Abstract
Papaya leaf curl China virus (PaLCuCNV) was previously reported as a distinct begomovirus infecting papaya in southern China. Based on molecular diagnostic survey, 13 PaLCuCNV isolates were obtained from tomato plants showing leaf curl symptoms in Henan and Guangxi Provinces of China. Complete nucleotide sequences of 5 representative isolates (AJ558116, AJ558117, AJ704604, FN256260, and FN297834) were determined to be 2738–2751 nucleotides, which share 91.7%–97.9% sequence identities with PaLCuCNV isolate G2 (AJ558123). DNA-β was not found to be associated with PaLCuCNV isolates. To investigate the infectivity of PaLCuCNV, an infectious clone of PaLCuCNV-[CN:HeNZM1] was constructed and agro-inoculated into Nicotiana benthamiana, N. tabacum Samsun, N. glutinosa, Solanum lycopersicum and Petunia hybrida plants, which induced severe leaf curling and crinkling symptoms in these plants. Southern blot analysis and polymerase chain reaction (PCR) indicated a systemic infection of test plants by the agro-infectious clone.
Keywords: Papaya leaf curl China virus (PaLCuCNV), Tomato, Infectivity
1. Introduction
Geminiviruses are plant viruses characterized by having a circular single-stranded DNA genome packaged within twinned icosahedral particles. The family Geminiviridae is divided into four genera (Mastrevirus, Curtovirus, Topocuvirus, and Begomovirus) (Fauquet et al., 2005). The majority of the economically important geminiviruses belong to the genus Begomovirus, which are transmitted by the whitefly, Bemisia tabaci, to a wide range of dicotyledonous plants. Begomoviruses have caused significant yield losses to many crops in tropical and subtropical regions worldwide (Fauquet et al., 2003; Moffat, 1999). Many begomoviruses have a bipartite genome, referred to as DNA-A and DNA-B, both of which are required for systemic infection. However, some begomoviruses have only a single genomic component, resembling DNA-A of the bipartite begomoviruses, which is sufficient for maintenance of the disease (Harrison and Robinson, 1999). Some monopartite begomoviruses were found to be associated with single-stranded DNA satellite, termed DNA-β (Saunders et al., 2001; Briddon et al., 2001; Zhou et al., 2003b).
In China, begomoviruses caused serious yield losses to tomato production and many species have been isolated (Li et al., 2004; Wu et al., 2006; Mugiira et al., 2008). Papaya leaf curl China virus (PaLCuCNV) was first detected in papaya in Guangxi Province of China (Wang et al., 2004). In this paper, PaLCuCNV was identified in tomato showing leaf curl disease in Guangxi and Henan Provinces and the complete nucleotide sequences of 5 PaLCuCNV isolates were determined and compared. Furthermore, an infectious clone of PaLCuCNV isolate was constructed and its infectivity in Nicotiana benthamiana, N. tabacum Samsun, N. glutinosa, Solanum lycopersicum and Petunia hybrida was examined.
2. Materials and methods
2.1. Source of virus
Tomato plants showing leaf curl symptoms were collected in fields of Henan and Guangxi Provinces of China, which were infected with PaLCuCNV.
2.2. Total DNA extraction, polymerase chain reaction (PCR) and cloning
Total DNA extraction from leaf samples was performed following the method as described by Xie et al. (2002). Partial DNA-A fragment was amplify by degenerate primers PA and PB and then purified, cloned and sequenced as described by Zhou et al. (2003a). The primer pair HeNfullF (5′-GGGATCCTTTACTAAACGAGTTTCC-3′) and HeNfullR (5′-AGGATCCCACATGTTTGACGTG-3′) were designed based on the determined sequence and used to amplify the complete viral DNA genome. PCR products of the expected size were recovered, cloned, and sequenced. Attempts were made to amplify the possible satellite DNA-β from the samples with primers β01 and β02 specific for DNA-β (Briddon et al., 2002) or with phi29 polymerase-based rolling circle amplification of circular single-stranded DNA molecules (Guo et al., 2009).
