Abstract
The gene PRT1 of Arabidopsis, encoding a 45-kD protein with two RING finger domains, is essential for the degradation of F-dihydrofolate reductase, a model substrate of the N-end rule pathway of protein degradation. We have determined the function of PRT1 by expression in yeast (Saccharomyces cerevisiae). PRT1 can act as a ubiquitin protein ligase in the heterologous host. The identified substrates of PRT1 have an aromatic residue at their amino-terminus, indicating that PRT1 mediates degradation of N-end rule substrates with aromatic termini but not of those with aliphatic or basic amino-termini. Expression of model substrates in mutant and wild-type plants confirmed this substrate specificity. A ligase activity exclusively devoted to aromatic amino-termini of the N-end rule pathway is apparently unique to plants. The results presented also imply that other known substrates of the plant N-end rule pathway are ubiquitylated by one or more different ubiquitin protein ligases.
Covalent addition of ubiquitin to cellular proteins is of central importance for protein turnover in eukaryotes (Hershko and Ciechanover, 1998; Pickart, 2001; Weissman, 2001). Ubiquitin-activating enzyme E1 forms a thioester to bind and activate free ubiquitin at its carboxyl-terminal Gly residue. Ubiquitin-conjugating enzymes (UBCs) E2 accept activated ubiquitin from E1 and, usually supported by ubiquitin protein ligases E3, are able to transfer ubiquitin to substrates by formation of Gly-Lys isopeptide bonds. Substrate proteins with four or more ubiquitin moieties bound to the substrate in the form of a Gly-76-Lys-48 multi-ubiquitin chain are recognized by the large protease complex, proteasome, and degraded to short peptides, with release of ubiquitin for further cycles of ubiquitylation. Both E2 and E3 enzymes occur as gene families, probably to serve a multiplicity of different substrates. Plants have particularly large gene families of E2 and E3 proteins (Bachmair et al., 2001; Vierstra, 2003). One might speculate that the range of substrates in plants is correspondingly diverse.
One conserved pathway of ubiquitin-dependent proteolysis is the N-end rule pathway. This pathway is apparently universal. The components that mediate substrate recognition and degradation, however, may well differ between phyla. The pathway has been used as a model for studying ubiquitin-dependent proteolysis. Most studies have concentrated on bakers' yeast (Saccharomyces cerevisiae; Varshavsky, 1996). Recently, components of the mammalian N-end rule pathway were also characterized on the molecular level (Grigoryev et al., 1996; Kwon et al., 1998). Existence of the N-end rule pathway in plants can be inferred from previously published data from our group and from others (Bachmair et al., 1993; Worley et al., 1998).
The degradation signal (degron) of the N-end rule pathway is a bulky first amino acid. Amino-termini with unacetylated bulky, but not with small or acetylated first residues, are recognized as a signal for ubiquitylation, which leads to the metabolic destabilization of the substrate by proteasomal degradation (Varshavsky, 1996, 2000). Although most proteins are synthesized starting with Met, the enzyme Met aminopeptidase cleaves off the start Met if—but only if—the second residue does not have a bulky side chain (Bradshaw et al., 1998). Frequently, Met cleavage is followed by acetylation (Polevoda and Sherman, 2003). These canonical processing events generate the bulk of cellular proteins that are not substrates of the N-end rule pathway. Clearly, a cell must possess other pathways that confer a different processing pattern onto a distinct subset of cellular proteins. Among these are processing events by endoproteolytic cleavage to generate substrates for the N-end rule pathway. Recently, the endoprotease Esp1 of yeast, a protein with homologs in all eukaryotes, was identified as such a processing protease (Rao et al., 2001).
Biological functions of the N-end rule pathway have been identified in fungi, animals, and plants. In yeast, the N-end rule pathway regulates peptide import (Turner et al., 2000). In mice (Mus musculus), N-end rule components are necessary for normal spatial memory, for cardiovascular development, and for embryogenesis (Kwon et al., 2000, 2001, 2002). In plants, the pathway has been implicated in senescence (Yoshida et al., 2002). We have analyzed the N-end rule pathway in Arabidopsis (Bachmair et al., 1993; Potuschak et al., 1998). PRT1, a component of the plant N-end rule pathway that was isolated by positional cloning, turned out to be dissimilar to the components of the well-known yeast N-end rule pathway or to any other gene from yeast. In this work, we investigate the biochemical function of PRT1 by expression in yeast and by assessment of substrate specificity in the plant. We find that PRT1 is a ubiquitin protein ligase acting on proteins with aromatic amino-termini, covering a fraction of the destabilizing residues known for the plant N-end rule. We also demonstrate that Arabidopsis has at least one additional ubiquitin protein ligase active in the N-end rule pathway.
