, the time of flowering was also approximately 1 week later in the Aco-1 plants, and the number of flowers was significantly lower at this time point; however, the total number of flowers was eventually the same across the genotypes.
Table I.
Phenotypic characteristics of the tomato genotypes
Flowering and biomass of the different organs were recorded from 20-week-old plants. Values presented as mean ± se, n = 6. Bold type indicates those that were determined by the Student's t test to be significantly different between genotypes.
| Genotype
|
Total Dry Wt
|
Parameter
|
||||
|---|---|---|---|---|---|---|
| Leaf | Stem | Fruit | Root | Start of floweringa | No. of flowersb | |
| g ± se | ||||||
| Lp | 20.76 ± 0.9 | 20.29 ± 0.5 | 0.56 ± 0.3 | 18.7 ± 1.6 | 71 ± 3 | 138.3 ± 15 |
| Aco-1 | 23.58 ± 0.3 | 18.55 ± 0.2 | 3.58 ± 0.9 | 15.2 ± 0.6 | 79 ± 4 | 90.9 ± 11 |
a Values for flowering time are presented as days after planting that the first flower appears.
b No. of flowers was assessed as a per plant basis approximately 25 d into the flowering period.
Photosynthetic Carbohydrate Metabolism of the Tomato Genotypes
As a first experiment, we analyzed the carbohydrate content of leaves from 6-week-old plants during a diurnal cycle (Fig. 3, C and D). There was a significant increase both in the starch content and the rate of starch accumulation in the Aco-1 plants when compared with the Lp plants; however, in both cases, the amount of starch accumulated by the end of the day was approximately twice that at the beginning of the light period. A similar if less dramatic pattern was observed in the Suc content of Aco-1 and Lp plants with a minor, yet significantly higher Suc content in the Aco-1 plants throughout the majority of the light period. These changes were accompanied by an elevated Glc content (data not shown) and a small reduction in the total levels of amino acids (especially prominent at the middle of both light and dark periods; Fig. 3E). Despite the changes observed in amino acid content, there were no differences in total protein content between the accessions (data not shown).
Photosynthetic Parameters in the Tomato Genotypes
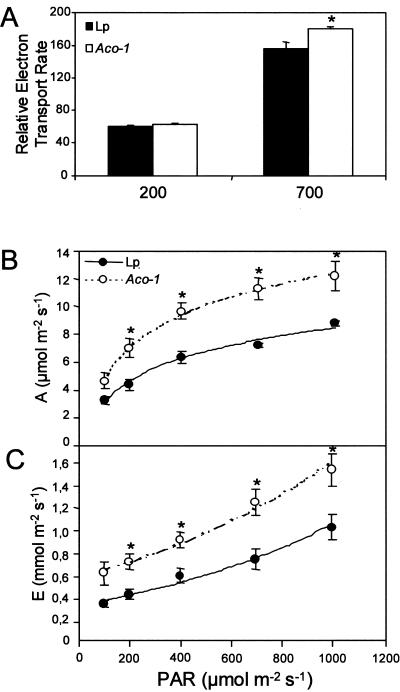
Considering that both Suc and starch levels increased within the Aco-1 plants, we decided to investigate whether they exhibited an altered photosynthetic rate. First, fluorescence emission was measured in vivo using a pulse amplitude modulation (PAM) fluorometer, and relative electron transport rates (ETRs) were calculated. When exposed to higher irradiance (photon flux density [PFD] of 700 μmol m–2 s–1), the Aco-1 plants exhibited a significantly elevated rate of electron transport (Fig. 4A). Second, gas exchange was measured in the two genotypes under PFDs that ranged from 100 to 1,000 μmol m–2 s–1 (Fig. 4, B and C). Aco-1 plants exhibited assimilation rates that were significantly higher than the wild type under all conditions except the lowest irradiance (Fig. 4B). Analysis of other parameters of gas exchange revealed that the Aco-1 genotype also exhibited a higher transpiration rate (Fig. 4C) at all PFDs, and there were minor changes in the internal CO2 concentration (data not shown), suggesting that the enhanced photosynthesis may, in part, be due to a more efficient stomatal conductance.
Figure 4.
Photosynthetic parameters of the tomato genotypes. A, In vivo fluorescence emission was measured as an indicator of the ETR by use of a PAM fluorometer at PFDs of 200 and 700 μmol m–2 s–1. Assimilation rate (B) and transpiration rate (C) as a function of PFD. Values presented are the mean ± se of measurements from five plants per genotype at 7 weeks after planting. An asterisk indicates those that are significantly different between genotypes.
In a complementary approach, we determined the effects of reduced aconitase activity on photosynthetic fluxes by studying the metabolism of 14CO2 by excised leaf discs illuminated for 30 min. In keeping with the data presented above, the assimilation rate was markedly increased in plants of the Aco-1 genotype. Furthermore, this increased carbon fixation was coupled with an increase in the accumulation of both [14C]Suc and [14C]starch (Table II).
Table II.
Effect of decreased aconitase activity on photosynthetic carbon partitioning at the onset of illumination
Leaf discs were cut from four separate plants of each genotype at the end of the night and illuminated at a PFD of 400 μmol photons m-2 s-1 of photosynthetically active radiation in an oxygen electrode chamber containing air saturated with 14CO2. After 30 min, the leaf discs were extracted and fractionated. Values presented are the mean ± se of measurements from six plants per genotype. Asterisks and bold type indicates those that were determined by the Student's t test to be significantly different between genotypes.
| Parameter | Total Uptake | Starch | Soluble Sugars | Amino Acids | Organic Acids |
|---|---|---|---|---|---|
| Label incorporated (Bq) | |||||
| Lp | 550.4 ± 30.2 | 176.2 ± 16.1 | 266.8 ± 28.2 | 53.3 ± 5.0 | 54.0 ± 3.9 |
| Aco-1 | 666.7 ± 29.1* | 231.8 ± 20.5* | 339.2 ± 26.6* | 47.8 ± 4.8 | 47.7 ± 5.1 |
| Redistribution of radiolabel (as percentage of total assimilated) | |||||
| Lp | 32.9 ± 3.3 | 47.3 ± 3.2 | 9.7 ± 0.9 | 10.0 ± 0.7 | |
| Aco-1 | 35.2 ± 3.1 | 50.4 ± 2.4 | 7.3 ± 0.7* | 7.2 ± 0.8* |
When these data are expressed as a percentage of the total 14CO2 metabolized, decreased aconitase activity led to decreased partitioning into organic and amino acids. These data imply a reduced respiration rate in these plants. Further studies also revealed that dark respiration was also reduced in the lines (–0.36 ± 0.03 and –0.86 ± 0.28 μmol m–2 s–1 for Aco-1 and Lp plants, respectively; values presented as mean ± se, n = 6).
