Abstract
Objective: To investigate the distributions of human leukocyte antigen (HLA)-A and -B alleles and HLA-A-B haplotypes in the Yi ethnic minority of the Yunnan Province, situated in southwestern China. Methods: DNA typing for HLA-A and -B loci was performed using the polymerase chain reaction-sequence-based typing (PCR-SBT) method on 114 randomly selected healthy individuals of the Yi population. The allelic frequencies of HLA-A and -B loci were calculated by direct counting and HLA-A-B haplotypes were estimated using the expectation maximization algorithm. Results: A total of 17 HLA-A and 38 HLA-B alleles were found in the Yi population. The most frequent alleles were A*2402 (32.46%), A*1101 (26.32%), and A*0203 (10.09%) at the HLA-A locus and B*4601 (12.28%), B*1525 (10.09%), B*4001 (8.77%), and B*3802 (7.89%) at the HLA-B locus. The predominant HLA-A-B haplotypes were A*2402-B*1525 (7.86%) and A*0203-B*3802 (5.64%), followed by A*1101-B*4001 (4.69%). Phylogenetic analysis indicates that the Yi population in the Honghe, Yunnan Province of China basically belongs to groups of southeastern Asian origin, but shares some characteristics with northeastern Asian groups. Conclusion: The present study may add to the understanding of HLA polymorphism in the Yi ethnic group that was poorly defined previously, and provide useful information for bone marrow transplantation, anthropological research, and forensic sciences as well as for disease-association studies.
Keywords: Yi ethnic minority, Human leukocyte antigen (HLA)-A, HLA-B, Allele, Haplotype, Polymerase chain reaction-sequence-based typing (PCR-SBT)
1. Introduction
The major histocompatibility complex (MHC) is a genetic region defined initially by the rejection of skin grafts in genetically incompatible mouse strains. In humans, the human leukocyte antigen (HLA) region, equivalent to the MHC, comprises about 3 Mb located on the short arm of chromosome 6 (6p21.3) (Erlich et al., 2001). HLA genes are divided into three basic groups: class I, class II, and class III. HLA-A, -B, and -C are the major genes in MHC class I, the most polymorphic and variable one, with currently more than 2 678 alleles identified (situation on July 2009) (http://www.ebi.ac.uk/imgt/hla/stats.html).
It has been shown that the distributions of HLA alleles and haplotypes are different from one ethnic group to another or even between the members of the same ethnic group living in different geographical areas (Shen et al., 2008). Due to its high degree of polymorphisms and variability in allele/haplotype frequencies, HLA loci analyses have become a valuable tool to trace population migration and origin, and to study population relationships for anthropological purposes (Hu et al., 2007).
The Yi ethnic minority, previously named “Luoluo,” is a modern ethnic group seen in China, Vietnam, and Thailand. With 8 million people, the Yi ethnic minority represents the 7th largest of 55 ethnic minority groups officially recognized in China. There are 3300 Yi people (1999 census) living in northeastern Vietnam (http://www.chinakindnesstour.com/Chinainfo/minority/EthnicMinority/Chinainfo128.shtml). Nearly all the Yi people live in the mountainous areas of Sichuan, Yunnan, Guizhou, and Guangxi Provinces in southwestern China, usually inhabiting the sides of steep mountain slopes far from cities. In the period between the 2nd century B.C. and the early Christian era, the activities of the ancient Yi centered on the areas of Dianchi in Yunnan Province and Qiongdou in Sichuan Province. After the 3rd century B.C., the ancient Yi extended their activities from the Anning River valley, the Jinsha River, the Dianchi Lake, and the Ailao Mountains to northeastern and southern Yunnan, northwestern Guizhou, and northwestern Guangxi (Zhu et al., 2008).
Most Yi people are farmers, herders of cattle, sheep and goats, and nomadic hunters. Only about one third of the Yi are literate and most elders are not able to write. The Yi language, which is written in the Yi script, belongs to the Tibeto-Burman language group of the Chinese-Tibetan language family, and includes six dialects. Most Yi are animists, with believes combined with elements of Daoism, shamanism, and fetishism, except for those who have been influenced by Buddhism through the Han culture or converted to Christianism (Guo and Dong, 2000).