RESULTS
PRT1 of Arabidopsis was isolated by positional cloning (Potuschak et al., 1998). Arabidopsis plants with mutation in the PRT1 gene are unable to degrade F-dihydrofolate reductase (DHFR), a model substrate of the N-end rule pathway. Database comparison with the genome of yeast indicates that bakers' yeast does not have an ortholog to this gene. The highest similarity is found to the DNA repair gene RAD18. However, this similarity is largely limited to the RING finger domain of RAD18, which mediates interaction with a UBC (Ulrich and Jentsch, 2000). A direct test of functional similarity by expression of PRT1 in a rad18 yeast mutant indicated that the plant gene cannot complement the radiation sensitivity phenotype of a rad18 mutant (data not shown).
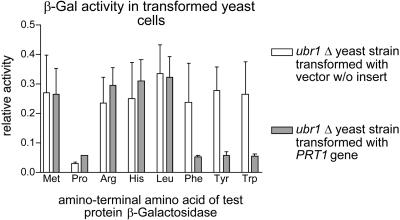
In one series of experiments, we expressed PRT1 in a yeast strain devoid of UBR1, the ubiquitin ligase of the yeast N-end rule pathway (Bartel et al., 1990). We found that yeast cells expressing PRT1 from a plasmid had a significantly lowered steady-state level of F-βgal, a model substrate of the yeast N-end rule pathway that is metabolically stable in ubr1Δ mutants (Figs. 1 and 2). This finding is suggestive of a role for PRT1 in degradation of N-end rule substrates. To rule out the possibility that selection of F-βgal occurs by a structure or sequence element other than the amino-terminal residue, we tested whether the steady-state level of test proteins with other amino-terminal residues is also changed in ubr1Δ PRT1 yeast cells as compared with ubr1Δ cells. We find that PRT1 lowers the concentration of proteins with an aromatic amino-terminus. Remarkably, βgal test proteins with aliphatic hydrophobic or basic amino-terminal residues are unchanged in their concentration (Fig. 1).
Figure 1.
PRT1 influences the amount of β-galactosidase test protein with amino acid X (one or three letter code) as a first amino acid residue (X-βgal) present in yeast cells lacking Ubr1, the ubiquitin protein ligase of the yeast N-end rule. A set of X-βgal test proteins with primary destabilizing amino-terminal residues according to the yeast N-end rule (Arg, His, Leu, Phe, Tyr, and Trp), one metabolically stable (Met-β-gal), and one metabolically unstable (ub-Pro-β-gal) control protein were assayed by enzyme activity measurements. N-end rule substrates with aromatic amino-termini (Phe, Tyr, and Trp) but not with the hydrophobic Leu or with basic residues (Arg or His) have significantly reduced steady-state levels in the presence of PRT1.
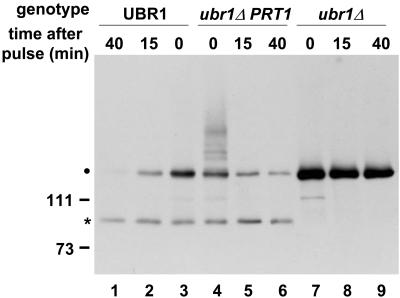
Figure 2.
PRT1 directs degradation of an F-β-gal test protein in yeast. Pulse chase experiments followed by immunoprecipitation of F-β-gal protein, electrophoretic separation, and detection by fluorography indicated that a decreased F-β-gal steady-state level is caused by metabolic instability. Lanes 1 to 3, Wild-type (UBR1) yeast cells were used to indicate metabolic instability of F-β-gal. Lanes 4 to 6, Expression of PRT1 in yeast cells with disrupted UBR1 results in instability of F-β-gal. Lanes 7 to 9, Yeast cells without UBR1 (and without PRT1) cannot degrade F-β-gal. Left, Positions of Mr marker bands. Dot, Position of mature F-β-gal on the gel. Asterisk, Metabolically stable β-gal fragment that is formed in yeast as a side product.
The lowered steady-state concentration is due to metabolic instability because pulse chase experiments demonstrate a short half-life of F-βgal in the ubr1Δ PRT1 yeast strain but not in the ubr1Δ yeast strain without the PRT1 gene (Fig. 2). The fact that UBR1 is the (only) recognition component of the yeast N-end rule and, thus, contains a binding site for the bulky first amino acid residue of the F-βgal test protein and initiates its degradation suggests that PRT1 also contains a binding site for the test protein and mediates its degradation.