2.3. DNA sequence analysis
DNA sequences were analyzed by the method as described by Zhang et al. (2009). DNA sequences used in this study for the phylogenetic analysis were: African cassava mosaic virus (ACMV-[CM], AF112352), Ageratum leaf curl virus (ALCV-[CN:G52], AJ851005), Ageratum yellow vein China virus (AYVCNV-[CN:Hn2.19], AJ564744), Beet curly top virus (BCTV-[US:Cal], M24597), Bhendi yellow vein mosaic virus (BYVMV-IN[IN:Mad], AF241479), Euphorbia leaf curl Guangxi virus (EuLCGXV-[CN:GX], AM411424), Indian cassava mosaic virus (ICMV-[IN:Mah], AJ314739), Papaya leaf curl China virus isolates (PaLCuCNV-[CN:LC1-2], AM691553; PaLCuCNV-[CN:FQ1], AM691554; PaLCuCNV-[CN:F25], AM691552; PaLCuCNV-[CN:G4], AJ811914; PaLCuCNV-[CN: G2], AJ558123; PaLCuCNV-[CN:ZM1], EU874386; PaLCuCNV-[CN:G10], AJ558125; PaLCuCNV-[CN: G43], AJ876548; PaLCuCNV-[CN:G7], AJ811439; PaLCuCNV-[CN:G8], AJ558124; PaLCuCNV-[VN: Hat], DQ641700), Papaya leaf curl Guangdong virus (PaLCuGUV-[CN:GD2], AJ558122), Tobacco curly shoot virus (TbCSV-[CN:Y35], AJ420318), Tobacco leaf curl Thailand virus (TbLCTHV-[TH], DQ871221), Tomato golden mosaic virus (TGMV-[BR:Com], K02029), Tomato leaf curl Java virus (ToLCJV-B[ID:Age], AB162141), Tomato leaf curl Taiwan virus (ToLCTWV-[CN:ZJ16], AM698111), Tomato leaf curl Vietnam virus (ToLCVNV-[VN:Co], AF264063), Tomato yellow leaf curl China virus (TYLCCNV-[CN:Y11], AJ319676), Tomato yellow leaf curl Guangdong virus (TYLCGDV-[CN:GD], AY602166), Tomato yellow leaf curl virus (TYLCV-[CN:SH2], AM282874), and Tomato yellow leaf curl Vietnam virus (TYLCVNV-[VN:Han], DQ641697).
2.4. Construction of an infectious clone and agro-inoculation of plants
The geminiviral circular single-stranded DNA was amplified by the commercial kit TempliPhi™ (GE Healthcare, Bucks, UK) following the manufacturer’s recommendations using the isolated total DNA as template. An approximately 0.4 mer (1 kb) fragment encompassing the intergenic region was released from the high molecular weight rolling circle amplification (RCA) DNA products by double digestion with BamHI/EcoRI, and cloned into BamHI/EcoRI double digested binary vector pBINPLUS (van Engelen et al., 1995) to produce clone pBINPLUS-HeNZM1-0.4A. The full length clone of PaLCuCNV-[CN:HeNZM1] was released from the RCA products by digestion with BamHI and cloned into the unique BamHI site in the pBINPLUS-HeNZM1-0.4A to yield pBINPLUS-HeNZM1-1.4A. Insert integrity and orientation were confirmed by PCR using primer pair HeNfullF and HeNfullR and digestion with EcoRI. pBINPLUS-HeNZM1-1.4A was mobilized into competent Agrobacterium tumefaciens strain EHA105 as described by Zhou et al. (2003b). Agro-inoculation of N. benthamiana, N. tabacum Samsun, N. glutinosa, S. lycopersicum cv. Hongbaoshi and P. hybrida was performed as described (Xie et al., 2006). Plants were kept at 25 °C with 70% relative humidity in an insect-free growth chamber with 16-h day light, and symptom development was daily examined.
2.5. Analysis of viral DNA in inoculated plants
Total nucleic acid extraction, electrophoresis and DNA blotting were performed as described by Zhang et al. (2009). PaLCuCNV specific probe was produced by PCR amplification of pBINPLUS-HeNZM1-1.4A with primer pair HeNfullF and HeNfullR, and labeled with digoxigenin (DIG) using the DIG DNA Labeling and Detection Kit (Roche Diagnostics Co., Mannheim, Germany).