Accounting for 61% of the total Yi population in China, the Yunnan Province ranks the first (Yang et al., 2004). The Honghe Hani and Yi Autonomous Prefecture is located in the south of the Yunnan Province of China and borders Vietnam to the south. According to the 2000 national census, Honghe has 973 732 Yi, accounting for 23.57% of the whole population in this area (China, National Bureau of Statistics, 2001).
Although there are several studies about the polymorphism of HLA genes of the Yi population using low resolution techniques (Chen W.M. et al., 2001; Lu et al., 2003a; 2003b; Wen et al., 2004; Xu et al., 2006; Chen S. et al., 2007), no high-resolution data using HLA class I gene polymorphisms of the Yi people are available now. In the present study we have investigated the distributions of HLA-A and -B alleles and HLA-A-B haplotypes in the Yi ethnic minority in the Honghe Prefecture of the Yunnan Province, southwestern China, using the polymerase chain reaction-sequence-based typing (PCR-SBT) method, and compared these data with ones recorded in other populations in order to enrich the knowledge of HLA polymorphism in Chinese ethnic groups and to better understand the genetic relationships among different populations. This is the first HLA study of the Yi ethnic minority of the Honghe Autonomous Prefecture. Moreover, the distribution of HLA-A-B haplotypes was also investigated for the first time in the Yi in this study. The results from these analyses therefore will provide useful information for blood and bone marrow transplantation, anthropological researches, and forensic science as well as for disease-association studies.
2. Materials and methods
2.1. Samples and DNA extraction
After informed consent, non-coagulated ethylenediaminetetraacetic acid (EDTA) blood samples were obtained from 114 unrelated healthy Yi individuals living in the Honghe Hani and Yi Autonomous Prefecture, Yunnan, China. All participants were randomly chosen among individuals whose ancestors have lived in the region for at least three generations and were self-identified as Yi. All blood samples were stored at −20 °C before DNA extraction. Genomic DNA was extracted using standard techniques (Miller et al., 1988).
2.2. Typing of HLA class I genes
All subjects were typed for HLA-A and -B loci. SBT of the HLA-A and -B loci was performed on exons 2 and 3, according to Kurz et al. (1999) and Pozzi et al. (1999) with minor modifications. Amplification by polymerase chain reaction (PCR) was accomplished on GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA, USA). PCR-amplified DNA fragments were purified and sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kits (Applied Biosystems, Foster City, CA, USA) using an ABI 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. Sequencing was always performed in forward and reverse directions and processed using Sequencing Analysis, MatchTools and Navigator (MatchTools Allele Identification package, Applied Biosystems, Foster City, CA, USA). This software was used to detect the heterozygous positions within each electropherogram and to assess the typing, based on an alignment of the processed sequence with an updated library of HLA sequences and alleles defined up to October 2007. Ambiguous types were resolved to four digits according to the updated database (IMGT release 2.19.0).
2.3. Statistical analysis
Allele frequencies of HLA-A and -B loci were estimated using SPSS 11.0 software (SPSS Inc., Chicago, Illinois). Haplotype frequencies were calculated from genotype data by expectation maximization (EM) algorithm using Arlequin software package version 3.0 (Laurent Excoffier, CMPG, Zoological Institute, University of Bern, Switzerland) (Excoffier et al., 2005). Forensic parameters including observed heterozygosity (Ho), expected heterozygosity (He), power of discrimination (PD), polymorphism information content (PIC), and probability of paternity exclusion (PPE) were computed using the PowerStat version 1.2 spreadsheet (Promega Corporation, USA) as described (Tereba, 1999). The exact test of Guo and Thompson (1992) was used to evaluate deviation from expected Hardy-Weinberg genotypic proportions. Absolute linkage disequilibrium (ALD), maximal linkage disequilibrium (MLD), and relative linkage disequilibrium (RLD) between two alleles at the HLA-A and -B loci were calculated according to Weir (1996). Analysis of molecular variance (AMOVA) was performed by Arlequin to estimate the population differentiation and to compare HLA allele frequencies between Yi and other populations, and the results are presented as the fixation index. A neighbor-joining (NJ) tree from allele frequencies of HLA-A and -B loci was constructed using the Mega3.1 software package (Center for Evolutionary Functional Genomics, the Biodesign Institute Tempe, AZ, USA).