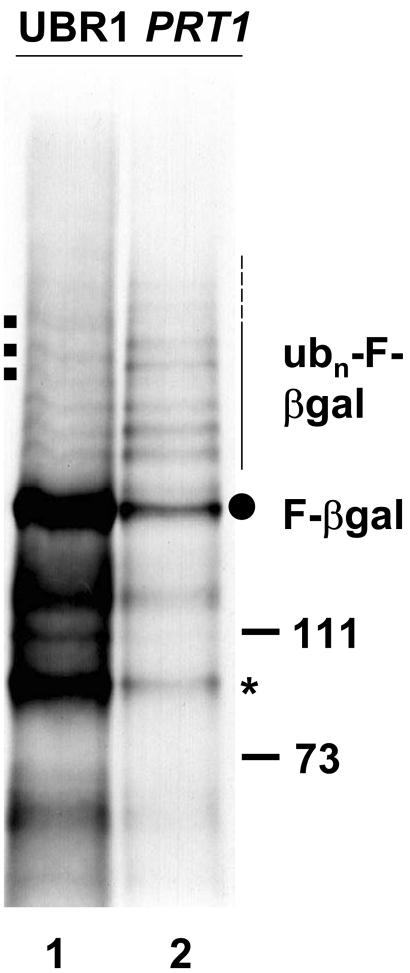
Long exposure of a fluorogram with immunoprecipitate from ubr1Δ PRT1 yeast cells shows a characteristic “ladder” of bands that indicates involvement of ubiquitin in PRT1-mediated degradation of F-βgal (Figs. 2 and 3). The presence of two RING finger domains in PRT1 (Potuschak et al., 1998) suggests that this protein can interact with UBC(s). Interestingly, some bands of the F-βgal ubiquitylation ladder differ either in intensity or in position from those observed in the UBR1 wild-type yeast strain. The overall increase in the steady-state level of ubiquitin ladder bands in the ubr1Δ PRT1 yeast strain could be explained by less efficient channeling of the ubiquitylated substrate protein to the proteasome (Ubr1 apparently delivers ubiquitylated substrates efficiently by direct binding to the proteasome, a property that might not be shared by PRT1; Xie and Varshavsky, 2000). Taken together, these data strongly suggest that PRT1 is a ubiquitin protein ligase.
Figure 3.
UBR1 of yeast and PRT1 of Arabidopsis mediate degradation of the F-β-gal test protein in yeast with differences in multiubiquitylated intermediates. Immunoprecipitation of radioactively labeled F-β-gal protein from wild-type yeast cells (lane 1) or from cells without UBR1 that express PRT1 (ubr1Δ PRT1 cells, lane 2) indicates that the ladder of multi-ubiquitylated species is more intense in ubr1Δ PRT1 cells. Furthermore, a few higher Mr species (square dots to the left) are not visible in ubr1Δ PRT1 cells. Dot to the right, Position of mature F-β-gal on the gel; asterisk, stable β-gal fragment. Mr marker bands are shown to the right. Thin and dashed line to the right, Position of ubiquitylated F-β-gal species.
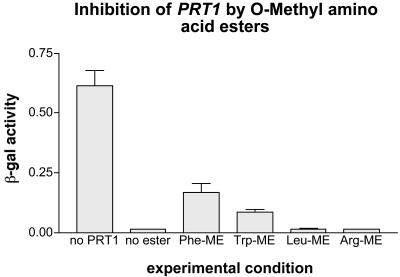
The binding of hydrophobic amino-termini to UBR1 can be inhibited by dipeptides with a hydrophobic first residue (Baker and Varshavsky, 1991). We find that aromatic, but not hydrophobic aliphatic, or basic amino acid methyl esters, are good in vivo inhibitors of PRT1 when expressed in ubr1Δ yeast cells (Fig. 4). This finding is another indication that PRT1 is a recognition element of the plant N-end rule pathway, and it confirms the specificity of PRT1 for exclusively the aromatic amino-termini of the N-end rule pathway.
Figure 4.
Activity of PRT1 is inhibited by O-methyl esters of destabilizing amino acids. Enzyme activity measurements similar to those of Figure 1 were carried out after growth of yeast cells in the presence of Phe-O-methyl ester, Trp-O-methyl ester, Leu-O-methyl ester, and Arg-O-methyl ester. Only those methyl esters which correspond to an amino-terminus destabilized by PRT1 expression can inhibit the degradation process.