3. Results
3.1. DNA sequence analysis
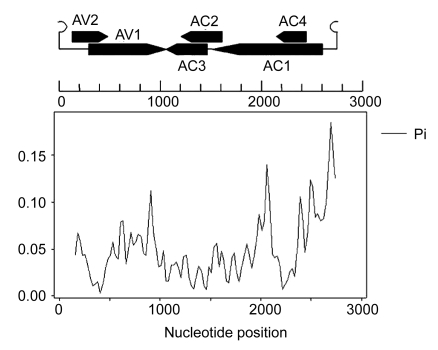
All the 13 tomato samples (10 from Guangxi and 3 from Henan) showing typical leaf curl symptoms were found to be infected by begomoviruses by PCR using primers PA and PB specific for begomoviruses. These fragments were cloned and sequenced. Alignment of the 500-bp sequences showed that these samples were infected by PaLCuCNV (data not shown). The complete DNA sequences of 5 representative isolates (AJ558116, AJ558117, AJ704604, FN256260, and FN297834) were determined to be 2738–2751 nucleotides. They have a typical genome organization of begomoviruses DNA-A, with two open reading frames (ORFs) [AV1 (coat protein) and AV2] in virion-sense DNA and four ORFs [AC1 (replicase), AC2, AC3 and AC4] in complementary sense DNA, separated by an intergenic region (IR). Sequence similarity search results revealed that the 5 isolates share 91.7% to 97.9% identity with PaLCuCNV-[CN:G2]. The sequences of the 5 isolates and previously reported sequences of PaLCuCNV isolates were aligned using DNAMAN software package. Nucleotide sequence identity ranged from 89.9% to 99.9% (Table 1). The level of variation of PaLCuCNV isolates was estimated by the average number of nucleotide differences per site (Pi) at all sites along the genome. Sliding window plot of Pi index revealed the uneven distribution of genetic variation of viral DNA (Fig. 1). The highest peaks resided in the IR region. There were other two obvious peaks, one in the 3′-terminal of AC1 ORF and another in the 3′-terminal of AV1 ORF. The most conserved regions reside in the overlapping region of AV2 and AV1 ORFs and the overlapping region of AC1 and AC4 ORFs.
Table 1.
Percentage nucleotide sequence identities of PaLCuCNV isolates from different locations and hosts
| Location | Host | Isolate | FQ1 | LC1-2 | F25 | G4 | G2 | G22 | HeNZM1 | ZM1 | G30 | GX4 | G10 | G43 | G7 | G8 | G12 | Hat |
| Fujian | Tomato | FQ1 | 100 | |||||||||||||||
| Fujian | Eclipta | LC1-2 | 99.9 | 100 | ||||||||||||||
| Fujian | Eclipta | F25 | 100 | 99.9 | 100 | |||||||||||||
| Guangxi | Papaya | G4 | 99.3 | 99.1 | 99.3 | 100 | ||||||||||||
| Guangxi | Papaya | G2 | 98.2 | 98.1 | 98.2 | 98.0 | 100 | |||||||||||
| Guangxi | Tomato | G22 | 98.5 | 98.3 | 98.5 | 98.4 | 97.9 | 100 | ||||||||||
| Henan | Tomato | HeNZM1 | 95.9 | 95.7 | 95.9 | 95.8 | 96.0 | 96.3 | 100 | |||||||||
| Henan | Tomato | ZM1 | 95.8 | 95.6 | 95.8 | 95.6 | 95.9 | 96.1 | 99.6 | 100 | ||||||||
| Guangxi | Tomato | G30 | 96.0 | 95.8 | 96.0 | 95.9 | 96.2 | 96.4 | 99.7 | 99.5 | 100 | |||||||
| Guangxi | Tomato | GX4 | 91.9 | 91.7 | 91.9 | 92.2 | 91.7 | 92.1 | 94.1 | 93.9 | 94.2 | 100 | ||||||
| Guangxi | Ageratum | G10 | 91.2 | 91.1 | 91.2 | 91.1 | 91.2 | 92.1 | 91.7 | 91.6 | 91.9 | 90.9 | 100 | |||||
| Guangxi | Ageratum | G43 | 91.2 | 91.1 | 91.2 | 91.2 | 91.3 | 92.1 | 91.8 | 91.8 | 92.0 | 91.0 | 99.3 | 100 | ||||
| Guangxi | Ageratum | G7 | 92.1 | 91.9 | 92.1 | 92.0 | 91.9 | 92.9 | 92.6 | 92.6 | 92.7 | 91.8 | 99.0 | 99.0 | 100 | |||
| Guangxi | Ageratum | G8 | 93.2 | 93.1 | 93.2 | 93.1 | 93.3 | 94.1 | 93.4 | 93.3 | 93.5 | 89.9 | 94.4 | 94.4 | 93.9 | 100 | ||
| Guangxi | Tomato | G12 | 93.4 | 93.2 | 93.4 | 93.3 | 93.5 | 94.4 | 93.6 | 93.5 | 93.8 | 90.0 | 94.3 | 94.4 | 93.9 | 98.9 | 100 | |
| Vietnam | Tobacco | Hat | 94.4 | 94.3 | 94.4 | 94.3 | 94.4 | 95.3 | 95.1 | 95.0 | 95.2 | 91.7 | 94.0 | 94.1 | 94.8 | 95.9 | 96.3 | 100 |
Fig. 1.