3. Results
3.1. Study design
Allele frequencies of HLA class I genes were investigated by PCR-SBT in 114 unrelated subjects from the Yi ethnic minority in the Yunnan Province of southwestern China. The Ewens-Watterson homozygosity test showed no evidence for a departure from selective neutrality at HLA-A and -B loci. No significant deviation from the Hardy-Weinberg equilibrium was observed at the HLA-A and HLA-B loci (P=0.05249 and 0.103721, respectively).
3.2. Allele distribution
In total, we detected 17 HLA-A and 38 HLA-B alleles in the subjects. The allelic distributions are summarized in Table 1. The three most common HLA-A alleles with a frequency greater than 10% were HLA-A*2402 (32.46%), A*1101 (26.32%), and A*0203 (10.09%). The second in rank with frequencies greater than 3% were alleles A*0207, A*0201, and A*3101. At the HLA-B locus, B*4601 had the highest frequency (12.28%), which was followed by B*1525 (10.09%), B*4001 (8.77%), and B*3802 (7.89%).
Table 1.
Allelic frequency (AF) distributions of HLA-A and -B in Yi population living in the Yunnan Province of China (N=114)
| HLA-A | AF (%) | HLA-B | AF (%) | HLA-B | AF (%) |
| A*0101 | 0.44 | B*0702 | 1.32 | B*3802 | 7.89 |
| A*0201 | 6.14 | B*0705 | 0.44 | B*3901 | 2.19 |
| A*0203 | 10.09 | B*1301 | 3.95 | B*4001 | 8.77 |
| A*0205 | 0.44 | B*1501 | 7.02 | B*4002 | 1.75 |
| A*0206 | 1.75 | B*1502 | 7.46 | B*4006 | 0.88 |
| A*0207 | 7.02 | B*1504 | 0.44 | B*4402 | 0.44 |
| A*0258 | 0.44 | B*1507 | 1.75 | B*4403 | 1.32 |
| A*0301 | 0.88 | B*1511 | 0.44 | B*4601 | 12.28 |
| A*1101 | 26.32 | B*1512 | 2.63 | B*4801 | 1.32 |
| A*1102 | 0.44 | B*1518 | 0.44 | B*5101 | 3.51 |
| A*2402 | 32.46 | B*1525 | 10.09 | B*5102 | 0.44 |
| A*2403 | 2.63 | B*1527 | 0.44 | B*5121 | 0.44 |
| A*2407 | 2.19 | B*1532 | 0.44 | B*5201 | 4.82 |
| A*2601 | 2.19 | B*2705 | 0.88 | B*5401 | 0.88 |
| A*2901 | 0.44 | B*2706 | 0.44 | B*5501 | 0.44 |
| A*3101 | 3.51 | B*3501 | 3.07 | B*5502 | 5.26 |
| A*3303 | 2.63 | B*3505 | 1.32 | B*5601 | 2.63 |
| B*3543 | 0.44 | B*5602 | 0.44 | ||
| B*3701 | 0.44 | B*5604 | 0.88 |
3.3. Haplotype frequencies
As summarized in Table 2, of the 180 HLA-A-B haplotypes predicted by the EM algorithm, 37 were found at frequencies greater than 0.58%. Two HLA-A-B haplotypes occurred at frequencies greater than 5%, i.e., HLA-A*2402-B*1525 (7.86%) and A*0203-B*3802 (5.64%), followed by A*1101-B*4001 (4.69%), A*0207-B*4601 (4.64%), A*1101-B*5201 (4.37%), and A*2402-B*4601 (4.09%). Table 2 also shows the values of ALD, MLD, and RLD, which are commonly used in the evaluation of the linkage disequilibrium (LD) in population genetics. Twenty-seven HLA-A-B haplotypes are common haplotypes with significant linkage disequilibrium (both haplotype frequency and RLD are higher than 1%).