We wanted to confirm the in planta relevance of the PRT1 substrate specificity determined in yeast. To that end, we made ubiquitin protein reference (UPR) constructs (Varshavsky, 2000) for Arabidopsis. A single transgene-encoded polypeptide is probably cotranslationally cleaved into two proteins. One protein is the metabolically stable reference protein. The other protein carries a potential degradation signal. Its metabolic stability can be determined by comparing steady-state levels of test and reference protein. Test proteins used in Figure 5 carry N-end rule degrons. They differ from the N-end rule substrates used in previous work (Bachmair et al., 1993; Potuschak et al., 1998) by a carboxyl-terminal extension that makes their size more easily distinguishable from the reference protein on western blots. When using antibodies against the HA epitope for detection, the larger test polypeptide stains with 3-fold intensity due to three repeats of the epitope, compared with one such epitope in the reference protein.
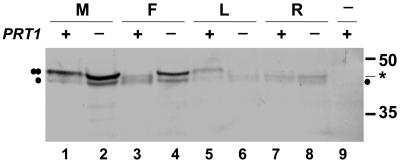
Figure 5.
Degradation of model substrates in PRT1 and prt1 plants. Plants transformed with UPR constructs were used for protein extraction from leaves. Test proteins were detected by western blotting using antibody directed against HA epitope tags. Odd-numbered lanes, PRT1 genotype (plus sign above lanes); even-numbered lanes, prt1 mutant genotype (minus sign above lanes). A single dot denotes the stable reference protein, and a double dot indicates position of the test protein. Asterisk, Metabolically stable fragment generated from unstable test proteins as side products. The test proteins are M-DHFR (“M,” lanes 1 and 2), F-DHFR (“F,” lanes 3 and 4), L-DHFR (“L,” lanes 5 and 6), and R-DHFR (“R,” lanes 7 and 8). Lane 9, Extract from untransformed plant. Mr marker sizes are indicated to the right. For further explanations, see text.
Expression of the UPR construct M-DHFR in wild-type and in prt1 mutant Arabidopsis plants allows the detection of two bands on a western blot with HA antibody, the larger band being about 3 times as intense as the smaller one (Fig. 5, lanes 1 and 2). The ratio between test protein and reference protein is the same both in wild type and in the prt1 mutant, consistent with the previous finding that M-DHFR is metabolically stable in both genotypes (Potuschak et al., 1998). In contrast, expression of R-DHFR from a UPR construct does not allow detection of the larger (R-DHFR) band (Fig. 5, lanes 7 and 8). This indicates fast turnover. Instead, lanes from R-DHFR-expressing plants have bands with a mobility intermediate between M-DHFR and the reference band, which are probably cleavage products of R-DHFR with amino-terminal truncation. Again, there is no obvious difference between wild type and prt1 mutant related to stability of R-DHFR, supporting the notion that a ubiquitin protein ligase distinct from PRT1 is instrumental in degradation of R-DHFR. As expected, we see a difference between the protein pattern of wild-type and prt1 plants when UPR construct F-DHFR is expressed (Fig. 5, lanes 3 and 4). F-DHFR is unstable in wild-type cells; therefore, the detected protein pattern closely resembles the R-DHFR lane. prt1 mutant cells, on the other hand, do not degrade F-DHFR, and the ensuing protein pattern resembles the M-DHFR lane of wild-type and mutant plants. To further characterize PRT1, we also expressed l-DHFR in plants. Wild-type plants apparently degrade this protein at a slower rate than R-DHFR or F-DHFR. Therefore, a weak band of l-DHFR can be visualized with HA antibody (Fig. 5, lane 5). Interestingly, in prt1 plants, degradation of l-DHFR is not only unimpeded, but it is actually increased compared with wild-type plants, suggesting that the PRT1 protein negatively influences L-DHFR degradation (Fig. 5, lane 6). Taken together, these data support the notion that the substrate specificity of PRT1 observed in yeast reflects the in planta specificity and that PRT1 is not the only ubiquitin protein ligase operating in the Arabidopsis N-end rule pathway.
DISCUSSION
In this work, we characterize PRT1, a component of the plant N-end rule pathway. Substrates for degradation by the N-end rule pathway are recognized by a bulky first amino acid (see introduction; Varshavsky, 1996). PRT1 was isolated previously by positional cloning, using a mutant with a transgene-dependent phenotype as a basis for gene identification (Bachmair et al., 1993; Potuschak et al., 1998). At the time of its isolation, the genetic data were the only connection of the PRT1 reading frame to protein turnover. However, we have noticed that similar to PRT1, a considerable number of proteins involved in ubiquitylation processes contain a so-called RING finger domain (Potuschak et al., 1998; for review, see Joazeiro and Weissman, 2000). RING finger domains have since been found to interact with UBCs and define a superfamily of ubiquitin protein ligases (Bachmair et al., 2001; Pickart, 2001; Weissman, 2001). In the work presented here, we show that PRT1 is a ubiquitin protein ligase. This conclusion is based on the following data. First, PRT1 can functionally replace a ubiquitin protein ligase of yeast, Ubr1 (Fig. 1). Second, protein substrates that are destabilized by expression of PRT1 in yeast show a characteristic ladder of higher Mr bands indicative of ubiquitin conjugation (Figs. 2 and 3). Third, mutations in the PRT1 gene cause specific stabilization of certain N-end rule substrates in plants (Fig. 5).