Sliding window plot showing the distribution of genetic variation estimated by nucleotide diversity (Pi) for PaLCuCNV isolates
A window size of 50 and a step of 25 nucleotides were used. The relative positions of the ORFs of viral DNA genome are illustrated above the plot in linear DNA format
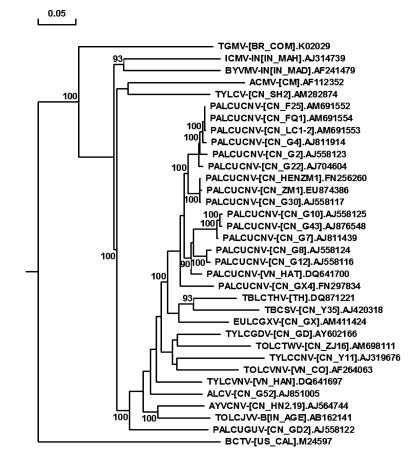
In phylogenetic tree, all the PaLCuCNV isolates were clustered together and closed to those begomoviruses isolated in southern China and adjacent regions (Fig. 2). Using primer pair β01 and β02, the satellite DNA-β molecule was not detected by PCR in selective samples. Attempts were also unsuccessful to amplify a DNA-B or DNA-β using RCA of circular single-stranded DNA molecules followed by restriction fragment length polymorphism.
Fig. 2.
Dendrogram showing the relationships among PaLCuCNV isolates and other representative geminiviruses based on multiple alignments of nucleotide sequences
The dendrogram was constructed by the neighbour-joining method of DNAMAN and bootstrapped 1000 times. Bootstrap scores exceeding 90% are placed at major nodes. Horizontal distances are proportional to sequence distances; vertical distances are arbitrary. The trees were rooted on the sequence of Beet curly top virus (BCTV)
3.2. Infectivity of PaLCuCNV-[CN:HeNZM1]
Agro-inoculation showed that the 1.4-mer clone of PaLCuCNV-[CN:HeNZM1] was highly infectious in N. benthamiana, N. tabacum Samsun, N. glutinosa, S. lycopersicum and P. hybrida plants (Table 2). In S. lycopersicum plants, inoculated plants produced downward leaf curling approximately 13 d after inoculation (dpi), which then progressed to severe downward leaf curling and crinkling at 20 dpi (Figs. 3b and 3c). The symptoms were identical to the ones observed in the tomato plant from which this clone was originally isolated. In N. benthamiana, upward leaf curling and crinkling symptoms were observed at 11 dpi (Figs. 3e and 3f). No symptoms were observed in mock-inoculated tomato or N. benthamiana plants (Figs. 3a and 3d). In N. tabacum Samsun, leaf curling and crinkling symptoms were induced at 17 dpi (Fig. 3g). In N. glutinosa, inoculated plants produced downward leaf curling and crinkling symptoms at 14 dpi (Fig. 3h). In P. hybrida, inoculated plants produced upward leaf curling and crinkling symptoms at 14 dpi (Fig. 3i).
Table 2.
Infectivity and symptoms induced by PaLCuCNV-[CN:HeNZM1]
| Plant | Infectivitya | Symptoms |
| Solanum lycopersicum | 15/15 | Leaf downward curling and crinkling |
| Nicotiana benthamiana | 15/15 | Leaf upward curling and crinkling |
| Nicotiana tabacum Samsun | 10/10 | Leaf curling and crinkling |
| Nicotiana glutinosa | 10/10 | Leaf downward curling and crinkling |
| Petunia hybrida | 10/10 | Leaf upward curling and crinkling |
Number of infected plants/number of inoculated plants
Results were derived from two independent experiments
Fig. 3.