Table 2.
Estimated haplotype frequencies of HLA-A-B in Chinese Yi population in Yunnan Province of China (haplotype frequency ≥0.58%)
| HLA-A-B haplotype | HF (%) | ALD (%) | MLD (%) | RLD (%) |
| A*2402-B*1525 | 7.86 | 4.54 | 6.77 | 67.04 |
| A*0203-B*3802 | 5.64 | 4.85 | 7.10 | 68.30 |
| A*1101-B*4001 | 4.69 | 2.38 | 6.46 | 36.80 |
| A*0207-B*4601 | 4.64 | 3.78 | 6.16 | 61.35 |
| A*1101-B*5201 | 4.37 | 3.10 | 3.55 | 87.31 |
| A*2402-B*4601 | 4.09 | 0.05 | 8.24 | 0.65 |
| A*1101-B*1502 | 3.30 | 1.34 | 5.49 | 24.39 |
| A*2402-B*4001 | 3.06 | 0.17 | 5.89 | 2.89 |
| A*2402-B*1501 | 2.63 | 0.32 | 4.71 | 6.86 |
| A*1101-B*1301 | 2.60 | 1.56 | 2.91 | 53.63 |
| A*2402-B*5502 | 2.41 | 0.68 | 3.53 | 19.32 |
| A*1101-B*5502 | 2.41 | 1.03 | 3.88 | 26.46 |
| A*2403-B*1512 | 1.75 | 1.69 | 2.56 | 65.77 |
| A*0201-B*1501 | 1.71 | 1.28 | 5.71 | 22.38 |
| A*0201-B*4601 | 1.60 | 0.84 | 5.39 | 15.63 |
| A*1101-B*5101 | 1.58 | 0.66 | 2.59 | 25.36 |
| A*0201-B*1502 | 1.36 | 0.90 | 5.68 | 15.91 |
| A*2402-B*1301 | 1.35 | 0.05 | 2.65 | 1.890 |
| A*2402-B*4801 | 1.32 | 0.88 | 0.88 | 100.00 |
| A*1101-B*0702 | 1.32 | 0.97 | 0.97 | 100.00 |
| A*3303-B*4403 | 1.32 | 1.28 | 1.28 | 100.00 |
| A*1101-B*4002 | 1.32 | 0.85 | 1.29 | 66.07 |
| A*2402-B*1507 | 1.32 | 0.74 | 1.18 | 62.75 |
| A*2402-B*3501 | 1.30 | 0.29 | 2.06 | 13.90 |
| A*0203-B*3901 | 1.27 | 1.04 | 1.97 | 52.95 |
| A*0203-B*1525 | 1.25 | 0.23 | 9.07 | 2.58 |
| A*0203-B*4001 | 1.03 | 0.14 | 7.89 | 1.83 |
| A*0207-B*5604 | 0.88 | 0.82 | 0.82 | 100.00 |
| A*2601-B*2705 | 0.88 | 0.86 | 0.86 | 100.00 |
| A*0206-B*4006 | 0.88 | 0.86 | 0.86 | 100.00 |
| A*0206-B*1512 | 0.88 | 0.83 | 1.71 | 48.65 |
| A*2601-B*5601 | 0.88 | 0.82 | 2.14 | 38.38 |
| A*2407-B*3501 | 0.88 | 0.81 | 2.13 | 38.10 |
| A*3101-B*5601 | 0.88 | 0.78 | 2.54 | 30.91 |
| A*3101-B*1501 | 0.88 | 0.63 | 3.26 | 19.34 |
| A*2402-B*5401 | 0.72 | 0.43 | 0.59 | 73.11 |
| A*0207-B*3802 | 0.62 | 0.07 | 6.46 | 1.09 |
HF: haplotype frequency; ALD: absolute linkage disequilibrium; MLD: maximal linkage disequilibrium; RLD: relative linkage disequilibrium
3.4. Forensic parameters
The values of common forensic parameters of HLA-A and -B loci in the Yi population are listed in Table 3. The Ho, He, PD, PIC, and PPE were 0.7997, 0.7456, 0.9289, 0.7762, 0.5023 for HLA-A and 0.9382, 0.8596, 0.9846, 0.9349, 0.7140 for HLA-B, respectively.