The finding that PRT1 can replace Ubr1 of yeast is somewhat surprising, considering the small size of PRT1 of Arabidopsis compared with Ubr1 (approximately 45 versus 220 kD). Moreover, apart from a RING finger domain, there is no obvious sequence similarity between the two proteins. Because of the size difference, it may not come as a surprise that PRT1 can replace Ubr1′s function only for a subset of the N-end rule substrates known in yeast. Ubr1 has a binding site for all hydrophobic residues and, in addition, another binding site for basic amino-termini. The experiments presented demonstrate that PRT1 can only ubiquitylate proteins with aromatic first residues (Figs. 2, 3, and 5). However, it is unclear whether PRT1 can catalyze multi-ubiquitylation of substrate proteins. Our data do not rule out the possibility that PRT1 catalyzes mono-ubiquitylation and that other ubiquitin protein ligases, presumably those of the UFD pathway (Johnson et al., 1992, 1995) attach additional ubiquitin moieties to build the multi-ubiquitin chain recognized by the proteasome. Examples for such an E3/E4 relationship among different ubiquitin ligases are known from fungi and from animals (Koegl et al., 1999; Grossman et al., 2003).
Substrate binding by the yeast ubiquitin protein ligase Ubr1 can be inhibited by the presence of dipeptides (Baker and Varshavsky, 1991). Binding of small peptides has a biological function in regulation of peptide import (Turner et al., 2000). We find that PRT1 activity can be inhibited by methyl esters of Phe and Trp but not by methyl esters of other amino acids such as Leu or Arg (Fig. 4). The choice of methyl esters over dipeptides was motivated largely by technical considerations (dipeptides are not transported into yeast cells in a ubr1Δ mutant). Although this specific inhibition of PRT1 does not imply a biological function in plants, it is a further proof that PRT1 has a binding site for aromatic residues and, therefore, that it recognizes substrates by their amino-termini.
In addition to the two RING finger domains, PRT1 contains another Zn2+-binding domain, a so-called ZZ domain. This domain can mediate protein-protein interactions (Anderson et al., 1996; Ponting et al., 1996; Cariou et al., 2002). Therefore, the ZZ domain could be involved in binding of the N-end rule degron, in oligomerization, or in binding of another protein.
The restricted substrate range of PRT1 as compared with Ubr1 raised the question of whether the N-end rule of plants is restricted to aromatic residues as amino-terminal degradation signals. Previous experiments presented elsewhere (Schlögelhofer and Bachmair, 2002) and additional findings by others (Worley et al., 1998) indicate that the plant N-end rule does at least include basic amino-termini. Experiments shown in Figure 5 directly address this question by expression of different model substrates in wild-type versus prt1 mutant plants. These experiments confirm the data obtained by heterologous expression of PRT1 in yeast. A model substrate with Arg as first residue is metabolically unstable in both wild-type and mutant plants. In contrast, Phe as a first residue results in a protein that is stable in mutant plants but unstable in wild-type plants. Another construct expressed a test protein with Leu as first residue. Interestingly, although the L-DHFR test protein is apparently of intermediary stability in wild-type leaves, it is actually less stable in the prt1 mutant. A possible interpretation of the decreased half-life of L-DHFR in mutant plants is that PRT1 has a second function as regulator of a Leu-specific ligase activity. However, this issue needs further clarification. In summary, we conclude that PRT1 is specific for aromatic amino-termini and that Arabidopsis contains at least one additional ubiquitin protein ligase that works in the N-end rule pathway.
Interestingly, the Arabidopsis genome encodes a protein with similarity to UBR1, At5g02300. This hypothetical reading frame has similarity to the amino-terminal one-half of UBR1 and is located next to a reading frame annotated as CER3, which has similarity to the carboxyl-terminal one-half of UBR1. Therefore, it is possible that the two genes can be cotranscribed to give one protein with similarity to UBR1 over its entire length. Experiments to understand these facets are under way. Because Arabidopsis prt1 mutants are entirely deficient in degradation of F-DHFR (Potuschak et al., 1998; Fig. 5), we would expect that any additional N-end rule ubiquitin protein ligase of Arabidopsis does not recognize aromatic amino-termini.