Symptoms induced by PaLCuCNV-[CN: HeNZM1]
(a)–(c) Healthy (a) or PaLCuCNV-[CN:HeNZM1] infected Solanum lycopersicum plant (b) and single leaf (c) at 20 d after agro-inoculation; (d)–(f) Healthy (d) or PaLCuCNV-[CN:HeNZM1] infected Nicotiana benthamiana plant (e) and single leaf (f) at 20 d after agro-inoculation; (g)–(h) PaLCuCNV-[CN:HeNZM1] infected N. tabacum Samsun (g), N. glutinosa (h) and Petunia hybrida (i) at 30 d after agro-inoculation
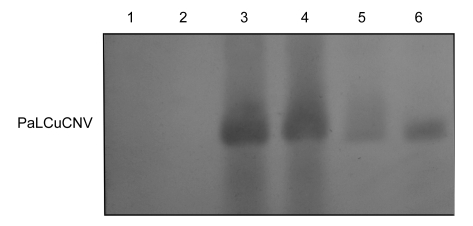
To further confirm a systemic infection of S. lycopersicum, N. tabacum Samsun, N. glutinosa, N. benthamiana and P. hybrida plants, total DNA isolated from newly emerged symptomatic leaves at 35 dpi was analyzed by PCR using primer pair HeNfullF and HeNfullR, and 2.8-kb PCR products were successfully amplified from all tested plant samples. Southern hybridization analysis of agro-inoculated S. lycopersicum plants showed that PaLCuCNV-[CN: HeNZM1] was present in the inoculated plants (Fig. 4).
Fig. 4.
Southern blot analysis of nucleic acids extracted from Solanum lycopersicum
Lanes 1–2: healthy plants; lanes 3–4: agro-inoculated plants with infectious clone PaLCuCNV-[CN:HeNZM1] at 35 d after inoculation; lanes 5–6: naturally infected plants
4. Discussion
PaLCuCNV, a distinct begomovirus, was originally found in Guangxi Province of China (Wang et al., 2004), which caused a severe leaf curl disease on papaya in southern China. Based on PCR detection and sequence analysis, we demonstrated that tomato samples showing leaf curl symptoms collected from Guangxi and Henan Provinces of China were infected by PaLCuCNV. Nucleotide sequence comparison showed that the PaLCuCNV isolates infecting tomato and other hosts in Guangxi, Fujian and Henan Provinces share high sequence identity and cannot be sub-clustered based on host and distribution. Most of begomoviruses detected in China are monopartite begomoviruses. In this study, we have not detected DNA-B or DNA-β component associated with PaLCuCNV, and infectivity tests confirmed that PaLCuCNV is a monopartite begomovirus. The result is consistent with our previous report (Wang et al., 2004).
It was found that begomoviruses have been infecting tomato in southern China for many years (Li et al., 2004). Since 2006, Tomato yellow leaf curl virus (TYLCV) and Tomato leaf curl Taiwan virus were found in Shanghai and Zhejiang (Wu et al., 2006; Mugiira et al., 2008), and TYLCV has spread to many parts of China including Jiangsu, Shandong, and Hebei Provinces (Zhang et al., 2009). Mix-infection detections showed that PaLCuCNV is only detected in tomato in Henan and Guangxi Provinces, but not in other regions of China (data not shown). There is a large (more than 1 000 km) separation between Henan and Guangxi Provinces where PaLCuCNV was detected in tomato. Commercial activities involving trade in tomato plants and some other living plants infected with PaLCuCNV may contribute to the spread of PaLCuCNV from Guangxi to Henan. As TYLCV has been spread to many regions in China, so it is urgent to take practical intervention measures, such as enforcement of quarantine regulations in the trade and movement of live plant materials, to prevent PaLCuCNV from being spread to other regions.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30530520), the Zhejiang Agricultural Science and Technology Key Research Projects (No. 2007C12054), and the Natural Science Foundation of Zhejiang Province, China (No. Y307397)
References
- 1.Briddon RW, Mansoor S, Bedford ID, Pinner MS, Saunders K, Stanley J, Zafar Y, Malik KA, Markham PG. Identification of DNA components required for induction of cotton leaf curl disease. Virology. 2001;285(2):234–243. doi: 10.1006/viro.2001.0949. [DOI] [PubMed] [Google Scholar]