Table 3.
Common forensic parameters of HLA-A and -B loci in the Yi population
| Locus | Ho | He | PD | PIC | PPE |
| HLA-A | 0.7997 | 0.7456 | 0.9289 | 0.7762 | 0.5023 |
| HLA-B | 0.9382 | 0.8596 | 0.9846 | 0.9349 | 0.7140 |
Ho: observed heterozygosity; He: expected heterozygosity; PD: power of discrimination; PIC: polymorphism information content; PPE: probability of paternity exclusion
3.5. Phylogenetic tree analysis
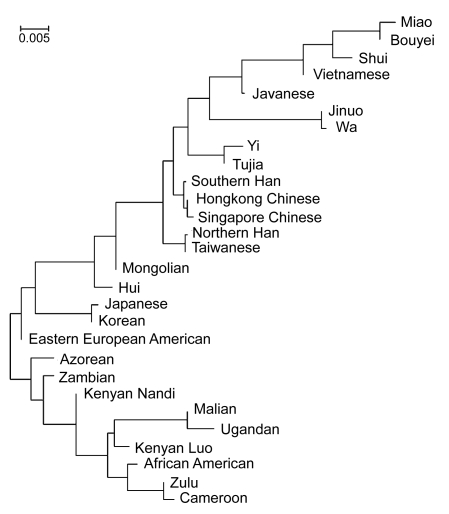
As shown in Fig. 1, an NJ phylogenetic tree was constructed using the allele frequencies of HLA-A and -B loci of 28 representative populations all over the world. Two main clusters were obtained: African and Asian/European. Located in the southeastern Asian cluster, the Yi population was first grouped with Tujia ethnic group and followed by Jinuo, Wa, Shui, Miao and Bouyei ethnic minorities living in China, as well as Vietnamese and Javanese groups. Subsequently, they were joined by the southern Chinese groups, including the Taiwanese, Hong Kong Chinese, Singapore Chinese, and southern Canton Chinese. Compared to other ethnic minorities like Jinuo, Wa, Shui, Miao, and Bouyei, the genetic structure of the Yi is the closest to the northeastern Asian populations.
Fig. 1.
Dendrogram constructed by the neighbor-joining method showing the relationship between Yi population with other 27 populations based on the allelic frequencies of HLA-A and -B loci
4. Discussion
The HLA is the most polymorphic genetic system of human genome, consisting of a number of closely linked genes (Marsh et al., 2002). The distributions of HLA alleles and haplotypes have been widely used in the investigation of the origin and migration of human populations, transplantation matching, forensic sciences, and disease-association studies (Li et al., 2009; Wang et al., 2009).
The technology of HLA typing has significantly evolved since the PCR method was introduced by Mullis and Faloona (1987). Nowadays, there are several kinds of PCR-based HLA typing being used: DNA amplification with sequence specific primers (SSP) (Zetterquist and Olerup, 1992), sequence-specific oligonucleotide probes (SSOP) (Petersdorf et al., 1991), single-stranded conformation polymorphism (SSCP) (Lo et al., 1992), sequence-based typing (SBT) (Santamaria et al., 1993), and DNA chip technology (Jiang et al., 2006). These HLA typing techniques have much higher accuracy as well as reliability than the classical serologic typing method, and also facilitate the standardization of HLA typing processes. Among all the PCR-based HLA typing, SBT is considered the gold standard (Danzer et al., 2007) and the most comprehensive method for characterizing MHC gene polymorphisms (Takeshima et al., 2009). Moreover, new HLA alleles can be easily identified by SBT and confirmed after cloning (Scheltinga et al., 1997).