Metabolite Levels in the Tomato Genotypes
Although the results of the above experiments indicate an enhanced rate of photosynthesis, we decided to study this in more detail by examining the levels of metabolic intermediates of the major pathways of plant photosynthetic metabolism. We began by evaluating the consequence of altered aconitase activity on the levels of Krebs cycle intermediates utilizing a recently established gas chromatography (GC)-mass spectrometry (MS) protocol. Several of these intermediates (namely fumarate, succinate, and α-ketoglutarate) were lower in the Aco-1 plants, and the levels of citrate were higher, as would be expected given the reduction in aconitase activity and assumed reduction in the rate of respiration (Table II) but surprisingly so was the level of isocitrate. Although the reason for this remains unclear from the present study it suggests that the utilization of isocitrate (and its production) has been reduced. Given that the reactions of the Krebs cycle have been postulated to form highly coordinated metabolic channels in yeast and E. coli (Sumegi et al., 1992; Velot et al., 1997; Haggie and Verkman, 2002), it is feasible to postulate that similar coordinated regulation also occurs in plants; however, many further studies are required to test this hypothesis.
Organic acids that are not associated with the Krebs cycle display no discernable trend; for example, saccharic acid is 2-fold higher in Aco-1, whereas threonic acid is only about 30% of the level determined in Lp plants (data not shown). Although it is technically unfeasible to obtain the concentration of metabolites within the mitochondria, at least one metabolite, succinate, reflects the mitochondrial situation because in plant cells it is present exclusively within the mitochondria (Hill, 1997). Further analysis of the GC-MS chromatograms indicated differences between genotypes in the levels of individual amino acid. In a separate experiment, we evaluated the sizes of the individual amino acid pools in the ethanolic extracts prepared for the diurnal carbohydrate analysis. Although the majority of amino acids were tendentially decreased in the Aco-1 plants (in keeping with the results presented in Fig. 3E) due to the large natural variation in amino acid content, these changes were generally not statistically significant. Those that are significantly different at either time point (10 h into the light period and 2 h into the dark period) are presented in Table III. We observed little change in the level of Glu in the day (although this amino acid was markedly decreased in the dark) but a significantly lower level of Gln in the Aco-1 plants at both time points. In the light, the level of Asp was also significantly lower in samples taken from the Aco-1 accession, but, in contrast, the level of Asn was higher. Thus, deduced Glu to Gln ratios were significantly elevated in the Aco-1 plants both during the light and the dark period, whereas the ratio of Asp to Asn was depressed in the Aco-1 plants in the light but unaltered in the dark (data not shown). Other changes of note were the large increases in the photorespiratory metabolite Gly (and a minor increase in Ser) in the Aco-1 plants (in the light) and lower levels of Arg, Phe, and Leu.
Table III.
Metabolite levels in illuminated leaves of 6-week-old plants of the tomato genotypes
Leaves were harvested 6 h into the photoperiod and irrespective of the method of analysis, all metabolite analyses were performed from the same tissue samples. Values presented are the mean ± se of measurements from six plants per genotype. Amino acid values in parentheses were determined in samples taken in the dark period. Organic acid contents, normalized as described in “Materials and Methods,” are also presented as the mean ± se of measurements from six plants per genotype. Asterisks and bold type indicates those that were determined by the Student's t test to be significantly different between genotypes.
| Metabolite | Lp | Aco-1 |
|---|---|---|
| Organic acids | ||
| Succinate | 1.0 ± 0.3 | 0.4 ± 0.1* |
| Isocitrate | 1.0 ± 0.3 | 1.4 ± 0.3*a |
| Citrate | 1.0 ± 0.1 | 1.5 ± 0.1* |
| α-kt-glutarate | 1.0 ± 0.4 | 0.3 ± 0.4* |
| Malate | 1.0 ± 0.5 | 0.9 ± 0.2 |
| Fumarate | 1.0 ± 0.1 | 0.5 ± 0.1* |
| nmol g fresh wt-1 | ||
| Amino acids | ||
| Asp | 308.2 ± 29.0 (222.9 ± 3.3) | 149.9 ± 12.2* (182.9 ± 33.4) |
| Glu | 732.8 ± 207.5 | 729.5 ± 75.4 (476.3 ± 112.7)* |
| (737.9 ± 91.7) | ||
| Asn | 141.5 ± 16.5 (163.8 ± 24.3) | 228.2 ± 40.6* (105.9 ± 19.7) |
| Ser | 138.6 ± 22.8 (144.9 ± 19.3) | 166.1 ± 40.1 (105.8 ± 19.1) |
| Gln | 264.3 ± 59.9 (259.3 ± 50.8) | 206.1 ± 51.4* (164.6 ± 47.0)* |
| Gly | 98.4 ± 26.82 (n.d.b) | 184.3 ± 30.9* (n.d.) |
| Arg | 124.9 ± 29.1 (148.7 ± 4.5) | 100.1 ± 9.9 (72.2 ± 11.3)* |
| Phe | 49.6 ± 18.7 (46.6 ± 9.4) | 28.5 ± 1.9* (22.7 ± 6.3)* |
| Leu | 32.5 ± 11.1 (11.7 ± 1.9) | 19.3 ± 0.9* (8.5 ± 1.3) |
| Nucleotides | ||
| ATP | 11.7 ± 2.7 | 19.7 ± 2.0 |
| ADP | 21.3 ± 3.1 | 44.0 ± 11.5* |
| ATP/ADP | 0.6 ± 0.0 | 0.4 ± 0.0* |
| Σ Adenylate | 32.3 ± 7.0 | 59.7 ± 18.3* |
| UDP-Glc | 4.7 ± 0.8 | 15.6 ± 5.6* |
| UTP | 15.9 ± 2.5 | 17.7 ± 3.9 |
| UDP | 4.7 ± 0.9 | 0.37 ± 0.0* |
| UTP/UDP | 3.7 ± 0.5 | 30.0 ± 3.6* |
| ATP/UTP | 0.9 ± 0.2 | 1.0 ± 0.2 |
| Sugar phosphate | ||
| 3 phosphoglycerate (3PGA) | 262.0 ± 23.2 | 316.0 ± 16.8* |
| Glucose 6-phosphate | 106.4 ± 4.9 | 137.2 ± 5.6* |
| Glucose-1-phosphate | 19.6 ± 3.3 | 25.1 ± 2.2 |
| Fructose 6-phosphate | 19.8 ± 1.6 | 24.7 ± 4.3 |
| Σ hexose-P | 145.8 ± 8.5 | 187.0 ± 9.9* |
| Pi | 5.3 ± 1.8 | 5.8 ± 0.6 |
| Σ Phosphates (inorganic phosphate) Pi | 75.5 ± 1.3 | 86.9 ± 2.1* |
| 3PGA/Pi | 48.6 ± 5.7 | 54.6 ± 3.95* |
a Isocitrate was measured following the protocol described by Beutler (1989).