- 2.Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. Universal primers for the PCR-mediated amplification of DNAβ: a molecule associated with some monopartite begomoviruses. Mol Biotechnol. 2002;20(3):315–318. doi: 10.1385/MB:20:3:315. [DOI] [PubMed] [Google Scholar]
- 3.Fauquet CM, Bisaro DM, Briddon RW, Brown JK, Harrison BD, Rybicki EP, Stenger DC, Stanley J. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch Virol. 2003;148(2):405–421. doi: 10.1007/s00705-002-0957-5. [DOI] [PubMed] [Google Scholar]
- 4.Fauquet CM, Mayo MA, Maniloff J, et al. Geminiviridae. In: Fauquet CM, Mayo MA, Maniloff J, et al., editors. Virus Taxonomy 8th Report of the International Committee on Taxonomy of Viruses. New York, NY, USA: Academic Press; 2005. pp. 277–370. [Google Scholar]
- 5.Guo W, Yang XL, Xie Y, Cui XF, Zhou XP. Tomato yellow leaf curl Thailand virus-[Y72] is a monopartite begomovirus associated with DNAβ. Virus Genes. 2009;38(2):328–333. doi: 10.1007/s11262-009-0327-4. [DOI] [PubMed] [Google Scholar]
- 6.Harrison BD, Robinson DJ. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (Begomoviruses) Annu Rev Phytopathol. 1999;37(1):369–398. doi: 10.1146/annurev.phyto.37.1.369. [DOI] [PubMed] [Google Scholar]
- 7.Li ZH, Zhou XP, Zhang X, Xie Y. Molecular characterization of tomato-infecting begomoviruses in Yunnan, China. Arch Virol. 2004;149(9):1721–1732. doi: 10.1007/s00705-004-0334-7. [DOI] [PubMed] [Google Scholar]
- 8.Moffat AS. Plant pathology—Geminiviruses emerge as serious crop threat. Science. 1999;286(5446):1835. doi: 10.1126/science.286.5446.1835. [DOI] [Google Scholar]
- 9.Mugiira RB, Liu SS, Zhou X. Tomato yellow leaf curl virus and Tomato leaf curl Taiwan virus invade southeast coast of China. J Phytopathol. 2008;156(4):217–221. doi: 10.1111/j.1439-0434.2007.01345.x. [DOI] [Google Scholar]
- 10.Saunders K, Bedford ID, Stanley J. Pathogenicity of a natural recombinant associated with Ageratum yellow vein disease: implications for geminivirus evolution and disease aetiology. Virology. 2001;282(1):38–47. doi: 10.1006/viro.2000.0832. [DOI] [PubMed] [Google Scholar]
- 11.van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. pBINPLUS—an improved plant transformation vector based on pBIN19. Transgenic Res. 1995;4(4):288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- 12.Wang XY, Xie Y, Zhou XP. Molecular characterization of two distinct begomoviruses from papaya in China. Virus Genes. 2004;29(3):303–309. doi: 10.1007/s11262-004-7432-1. [DOI] [PubMed] [Google Scholar]
- 13.Wu JB, Dai FM, Zhou XP. First report of Tomato yellow leaf curl virus in China. Plant Dis. 2006;90(10):1359. doi: 10.1094/PD-90-1359C. [DOI] [PubMed] [Google Scholar]
- 14.Xie Y, Zhou XP, Zhang ZK, Qi YJ. Tobacco curly shoot virus isolated in Yunnan is a distinct species of Begomovirus. Chin Sci Bull. 2002;47(3):197–200. doi: 10.1360/02tb9047. [DOI] [Google Scholar]
- 15.Xie Y, Jiang T, Zhou XP. Agroinoculation shows Tobacco leaf curl Yunnan virus is a monopartite begomovirus. Eur J Plant Pathol. 2006;115(4):369–375. doi: 10.1007/s10658-006-9021-8. [DOI] [Google Scholar]
- 16.Zhang H, Gong HR, Zhou XP. Molecular characterization and pathogenicity of Tomato yellow leaf curl virus in China. Virus Genes. 2009;39(2):249–255. doi: 10.1007/s11262-009-0384-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhou XP, Xie Y, Peng Y, Zhang ZK. Malvastrum yellow vein virus, a new Begomovirus species associated with satellite DNA molecule. Chin Sci Bull. 2003;48(20):2205–2209. doi: 10.1360/03wc0272. [DOI] [Google Scholar]
- 18.Zhou XP, Xie Y, Tao XR, Zhang ZK, Li ZH, Fauquet CM. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J Gen Virol. 2003;84(1):237–247. doi: 10.1099/vir.0.18608-0. [DOI] [PubMed] [Google Scholar]