Taken together, our data indicate that PRT1 is one of at least two ubiquitin protein ligases of the plant N-end rule pathway and has specificity for aromatic amino-termini. Therefore, the existing data are consistent with increasing complexity of the N-end rule pathway in metazoans.
MATERIALS AND METHODS
Microbial Strains, Plant Lines, Growth, and Transformation
Escherichia coli strain DH5α (supE44 ΔlacU169 [Φ80lacZΔM15] hsdR17 recA1 endA1 gyrA96 thi1 relA1) was used for DNA manipulations. Agrobacterium tumefaciens C58C1pCV2260 was used in plant transformation. Yeast (Saccharomyces cerevisiae) strains K700 (MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-52 GAL+; a kind gift from Kim Nasmyth, Research Institute of Molecular Pathology, Vienna), BBY47 (Bartel et al., 1990), and SS13 (MATa trp1-1 his3-Δ200 leu2,3-112 ura3-52 can1-100 GAL+ ubr1-Δ1::LEU2; segregant from cross K700 × BBY47) were used in this work. Arabidopsis ecotype Columbia-0 was used for in planta experiments. Plant transformation was carried out by the infiltration method (Bechthold et al., 1993; G. Cardon, personal communication). Yeast growth and transformation were carried out as described by Kaiser et al. (1994).
Vector Constructs
A yeast vector for expression of PRT1 was made in the following way: Yeast expression vector pMA91 (Mellor et al., 1983), which contains, between two HindIII sites, promoter and terminator of yeast phosphoglycerate kinase separated by a unique BglII site, was modified by insertion of an NdeI linker (CCATATGG; New England Biolabs, Frankfurt) into the BglII site. A HindIII fragment with promoter and terminator from this vector was cloned into YCplac22 (Gietz and Sugino, 1988) to give vector YCplac22+. Using PCR, the 5′ region of the PRT1 cDNA was modified to contain an NdeI site at the start codon. An NdeI fragment, containing the PRT1 reading frame from the introduced NdeI site to a naturally occurring NdeI site 22 nucleotides downstream of the stop codon, was inserted in sense orientation into YCplac22+ to give YCplac22-PRT1.
Vectors for expression of X-βgal substrate proteins were described previously (Bachmair et al., 1986).
Vectors to express UPR constructs in plants were made in the following way: A DHFR containing EcoRI-AgeI (end filled in using Klenow enzyme) fragment from plasmid pRc/dUb-X-nsP4-bgal (Lévy et al., 1996; a kind gift of Dr. Frédéric Lévy, Ludwig Institute for Cancer Research, Epalinges, Switzerland) was inserted between EcoRI and SmaI sites of pSK. Using oligonucleotides, a part of the translation-enhancing Omega sequence from tobacco mosaic virus was inserted into EcoRI-XhoI-digested vector to give plasmid pSKDHFRo2. Using primer oligonucleotides TGC TCT AGA CGG GTC GAC TCC TTC TGA ATG TTG T and CCG CTC GAG ATG CAG ATT TTC GTC AAG ACT, a PCR fragment was generated with plasmid pRc/dUb-X-nsP4-bgal as a template. After digestion with XbaI and XhoI, the fragment was inserted into SalI- and XbaI-digested vector pSKDHFRo2 to give plasmid pSKDU. A fragment was amplified from Arabidopsis RNA by reverse transcriptase-PCR, containing part of the SUMO1 (At4g26840) reading frame. In this fragment, the last 10 amino acids of SUMO1 are replaced by Ser, followed by a stop codon and an XbaI site. After cloning into an appropriate vector, the amino-terminus of the SUMO reading frame was extended by a fragment containing a KpnI site, followed by three HA epitope tags. An NruI-KpnI fragment from vector pRc/dUb-X-nsP4-bgal that contains the DHFR sequence was linked between the KpnI site at the beginning of the extended SUMO1 reading frame and a unique SmaI site of the vector to give plasmid pDHFRSUMO. An SalI linker (New England Biolabs) was inserted into the SnaBI site of pDHFRSUMO to give plasmid pDHFRSUMOSal. This plasmid was digested with SalI and BstXI, and SalI-BstXI fragments as contained in constructs M-DHFR and F-DHFR (Potuschak et al., 1998) were inserted. The ensuing plasmids, pUFDS and pUMDS, contained a short segment from ubiquitin joined to Phe and Met, respectively, followed by an oligo-Gly and lacI extension and DHFR-SUMO. An SalI-XbaI fragment from pUFDS and pUMDS, respectively, was inserted into SalI-XbaI-digested vector pSKDU to give pUPR-F and pUPR-M, respectively. In these constructs, a single open reading frame codes for murine DHFR cDNA followed by one HA epitope tag, followed by variant yeast ubiquitin with the changes D52G and K48R. The ubiquitin sequence is followed by amino acids Phe or Met, respectively, and by an oligo-Gly extended lacI fragment, fused to DHFR sequence identical to the amino-terminal sequence, followed by three HA epitope tags. The construct ends with the SUMO domain. pUPR-M was digested with SalI and BglII, and double-stranded oligonucleotides were inserted to change the Met to an Arg (codon AGA) and a Leu residue (codon CTT) to give pUPR-R and pUPR-l, respectively. XhoI-XbaI fragments from pUPR-F, -M, -R, and -l were inserted into XbaI-XhoI-digested vector pHi (Schlögelhofer and Bachmair, 2002) to give vectors pHiUPR-F, -M, -R, and -L.