b n.d., Not detectable.
Next, we determined the levels of adenylates and uridinylates by HPLC in tricarboxylic acid extracts from the two accessions (Table III). Perhaps it is surprising, given that the Aco-1 plants are deficient in the activity of an enzyme of the Krebs cycle, that they display increased levels of total adenylates containing a minor increase in the level of ATP but almost a 2-fold higher ADP content than the Lp accession, with a resultant significant decrease in the deduced ATP to ADP ratio. Notably, however, the ATP to UTP ratio remains the same across the genotypes. Interestingly, the content of UDP is markedly lower in the Aco-1 lines, resulting in a large change in the deduced UTP to UDP ratio. Also, these plants were further characterized by large accumulation of UDP-Glc. In addition to the nucleotide measurements, we determined the levels of the key photosynthetic metabolites Glc 6-phosphate, Glc 1-phosphate, Fru 6-phosphate, and 3-phosphoglycerate using sensitive cycling assays that have been established recently (Gibon et al., 2002) and inorganic phosphate. These assays revealed significantly higher levels of 3-phosphoglycerate, Glc 6-phosphate, and total hexose phosphates but no significant differences in the levels of Glc 1-phosphate, Fru 6-phosphate, and phosphate. As a consequence, there were significantly higher phosphate ester to inorganic phosphate ratios and 3PGA to inorganic phosphate ratios in the Aco-1 plants.
Measurement of Activities of Key Enzymes of Carbohydrate Metabolism
To better understand the enhanced photosynthesis observed in Aco-1, we determined the activities of a broad range of enzyme activities important in photosynthesis, starch and Suc synthesis, and tricarboxylic acid cycle function (Table IV). There were no significant differences in the maximal catalytic activities of UDP-Glc pyrophosphorylase, Suc phosphate synthase, Rubisco, NADP+-dependent glyceraldehyde phosphate dehydrogenase, transaldolase, transketolase, phosphoribulokinase, phosphoglycerate kinase, both pyrophosphate-dependent and ATP-dependent PFPs, or pyruvate kinase. Likewise, the initial Rubisco activities and the deduced activation state of Rubisco and the maximal catalytic activities of AGPase and phosphoglucomutase were unaltered.
Table IV.
Enzyme activities in the leaves of the tomato genotypes
Enzyme activities were determined in source leaves samples from 6-week-old plants. The data presented are the mean ± se of measurements from six plants per genotype.
| Enzyme
|
Activity
|
|
|---|---|---|
| Lp | Aco-1 | |
| nmol min-1 g fresh wt-1 | ||
| UDPGlucose pyrophosphorylase | 1,043.2 ± 210.1 | 1,231.9 ± 176.8 |
| Sucrose phosphate synthase (SPS) Vmaxa | 1.3 ± 0.7 | 1.0 ± 0.2 |
| Rubisco initial activity | 4,498.3 ± 448.4 | 5,488.6 ± 834.1 |
| Rubisco total activity | 9745.9 ± 1619.1 | 10,209.8 ± 933.3 |
| Rubisco activation state (%) | 50.0 ± 12.8 | 54.0 ± 8.5 |
| NADP-GAPDH | 1,790.4 ± 903.7 | 2,603.7 ± 948.1 |
| Transketolase | 2,099.6 ± 909.2 | 3,003.5 ± 1,084.2 |
| Transaldolase | 39.5 ± 9.5 | 51.2 ± 16.4 |
| Phosphoribulokinase | 2,811.9 ± 528.9 | 1,911.1 ± 265.8 |
| Pyruvate kinase | 404.4 ± 196.3 | 276.7 ± 61.6 |
| PFP | 146.8 ± 31.4 | 124.9 ± 8.5 |
| PFP | 322.3 ± 40.3 | 332.5 ± 42.7 |
| Phosphoglycerate kinase | 9,270.9 ± 1,513.8 | 12,225.9 ± 802.7 |
| ADPglucosepyrophosphaorlyase | 48.5 ± 12.4 | 57.2 ± 3.9 |
| Phosphogluromutave | 2404.4 ± 359.1 | 2642.3 ± 453.5 |
a SPS values are presented as micromoles Suc per gram fresh wt per hour.
DISCUSSION
Isolation and Characterization of a cDNA Encoding the Tomato and Aco-1 Alleles of Aconitase and Molecular Characterization of the Aco-1 Accession
Full-length 2.8-kb clones were obtained from both Lp and Aco-1 accessions of tomato utilizing a PCR-based approach using primers based on a consensus of the publicly available plant aconitase sequences. These cDNAs both encode proteins of molecular masses of 98 kD and exhibit high homology to all sequenced aconitases but particularly to bacterial and mammalian cytoplasmic aconitases (Peyret et al., 1995; Navarre et al., 1999; Sadka et al., 2000; Williams et al., 2002). Using TargetP, an in silico protein targeting prediction tool based on knowledge derived from Swiss-Prot sequence entries (Emanuelsson et al., 2000), we were unable to identify features characteristic of transit peptides in either sequence. However, western-blot analysis revealed that the Aco-1 accession, which carries a mutant aconitase allele, was deficient both in the mitochondrial and the cytosolic protein.
Searching a tomato EST collection for clones with significant homology to other reported plant aconitases resulted in the detection of a total of 45 clones within two highly homologous tentative consensus sequences, suggesting the possibility of a unique gene. However, further map-based localization studies using the clone described here were in close agreement with the previous annotations based on isozyme analysis (Tanksley and Rick, 1980), which suggested the presence of two aconitase genes in the tomato genome.