We found that there were significant differences between the studied Yi population living in Yunnan and the one living in Sichuan at HLA-A (P=0.037) (Lu et al., 2003a) and HLA-B (P<0.001) (Xu et al., 2006). Studies have shown that allelic distributions in the HLA-A and -B loci differ from the members of the Chinese Yi ethnic group living in different geographic areas. In Asians, HLA-A*1101, A*2402, and A*3303 were found at significantly higher frequencies than those observed in other populations (Cao et al., 2001). As the predominant HLA-A allele in the Yi population, A*2402 (32.46%) is also the most common one of other populations such as the Tibetan (Chen et al., 2006), Mongolian (Hong et al., 2007), Japanese (Saito et al., 2000), and Korean groups (Lee et al., 2005) in northern Asia and interestingly, also in north native Americans (Cao et al., 2001). A*1101, the second most common HLA-A allele in the Yi (26.32%), has the highest prevalence in Southwest Asia (Chen et al., 2007; Ogata et al., 2007; Hoa et al., 2008; Shi L. et al., 2008) and in many Chinese population groups (20.23%–28.70%) (Middleton et al., 2004; Yang et al., 2006; Trachtenberg et al., 2007; Wen et al., 2008). A*1101 and A*2402 together accounting for almost 60% of all HLA-A alleles were observed in the Yi population, whereas A*3303 had a relatively low frequency (2.63%) than that in other Asian populations such as the Korean (16.49%) (Lee et al., 2005), Wa ethnic minority living in China (16.10%) (Shi L. et al., 2008), Javanese (15.61%) (Yuliwulandari et al., 2009), Vietnamese (11.50%) (Hoa et al., 2008), and other Chinese groups (6.23%–11.70%) (Middleton et al., 2004; Yang et al., 2006; Trachtenberg et al., 2007; Wen et al., 2008). These observations suggest that the Yi people have some characteristics of northeastern Asian groups in terms of HLA-A allelic distribution.
In the Yi people, B*4601 had the highest prevalence (12.28%) at the HLA-B locus. In a disease association study between the HLA allele distribution and the epidemic of severe acute respiratory syndrome (SARS), Lin et al. (2003) showed that HLA-B*4601 was a possible susceptibility HLA allele for the infection of SARS. In previous studies, HLA-B*4601 was also reported to occur at high frequencies in Southeast Asian populations such as the Vietnamese population (11.50%) (Hoa et al., 2008) and in southern Chinese groups such as in the Taiwanese (Wen et al., 2008), Hong Kong Chinese (Middleton et al., 2004), Singapore Chinese (Middleton et al., 2004), and southern Canton Chinese groups (Trachtenberg et al., 2007) (11.90%–16.30%), where the highest occurrence of SARS cases of the world was found. By contrast, B*4601 is rarely seen in European populations (0–0.9%) (Cao et al., 2001; Spínola et al., 2005a; 2005b; Dunne et al., 2008; Mack et al., 2009), and never seen in African populations (Cao et al., 2001; 2004) or in native Americans (Cao et al., 2001), where very few cases of SARS infection were reported. HLA-B*1502, a common HLA-B allele in the Yi population (7.46%) and in Southeast Asia, was reported to have a strong association with cutaneous adverse drug reactions, including carbamazepine (CBZ)-induced Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) in Han Chinese and many other Asian populations (Chung et al., 2004; Hung et al., 2006; Lonjou et al., 2006; Man et al., 2007). So far, there have been no epidemiologic studies of SARS, CBZ-SJS, or TEN conducted in the Yi population.