Protein Analysis
Yeast pulse chase experiments were performed as described (Bachmair et al., 1986), except that Complete mini protease inhibitor tablets (Roche, Vienna) plus 20 μg mL–1 Pepstatin A were used for inhibiton of proteases, and no SDS was used during washing of immunoprecipitates. Monoclonal mouse anti-β-galactosidase antibody (Promega, Mannheim, Germany) was used for immunoprecipitation.
To analyze plant protein extracts, 5 mg of fresh leaf material was collected in a 1.5 mL reaction tube and frozen in liquid nitrogen. Two hundred microliters of prewarmed (37°) extraction buffer (50 mm Tris-Cl [pH 6.8], 4% [w/v] SDS, and 10% [v/v] β-mercaptoethanol) was added, and the samples were ground to a fine powder. After homogenization, samples were centrifuged (10,000 rpm for 1 min). The supernatant was transferred, heated to 95°C, and centrifuged at 14,000 rpm for 10 min. The supernatant was mixed with 1 volume 2× Laemmli sample buffer (50% [v/v] glycerol, 20 mm dithiothreitol, 2% [w/v] SDS, 125 mm Tris-Cl [pH 6.8], and 0.003% [w/v] bromphenol blue) and frozen for storage. Samples were heated to 55°C for 10 min before loading for SDS-PAGE (10% [w/v] acrylamide; Mini-Protean Cell, Bio-Rad, Munich). After electrophoresis, the gel was shaken at room temperature for 40 min in transfer solution (190 mm Gly, 25 mm Tris, 20% [v/v] methanol, and 0.05% [v/v] SDS). Transfer to Immobilon-P membrane (Millipore, Eschborn, Germany) was carried out in transfer solution at 100 V for 1.5 h (Transblot apparatus; Bio-Rad). After transfer, the membrane was incubated for 2 × 10 min in 1× alkaline NaCl-Tris buffer (ANT) (150 mm NaCl, 50 mm Tris-Cl [pH 8.0], and 0.002% [v/v] NaN3), and 1.5 h in blocking solution (1× ANT/20% [v/v] newborn calf serum) at room temperature. The first antibody incubation (1:3,000 [v/v] in blocking solution; rat anti-HA monoclonal antibody, Roche, Mannheim, Germany) was carried out overnight at 4°C. Thereafter, the membrane was washed 4 × 15 min with 1× ANT/0.05% (v/v) Tween 20 at room temperature, rinsed briefly with 1× ANT, and incubated with the second antibody (1:1,000 [v/v] in blocking solution; goat anti-rat IgG coupled to alkaline phosphatase, Sigma-Aldrich, Taufkirchen, Germany) for 2 h at room temperature. After further washing (3 × 15 min with 1× ANT/0.05% [v/v] Tween 20), the membrane was stained as recommended for the secondary antibody.
β-Galactosidase Activity Determination
A preculture of the β-galactosidase-expressing yeast strain was grown for 2 to 3 d in dropout medium containing 2% (w/v) raffinose as sole carbon source. For induction of β-galactosidase expression, the preculture was diluted approximately 1:30 (v/v) in Gal-containing medium. β-galactosidase activity was measured by the rate of ortho-nitrophenyl β-d-galactopyranoside cleavage in permeabilized cells (Kaiser et al., 1994) and normalized to equal cell numbers. Results of at least three independent assays were used to determine the sd of the mean (95% confidence interval).
Inhibition of the N-End Rule Pathway by Amino Acid Derivatives
A preculture of yeast strain SS13 expressing PRT1 and an F-βgal substrate under the control of a Gal-inducible promoter was grown in raffinose dropout medium for 1 d. The preculture was diluted 1:20 (v/v) in Gal dropout medium containing either 10 mm phenylananine O-methyl ester, 10 mm Trp O-methyl ester, 10 mm Leu O-methyl ester, or 10 mm Arg O-methyl ester and was incubated at 30°C for 5 h. Thereafter, cells were harvested for β-galactosidase activity determination.