When taken together with the demonstration that the Aco-1 accession is deficient in aconitase protein and activity in both the cytoplasm and mitochondria, these data suggest that the both cytosolic and mitochondrial aconitases from tomato are encoded by the gene cloned here. A range of observations, some of them purely circumstantial, indicate that this is probably also the case in a range of other plant species. First, cytoplasmic and mitochondrial isoforms of aconitase have been separated both from Acer pseudoplatanus cell suspension cultures (Brouquisse et al., 1987) and from etiolated pumpkin cotyledons (De Bellis et al., 1993) with the separated isoforms displaying very similar kinetic properties. More recently, despite early indications that there is only one gene encoding aconitase in Arabidopsis that produces both a cytosolic and a mitochondrial isoform (Peyret et al., 1995), genome sequencing has revealed three genes encoding aconitase, all of which have been localized to the mitochondria using proteomic techniques (Kruft et al., 2001; Millar et al., 2001). Intriguingly, as was the case with the aconitases cloned in the present study, none of the plant aconitases in the databases contain a mitochondrial recognizable target sequence. A similar finding has been noted previously in yeast, where a single nuclear gene encodes aconitase, and “inefficient import” of this protein is believed to be responsible for its dual location (Gangloff et al., 1990).
Effect of the Reduction of Aconitase Activity on the Rate of Photosynthesis and on Fruit Yield
Reduction of aconitase activity led to a stunted phenotype at early stages of development; however, after a period of 4 to 5 weeks, the Aco-1 plants were equivalent in height to the control accession. However, leaves from the Aco-1 plants did display a darker pigmentation characterized by elevated levels of chlorophylls and carotenoids and exhibited higher rates of assimilation, transpiration, and electron transport through the PSs. In keeping with elevated rates of both carbon assimilation and photosynthesis, the Aco-1 plants were characterized by elevated accumulation of Suc and starch during the light period and an increased fruit yield. Although it seems likely that the increased fruit yield results from increases in leaf photoassimilate supply, the data presented in the current study do not preclude the possibility that the increased fruit yield is, at least in part, a direct effect of modulating the aconitase activity in the fruit. The enhancement of photosynthesis is somewhat surprising, particularly given the limited success in achieving this result through direct modification of the Calvin cycle (for review, see Raines, 2003). The only successful example published to date was that obtained after overexpression of a bifunctional cyanobacterial Fru bisphosphatase/sedoheptulose bisphosphatase in tobacco (Miyagawa et al., 2001).
Although the reduction in aconitase activity clearly results in an elevated rate of photosynthesis in the Aco-1 plants, the exact mechanism by which this comes about remains unresolved. However, a broad screen of other key enzymes of photosynthetic carbohydrate metabolism revealed that only the activity of aconitase was altered in the Aco-1 accession. A number of other possible explanations for the increased rate of photosynthesis exist—the most likely of these is that it is a consequence of a reduction of partitioning of assimilate toward the Krebs cycle. Results from the 14CO2 feeding experiment demonstrate a clear reduction in labeling of organic and amino acids. Although some of these acids may well be formed by fixation of carbon dioxide by phosphoenolpyruvate carboxylase, and the data do not constitute a direct measurement of flux, we nevertheless believe that these results imply that the reduction in aconitase activity results in a reduced operation the Krebs cycle. Furthermore, the data from these feeding experiments are in accordance with the metabolite levels determined in this study—with the Aco-1 plants displaying decreased levels of fumarate, succinate, and α-ketoglutarate (and total amino acid levels) and increased levels of Suc and starch. In addition, the Aco-1 plants exhibited significantly elevated levels of 3PGA, hexose phosphates, and UDP-Glc, which are the intermediates of photosynthetic Suc biosynthesis. The changes in the levels of these key metabolites may explain the altered carbon partitioning in the Aco-1 accession because the results of studies of both covalent and allosteric properties of Suc phosphate synthase (for review, see Winter and Huber, 2000) suggest that a combined elevation of the hexose phosphates and UDP-Glc would stimulate the in vivo activity of this enzyme. This is in keeping with suggestions that overexpression of a maize (Zea mays) Suc phosphate synthase in tomato increase photosynthesis (Lunn et al., 2003) and fruit yield (Laporte et al., 1997). In addition, the increase in starch levels in Aco-1 accession may well be a consequence of the elevated 3PGA to Pi ratio, which has been demonstrated to enhance the in vivo activity of AGPase, the first committed enzyme of starch biosynthesis (Preiss, 1988). A further possibility is that the low levels of one of the products of Suc synthesis (UDP) in the Aco-1 accession alters the equilibrium position of SPS in the favor of Suc synthesis. However, this seems unlikely because the second product of SPS, Suc phosphate, only accumulates to very low levels in the cell and is cleaved readily by Suc phosphate phosphatase (Lunn and ap Rees, 1990), which would suggest that it is unlikely that the level of UDP has a major influence on the rate of flux through this enzyme.
Interestingly, despite a probable reduction in the flux through the Krebs cycle, the Aco-1 plants are characterized by enhanced adenylate levels but a decrease in the cellular ATP to ADP ratio. There are two possibilities that can be proposed to explain the enhanced levels of adenylates. First, it is highly likely that a large proportion of the increase in adenylate pool size is met by the increased rate of photosynthesis discussed above. However, it is also conceivable that the increased size of the adenylate pools is the consequence of a compensatory induction of photorespiration in this accession. In keeping with the second explanation, the levels of the photorespiratory intermediates Gly and to a lesser extent Ser accumulate in the Aco-1 accession, and experimental evidence has been provided that the mitochondrial electron transport chain oxidizes not only NADH produced by the Krebs cycle but also that produced by photorespiration (Krömer, 1995; Aitkin et al., 2000b). Thus, these data demonstrate that although operation of the Krebs cycle in the light may normally contribute to the energy requirements of photosynthesis, it is by no means essential for this function.
Although the above mechanism is the one we favor to explain the increase in photosynthesis, other plausible explanations exists. First, although the increase in pigment contents cannot be explained directly by the metabolic changes we describe here, it is conceivable the reduction in aconitase protein levels could result in a surplus of cellular iron, which could in turn explain the increase pigment contents in the Aco-1 plants. A second plausible explanation, albeit purely speculative, is that the reduction in the aconitase activity disrupts a cellular signaling pathway. Such a role for aconitase is not without precedence because it has been demonstrated to be a key component of cellular iron homoeostasis in animal systems (for review, see Cairo and Pietrangelo, 2000) and in nitric oxide signaling pathways in tobacco (Navarre et al., 2000).
Effect of Reduced Aconitase Activity on the Minor Products of Photosynthesis
The pattern of change in the amino acid pool sizes in the tomato accessions between the day and night was similar to those previously reported in tobacco (Scheible et al., 2000; Matt et al., 2001a; Masclaux-Daubresse et al., 2002). However, an important distinction can be made between this and previous studies wherein the levels of amino acids were determined in leaves of different genotypes during diurnal rhythms and in response to changes in the rate of photosynthesis and photorespiration (see also Matt et al., 2001b; Novistskaya et al., 2002; Stitt et al., 2002). This distinction is the fact that in all the other studies, the levels of Glu and α-ketoglutarate are relatively stable. Because α-ketoglutarate is the immediate acceptor in the GOGAT pathway and Glu is the immediate amino donor for the synthesis of most amino acids, this fact has been interpreted to suggest that sophisticated control mechanisms exist to control the levels of these central metabolites (Stitt and Fernie, 2003). In the present study, however, the α-ketoglutarate is markedly reduced in the Aco-1 accession, and although the level of Glu is relatively unaltered in these plants, the levels of Gln and several other amino acids including Asp, Phe, and Leu are also significantly reduced. These data are consistent with those of previous studies in suggesting an important role for α-ketoglutarate in ammonium assimilation (Lancien et al., 2000; Hodges, 2002). The formation of α-ketoglutarate is dependent on its production from isocitrate by either cytosolic or mitochondrial isoforms of isocitrate dehydrogenase (Galvez et al., 1999; Miflin and Lea, 1980) but equally on the essential production of isocitrate by aconitase. Interestingly, a 92% reduction in cytosolic NADP-dependent isocitrate dehydrogenase activity had no major effect on CN metabolism in potato leaves, suggesting that this isoform is not essential for normal functioning of the GS/GOGAT cycle (Kruse et al., 1998). However, it should be noted that the residual cytosolic NADP-dependent isocitrate dehydrogenase activity in these lines might be sufficient to allow ammonium assimilation. However, the results of the present study reveal that a relatively minor (approximately 60%) reduction in total aconitase activity has a large effect on amino acid levels. Unfortunately, given that the Aco-1 plants are deficient in both mitochondrial and cytoplasmic aconitase activities, the relative importance of the compartmented pathways to ammonium assimilation cannot be assessed.
CONCLUSION
In this paper, we have shown that repression of both mitochondrial and cytosolic aconitase activities in wild tomato has dramatic effects on the photosynthetic metabolism and fruit yield of the plant. This repression of activity resulted in a reduction in the levels of α-ketoglutarate and, hence, in Gln, but despite an apparent reduction in flux through the Krebs cycle, it did not result in a repression but rather an increase in the rate of photosynthetic Suc synthesis. When taken together, these results suggest that the Krebs cycle normally competes with the Suc synthetic pathway for carbon but is not essential for the supply of energy to fuel the operation of this pathway. That said, these changes are observed during a point at which the plants have achieved developmental equivalence, and further experimentation is required to elucidate factors involved in the dramatic phenotype observed during early development.
MATERIALS AND METHODS
Plant Material and Growth of Plants
Tomato (Lycopersicon pennellii) seeds of accession numbers LP1940 (Lp) and LP2901 (Aco-1) were obtained from the true-breeding monogenic stocks maintained by the Tomato Genetics Stock Center (University of California, Davis). The seeds were germinated on Murashige and Skoog media (Murashige and Skoog, 1962) containing 2% (w/v) Suc and were grown in a growth chamber (250 μmol photons m–2s–1, 22°C) under a 16-h-light/8-h-dark regime before transfer into the greenhouse, where they were grown in parallel with a minimum of 250 μmol photons m–2s–1, 22°C under a 16-h/8-h day light. Experiments were carried out on mature fully expanded source leaves from 6- to 8-week-old plants. Root, flower, and fruit samples and morphological measurements were taken at the time points indicated in the text.
Cloning of Aconitase Alleles
A pair of primers was designed based on the consensus sequence of all cloned plant aconitase genes (potato [Solanum tuberosum], tobacco (Nicotiana tabacum), pumpkin [Cucurbita maxima], Arabidopsis, and lemon [Citrus limon]), and these were named ACO-15 (5′ CCATGGCTGCAGAGAACCC 3′) and ACO-13 (5′ ATGTGGATTCTCATTGCTG 3′), respectively. Total RNA (3 μg) was obtained from LP1940 and Aco-1 leaves with Trizol (Gibco BRL, Karlsruhe, Germany), followed by DNAse treatment, and converted into cDNA with SuperScript II reverse transcriptase (Gibco BRL). The resultant cDNAs were then used as template for PCR amplification using 1 unit of Pfu DNA polymerase (Stratagene, Amsterdam) in 50 μL of the recommended buffer (0.1 mm dNTPs and 0.25 μm of each primer). Conditions were: initial denaturation at 94°C for 2 min and 30 cycles, each consisting of denaturation at 94°C for 45 s, annealing at 48°C for 1 min, and elongation at 72°C for 2 min. The last round of elongation was for 10 min at 72°C. PCR products were separated by 1% (w/v) agarose gel electrophoresis and purified by using the NucleoSpin Extract kit (Macherey-Nagel, Düren, Germany) and subjected to DNA sequence analysis at AGOWA GmbH (Berlin). Two independent PCR products from each genotype were sequenced and verified by comparison with sequences present in GenBank.
Southern- and Northern-Blot Analysis
Genomic DNA (10 μg) from LP1940 and Aco-1 was isolated and Southern blotted as described by Hoisington et al. (1994). The blot was hybridized with four different tomato EST clones (cLEI2O12, cLEM22H8, cLET3J24, and cTOE5G1; Clemson State University collection, SC) and with the aconitase cDNA cloned from the accession LP1940 using standard conditions (Sambrook et al., 1989) at 60°C. Washes were done at low stringency (2× SSC and 1× SSC, 0.1% [w/v] SDS at 60°C). Chromosomal mapping was done according to Eshed and Zamir (1994) using tomato introgression lines.