Acknowledgments
We want to thank Bonnie Bartel (Rice University, Houston) and Kim Nasmyth (IMP, Vienna) for yeast strains and Kerstin Luxa (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for help with DNA cloning and plant transformation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.029272.
This work was supported by the Austrian Science Foundation FWF (grant no. P 13927) and by the Max Planck Society.
References
- Anderson TJ, Robers RP, Jarrett HW (1996) Ca2+-Calmodulin binds to the carboxyl-terminal domain of dystrophin. J Biol Chem 271: 6605–6610 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Becker F, Schell J (1993) Use of a reporter transgene to generate Arabidopsis mutants in ubiquitin-dependent protein degradation. Proc Natl Acad Sci USA 90: 418–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F (2001) Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Baker R, Varshavsky A (1991) Inhibition of the N-end rule pathway in living cells. Proc Natl Acad Sci USA 88: 1090–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Wünnung I, Varshavsky A (1990) The recognition component of the N-end rule pathway. EMBO J 9: 3179–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechthold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Bradshaw RA, Brickey WW, Walker KW (1998) N-terminal processing: the methionine aminopeptidase and N-acetyl transferase families. Trends Biochem Sci 23: 263–267 [DOI] [PubMed] [Google Scholar]
- Cariou B, Perdereau D, Cailliau K, Browaeys-Poly E, Béréziat V, Vasseur-Cognet M, Girard J, Burnol A-F (2002) The adapter protein ZIP binds Grb14 and regulates its inhibitory action on insulin signaling by recruiting protein kinase C zeta. Mol Cell Biol 22: 6959–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Grigoryev S, Stewart AE, Kwon YT, Arfin SM, Bradshaw RA, Jenkins NA, Copeland NG, Varshavsky A (1996) A mouse amidase specific for N-terminal asparagine. J Biol Chem 271: 28521–28532 [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM (2003) Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300: 342–344 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Joazeiro CAP, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Bartel B, Seufert W, Varshavsky A (1992) Ubiquitin as a degradation signal. EMBO J 11: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PCM, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644 [DOI] [PubMed] [Google Scholar]
- Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, Gaur A, Hyde L, Deneberg VH, Varshavsky A (2000) Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol Cell Biol 20: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Davydov IV, Hu R-G, An JY, Seo JW, Du F, Varshavsky A (2002) An essential role of N-terminal arginylation in cardiovascular development. Science 297: 96–99 [DOI] [PubMed] [Google Scholar]
- Kwon YT, Reiss Y, Fried VA, Hershko A, Yoon JK, Gonda DK, Sangan P, Copeland NG, Jenkins NA, Varshavsky A (1998) The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci USA 95: 7898–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Xia Z-X, Davydov IV, Lecker SH, Varshavsky A (2001) Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol Cell Biol 21: 8007–8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy F, Johnsson N, Rümenapf T, Varshavsky A (1996) Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci USA 93: 4907–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Dobson MJ, Roberts NA, Tuite MF, Emtage JS, White S, Lowe PA, Patel T, Kingsman AJ, Kingsman SM (1983) Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene 24: 1–14 [DOI] [PubMed] [Google Scholar]
- Pickart C (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Polevoda B, Sherman F (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol 325: 595–622 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ (1996) ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci 21: 11–13 [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlögelhofer P, Becker F, Nejinskaia V, Bachmair A (1998) PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci USA 95: 7904–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Uhlmann F, Nasmyth K, Varshavsky A (2001) Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410: 955–959 [DOI] [PubMed] [Google Scholar]
- Schlögelhofer P, Bachmair A (2002) A test of fusion protein stability in the plant, Arabidopsis thaliana, reveals degradation signals from ACC synthase and from the plant N-end rule pathway Plant Cell Rep 21: 174–179 [Google Scholar]
- Turner GC, Du F, Varshavsky A (2000) Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405: 579–583 [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Jentsch S (2000) Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J 19: 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (1996) The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A (2000) Ubiquitin fusion technique and its descendants. In J Thorner, SD Emr, JN Abelson, eds, Methods in Enzymology, Vol 327 Academic Press, New York, pp 578–593 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Worley CK, Ling R, Callis J (1998) Engineering in vivo instability of firefly luciferase and Escherichia coli β-glucuronidase in higher plants using recognition elements from the ubiquitin pathway. Plant Mol Biol 37: 337–347 [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A (2000) Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci USA 97: 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Callis J, Nishida I, Watanabe A (2002) A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J 32: 129–137 [DOI] [PubMed] [Google Scholar]