Total RNA was isolated using the commercially available Trizol kit (Gibco BRL) according to the manufacturer's suggestions for extraction from plant material. The RNA (15 μg) was then size fractionated on a 1% (w/v) agarose MOPS-formaldehyde gel before transfer to a nylon membrane filter. This filter was subsequently probed using the same clone as was used in the Southern experiment described above. Hybridization was carried out using standard protocols (Sambrook et al., 1989). Filters were washed once for 20 min at 42°C in 2× SSC and 0.1% (w/v) SDS, once for 20 min at 42°C in 0.5× SSC and 0.1% (w/v) SDS, and finally for 20 min at 65°C in 0.1× SSC and 0.1% (w/v) SDS. After washing, the filters were exposed to x-ray films (Xomat, Eastman-Kodak, Rochester, NY) for 1 to 2 d.
Immunodetection of Aconitase Protein and Gel Stains of Aconitase Activity
Western analysis of aconitase protein was carried out either on crude protein extract or on mitochondrial protein extract, exactly as described by Hayashi et al. (1995) using antiserum kindly provided by the authors. Activity gels were performed on 40 and 80 μg of crude protein extract from two different plants, using the protocol described by Slaughter et al. (1977).
Analysis of Enzyme Activities
With the exceptions of the enzymes mentioned below, all enzymes were determined following the extraction and assay procedures detailed by Tauberger et al. (2000) or Fernie et al. (2001a). Total aconitase activity was determined in leaf crude extract following the method of Jenner et al. (2001). The mitochondrial activity was subsequently determined applying the same method to mitochondrial fractions following the protocol for mitochondrial isolation described by Sweetlove et al. (2002). Activities are given per gram fresh weight based on the amount of tissue from which the mitochondria were prepared. The purity of the mitochondrial preparations was confirmed by measurement of CCO (following the protocol of Neuberger, 1985) and pyrophosphate-dependent PFP (Fernie et al., 2001b), which serve as marker enzymes for the mitochondria and cytoplasm, respectively. In all instances, the contamination of the mitochondria was less than 2.4%, and recoveries of both marker enzymes were 90.6% for PFP and 92.4% for CCO. Rubisco was assayed as detailed by Sharkey et al. (1991), UDPGlucose pyrophosphorylase as described by Sweetlove et al. (1996), NADP+ GAPDH as detailed by Leegood and Walker (1980), phosphoribulose kinase as described by Haake et al. (1998), and transaldolase and transketolase as detailed by Debnam and Emes (1999).
Measurements of Photosynthetic Parameters
Fluorescence emission was measured in vivo using a PAM fluorometer (Walz, Effeltrich, Germany) on 6-week-old plants maintained at fixed irradiance (200 and 700 μmol photons m–2 s–1) for 30 min previous to measure chlorophyll fluorescence yield, and relative ETR was calculated using the WinControl software package (Walz). Gas-exchange measurements were performed in a special custom-designed open system (Walz; for detailed description, see Muschak et al., 1997). Relative air humidity was controlled by saturating the air flow with water before passing it through a cold trap for dew point adjustment, whereas air temperature was regulated in the measuring cuvette by a Peltier element and held constant at 24.0°C ± 0.1°C during all experiments. Leaf temperature was detected using a NiCr-Ni thermocouple, whereas leaf size determination was performed graphically by copying the leaf shape onto graded paper. The Diagas software package (Walz) was used to calculate the assimilation rates and transpiration rates according to von Caemmerer and Farquhar (1981).
Incubation of Leaf Material with 14CO2
The 14C-labeling pattern of Suc, starch, and other cellular constituents was performed by illuminating leaf discs (10-mm diameter) in a leaf disc oxygen electrode (Hansatech, Kings Lynn, Norfolk, UK) at 250 μmol photosynthetically active radiation m–2 s–1 at 20°C for 30 min. The carbon dioxide was supplied from 400 μL of 1 m NaH14CO3 (specific activity 1.96GBq mmol–1 [pH 9.3]) and placed on a felt mat at the base of the oxygen electrode chamber. After this experiment, the leaf discs were frozen in liquid nitrogen until further analysis. Frozen leaf tissue was fractionated to allow the determination of the metabolic fate of the assimilated 14CO2 exactly as detailed by Lytovchenko et al. (2002).
Metabolite Analysis
Tissue samples were rapidly frozen in liquid nitrogen. Subsequently, the samples were extracted in either ethanol (for determination of carbohydrate and amino acid content) or in trichloroacetic acid (for the determination of triose and hexose phosphates, phosphate, 3PGA, and nucleotides) as detailed by Fernie et al. (2001a). The carbohydrate contents were determined spectrophotometrically as described by Fernie et al. (2001b), whereas the amino acid contents were determined using the HPLC protocol described by Geigenberger et al. (1996). The levels and recoveries of hexose phosphates and 3PGA were determined spectrophotometrically as detailed by Gibon et al. (2002)—recoveries were 94%, 92%, and 91% for Glc 6-phosphate, Fru 6-phosphate, and Glc 1-phosphate, respectively. Inorganic phosphate content was determined as described by Lytovchenko et al. (2002), and the levels of nucleotides and nucleosides were determined using the HPLC protocol defined by Regierer et al. (2002). The relative levels of organic acids were determined using a GC-MS protocol as described by Roessner et al. (2001a). Data are presented normalized to wild type as detailed by Roessner et al. (2001b). The recovery of small, representative amounts of each metabolite through the extraction, storage, and assay procedures of the chromatography methods has been documented previously (Roessner et al., 2000; Fernie et al., 2001b). Chlorophyll was measured in 80% (w/v) acetone extracts as described by Lichtenthaler (1987). Isocitrate determination was carried out spectrophotometrically following the method described by Beutler (1989).
Statistical Analysis
Student's t tests were performed using the algorithm embedded into Microsoft Excel (Microsoft Corporation, Seattle). The term significant is used in the text only when the change in question has been confirmed to be significant (P < 0.05) with the Student's t test.
Acknowledgments
Discussions and support of Prof. Lothar Willmitzer throughout this work are most gratefully acknowledged. We are very grateful to Dani Zamir for carrying out the mapping analysis described in this paper. We are also thankful to Prof. Mark Stitt for provision of technical facilities, to Dr. John Lunn for many helpful comments during the writing of this manuscript, and to Dr. Joachim Fisahn for help in organization of gas-exchange measurements and discussion of the results. We are also thankful to Helga Kulka for excellent care of the plants.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026716.
This work was supported by Max-Planck-Gesellschaft (to F.C. and A.N.-N.), by Conselho Nacional de Desenvolvimento Científìco e Technológico (Brazil; to A.N.-N. and A.R.F.), and by DIP (to A.N.-N. and A.R.F.).
References
- Aitkin OK, Evans JE, Ball MC, Lambers H, Pons TL (2000a) Leaf respiration of snowgum in light and dark: interactions between temperature and irradiance. Plant Physiol 122: 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitkin OK, Millar AH, Gardeström P, Day DA (2000b) Photosynthesis, carbohydrate metabolism and respiration in leaves from higher plants. In RC Leegood, TD Sharkey, S von Caemmerrer, eds, Photosynthesis: Physiology and Metabolism. Kluwer Academic Publishers, The Netherlands, pp 153–175
- Beevers H (1961) Respiratory Metabolism in Plants. Row, Peterson, Everson, UK
- Bernatzky R, Tanksley SD (1986) Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler H-O (1989) d-Isocitrate. In HU Bermeyer, J Bergmeyer, M Graβl, eds, Method of Enzymatic Analysis. VCH Verlagsgesellschaft, Weinheim, Germany, pp 13–19
- Brouquisse R, Nishimura M, Galliard J, Douce R (1987) Characterization of a cytosolic aconitase in higher plant cells. Plant Physiol 84: 1402–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde RJA, Randall DD (1990) Pea leaf mitochondrial pyruvate dehydrogenase complex is inactivated in vivo in a light dependent manner. Proc Natl Acad Sci USA 87: 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo G, Pietrangelo A (2000) Iron regulatory proteins in pathobiology. Biochem J 352: 241–250 [PMC free article] [PubMed] [Google Scholar]
- De Bellis L, Tsugeki R, Alpi M, Nishimura M (1993) Purification and characterization of aconitase isoforms from etiolated pumpkin cotelydons. Physiol Plant 88: 485–492 [Google Scholar]
- Debnam PM, Emes MJ (1999) Subcellular distribution of enzymes of the oxidative pentose phosphate pathway in root and leaf tissues. J Exp Bot 50: 1653–1661 [Google Scholar]
- Douce R, Neuberger M (1989) The uniqueness of plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol 40: 371–414 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79: 175–179 [Google Scholar]
- Fernie AR, Roessner U, Trethewey RN, Willmitzer L (2001a) The contribution of plastidial phosphoglucomutase to the control of starch synthesis within the potato tuber. Planta 213: 418–426 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ (2001b) Fructose 2, 6-bisphosphate activates pyrophosphate: fructose-6-phosphate:1-phosphotransferase and increases triose-phosphate to hexose-phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Galvez S, Lancien M, Hodges M (1999) Are isocitrate dehydrogenases and 2-oxoglutarate involved in the regulation of glutamate synthesis? Trends Plant Sci 4: 484–490 [DOI] [PubMed] [Google Scholar]
- Gangloff SP, Marguet D, Lauquin GJM (1990) Molecular cloning of the yeast aconitase gene (ACO-1) and evidence of the synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol 10: 3551–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U (1996) Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19: 43–55 [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M (2002) Sensitive and high throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J 30: 221–235 [DOI] [PubMed] [Google Scholar]
- Graham D (1980) Effects of light on “dark” respiration. In DD Davies, ed, Biochemistry of Plants, Vol. 2. Academic Press, New York, pp 525–579 [Google Scholar]
- Haake V, Zrenner R, Sonnewald U, Stitt M (1998) A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant J 14: 147–157 [DOI] [PubMed] [Google Scholar]
- Haggie PM, Verkman AS (2002) Diffusion of tricarboxylic acid cycenzymes in the mitochondrial matrix in vivo: evidence for restricted mobility of a multi-enzyme complex. J Biol Chem 277: 40782–40788 [DOI] [PubMed] [Google Scholar]
- Hanning I, Heldt HW (1993) On the function of mitochondrial metabolism during photosynthesis in spinach (Spinacia oleracea L.) leaves: partitioning between respiration and export of redox equivalents and precursors for nitrate assimilation products. Plant Physiol 103: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Debellis L, Alpi A, Nishimura M (1995) Cytosolic aconitase participates in the glyoxylate cycle in etiolated pumpkin cotyledons. Plant Cell Physiol 36: 669–680 [PubMed] [Google Scholar]
- Hill SA (1997) Carbon metabolism in mitochondria. In DT Dennis, DH Turpin, DD Lefebvre, DB Layzell, eds, Plant Metabolism. Longman, Harlow, UK
- Hodges M (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation J Exp Bot 53: 905–916 [DOI] [PubMed] [Google Scholar]
- Hoisington D, Khairallah M, Gonzalez de León D (1994) Laboratory Protocols. El Centro Internacional de Mejoramiento al Maize y Trigo Applied Molecular Genetics Laboratory, El Baton, Mexico
- Jenner HL, Winning BM, Millar AH, Tomlinson KM, Leaver CJ, Hill SA (2001) NAD Malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Kawamura A, Kihara T, Hara T, Takita E, Shibata D (2000) Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol 41: 1030–1037 [DOI] [PubMed] [Google Scholar]
- Krömer S (1995) Respiration during photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 46: 45–70 [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol, 127: 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Kruse A, Fieuw S, Heineke D, Müller-Röber B (1998) Antisense inhibition of cytosolic NADP-dependent isocitrate dehydrogenase in transgenic potato plants. Planta 205: 82–91 [Google Scholar]
- Lancien M, Gadal P, Hodges M (2000) Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol 123: 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschütze V, Willmitzer L, Müller-Röber B (1995) Inhibition of flower formation by antisense repression of mitochondrial citrate synthase in transgenic potato plants leads to a specific disintegration of the ovary tissues of flowers. EMBO J, 14: 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte MM, Galagan JA, Shapiro JA, Boersig MR, Shewmaker CK, Sharkey TD (1997) Sucrose-phosphate synthase activity and yield analysis of tomato plants transformed with maize sucrose-phosphate synthase. Planta 203: 253–259 [DOI] [PubMed] [Google Scholar]
- Leegood RC, Walker DA (1980) Autocatalysis and light activation of enzymes in relation to photosynthetic induction in wheat chloroplast. Arch Biochem Biophys 200: 572–582 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids –pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lunn JE, ap Rees T (1990) Apparent equilibrium constant and mass-action ratio for sucrose-phosphate synthase in seeds of Pisum sativum. Biochem J 267: 739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Gillespie VJ, Furbank RT (2003) Expression of a cyanobacterial sucrose-phosphate synthase from Synechocyxtis sp. PCC6803 in transgenic plants. J Exp Bot, 54: 223–237 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A, Bieberich K, Willmitzer L, Fernie AR (2002) Carbon assimilation and partitioning in potato leaves deficient in plastidial phosphoglucomutase. Planta 215: 802–811 [DOI] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11: 571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Valadier MH, Carrayol E, Reisdorf-Cren M, Hirel B (2002) Diurnal changes in the expression of glutamate dehydrogenase and nitrate reductase are involved in the C/N balance of tobacco source leaves. Plant Cell Environ 25: 1451–1462 [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001a) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24: 1119–1137 [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001b) The immediate cause of the diurnal changes of nitrogen metabolism in leaves of nitrate-replete tobacco: a major imbalance between the rate of nitrate reduction and the rates of nitrate uptake and ammonium metabolism during the first part of the light period. Plant Cell Environ 24: 177–190 [Google Scholar]
- Miflin BJ, Lea PJ (1980) Ammonium assimilation. In BJ Miflin, ed, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 169–202 [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S (2001) Overexpression of a cyanobacterial fructose-1, 6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19: 965–969 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Muschak M, Hoffmann-Benning S, Fuss H, Kossmann J, Willmitzer L, Fisahn J (1997) Gas exchange and ultrastructural analysis of transgenic potato plants expressing mRNA antisense construct targeted to the cp-fructose-1,6-bisphosphate phosphatase. Photosynthetica 33: 455–465 [Google Scholar]
- Navarre DA, Wendehenne D, Durner J, Noad R, Klessig DF (2000) Nitric oxide modulates the activity of tobacco aconitase. Plant Physiol 112: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M (1985) Preparation of plant mitochondria, criteria for assessment of mitochondrial integrity and purity, survival in vitro. In R Douce, D Day, eds, Higher Plant Cell Respiration. Springer-Verlag, Berlin, pp 7–24
- Novistskaya L, Trevanion SJ, Driscoll S, Foyer CH, Noctor G (2002) How does photorespiration modulate leaf amino acid contents? A dual approach through modeling and metabolite analysis. Plant Cell Environ 25: 821–835 [Google Scholar]
- Padmasree K, Padmavathi L, Raghavendra AS (2002) Essentiality of mitochondrial oxidative metabolism for photosynthesis: optimization of carbon assimilation and protection against photoinhibition. Crit Rev Biochem Mol Biol 37: 71–119 [DOI] [PubMed] [Google Scholar]
- Peyret P, Perez P, Alric M (1995) Structure, genomic organisation, and expression of the Arabidopsis thaliana aconitase gene. J Biol Chem 270: 8131–8137 [DOI] [PubMed] [Google Scholar]
- Preiss J (1988) Biosynthesis of starch and its regulation. In The Biochemistry of Plants. Academic Press, New York, pp 181–254
- Raines CA (2003) The Calvin cycle revisited. Photosynth Res 75: 1–10 [DOI] [PubMed] [Google Scholar]
- Regierer B, Fernie AR, Springer F, Perez-Melis A, Leisse A, Koehl K, Willmitzer L, Geigenberger P, Kossmann J (2002) Starch content and yield increase as a result of altering adenylate pools in transgenic plants. Nat Biotechnol 20: 1256–1260 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie R (2001a) Metabolic profiling and phenotyping of genetically and environmentally modified systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Roessner U, Willmitzer L, Fernie AR (2001b) High-resolution metabolic profiling: identification of phenocopies. Plant Physiol 127: 749–764 [PMC free article] [PubMed] [Google Scholar]
- Sadka A, Dahan E, Cohen L, Marsh KB (2000) Aconitase activity and expression during the development of lemon fruit. Physiol Plant 108: 255–266 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scheible WR, Krapp A, Stitt M (2000) Reciprocal diurnal changes of phosphoenolpyruvate carboxylase expression and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ 23: 1155–1167 [Google Scholar]
- Sharkey TD, Savitch LV, Butz ND (1991) Photometric method for routine determination of kcat and carbamylation of Rubisco. Photosynth Res 28: 41–48 [DOI] [PubMed] [Google Scholar]
- Siedow J, Day DA (2000) Respiration and Photorespiration. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 676–728
- Slaughter CA, Hopkinson DA, Harris H (1977) Distribution and properties of aconitase isozymes in man. Ann Human Genet 40: 385–401 [PubMed] [Google Scholar]
- Stitt M, Fernie AR (2003) From measurements of metabolites to metabolomics: an “on the fly” perspective illustrated by recent studies of carbon-nitrogen interactions. Curr Opin Biotechnol 14: 136–144 [DOI] [PubMed] [Google Scholar]
- Stitt M, Muller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Sumegi B, McCammon M, Sherry AD, Keys DA, Mcalisterhenn L, Srere PA (1992) Metabolism of [3-C-13] pyruvate in TCA cycle mutants of yeast. Biochemistry 31: 8720–8725 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Burrell MM, apRees T (1996) Starch metabolism in tubers of transgenic potato (Solanum tuberosum) with increased ADPglucose pyrophosphorylase. Biochem J 320: 493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB et al. (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Rick CM (1980) Isozymic gene linkage map of the tomato: applications in genetics and breeding. Theor Appl Genet 57: 161–170 [DOI] [PubMed] [Google Scholar]
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J 23: 43–53 [DOI] [PubMed] [Google Scholar]
- Velot C, Mixon MB, Teige M, Srere PA (1997) Model of a quinary structure between Krebs TCA cycle enzymes: a model for the metabolon. Biochemistry 36: 1421–1427 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387 [DOI] [PubMed] [Google Scholar]
- Williams CH, Stillman TJ, Barynin VV, Sedelnikova SE, Tang Y, Green J, Guest JR, Artymiuk PJ (2002) E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition. Nat Struct Biol 9: 447–452 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes Crit Rev Biochem Mol Biol 35: 253–289 [DOI] [PubMed] [Google Scholar]