As one of the most important characteristics of the HLA system, the LD between the HLA-A and -B loci in the Yi population was measured through calculating ALD, MLD, and RLD. Six HLA-A-B haplotypes, including A*2402-B*4801, A*1101-B*0702, A*3303-B*4403, A*0207-B*5604, A*2601-B*2705, and A*0206-B*4006, had the most significant LD (RLD=1), which indicates that there is very little chance of the recombination between the A and B alleles for these haplotypes in the Yi population. The haplotypes in this study were estimated using EM algorithm, but the haplotypes presented were not based on family data; therefore, we need to acknowledge the potential errors inherent to haplotype estimation methods (Single et al., 2002).
In the NJ-constructed phylogenic tree, the Yi population has the closest genetic distance to the Tujia ethnic group that was sampled in the Hubei Province of China. The Tujia ethnic group may trace back their history to the ancient Ba people who occupied the area around modern-day Chongqing in southwestern China some 2500 years ago (http://en.wikipedia.org/wiki/Tujia). The close genetic relationship between Yi and Tujia is also supported by the origin of their languages, which have grammatical and phonological similarities (http://www.brassett.org.uk/tujia/elng/elintro.html) belonging to the Tibeto-Burman language family (often considered a subgroup of the Sino-Tibetan language family) (Shi M. et al., 2008), and are spoken in various central, eastern, southern, and southeastern Asian countries.
In a previous study of genetic structures in Chinese populations, it was shown that although the Tujia people belong to southern populations of China, they also contain certain lineages that are seen in northern populations. It was also shown that the Yi population was the first to be grouped with the Tujia group, followed by the Bouyei and Wa populations (Du et al., 1997), which is in accordance with the observation in this study. Some scholars consider that the Yi descended from the ancient Qiang people of today’s western China, who are also said to be the ancestors of the Tibetan, Naxi, and Qiang peoples. They migrated from southeastern Tibet through Sichuan into Yunnan, where their largest populations can be found today. This may explain that the northeastern Asian groups are closer in the phylogenetic tree to the Yi population (Fig. 1) than other southeastern Asian ethnic groups located in adjacent regions to the Yi, such as Miao, Bouyei, Shui, Jinuo, and Wa, which have different origins. In a previous study, it was concluded that the Yi originated from the north of the Yangtze River where a significant fraction of them still live, but they at present belong to the southern group (Du et al., 1997).
As a highly polymorphic genetic marker system, HLA has been applied to forensic science for a long time (Mayr, 1988; Lee et al., 1994) and is considered a useful tool for personal identification and paternity testing (Allen et al., 1993; Li et al., 2004). The polymorphisms of genetic markers can be evaluated by some forensic parameters such as Ho, He, PD, PIC, and PPE. Using these parameters, gene heterozygosity can objectively reflect the genetic variation level in a population. Polymorphism information content is often used to measure the indicative strength of genetic markers for linkage studies. A locus is considered highly polymorphic when its PIC is higher than 0.5. Also, PD and PPE are indicators for discrimination capability of a genetic marker and the PD and PPE values of a marker with high polymorphism are normally higher than 0.8 and 0.5, respectively. In our study, the PIC values of HLA-A and -B loci in the Yi population were both higher than 0.5 (0.7762 and 0.9349, respectively) and the PD and PPE at both HLA-A and -B loci were higher than the cutoff values mentioned above. Therefore, the HLA-A and -B polymorphism data obtained from the Yi people were valuable for forensic science research.
In conclusion, the HLA polymorphism data presented in this study indicate that the Yi population in Honghe, Yunnan, China, basically belongs to groups of southeastern Asian origin, but at the same time they share some characteristics with northeastern Asian group in terms of the distribution of allele frequencies. This study adds to our understanding of HLA polymorphisms in the ethnic groups in Asia. In addition, these results may provide useful information for blood and bone marrow transplantation, forensic science, and disease-association studies. However, it will be necessary to perform further HLA analysis on class II and other class I loci in order to describe the complete profiles of the Yi population.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 30700470 and 30871348), and the Shaan’xi Provincial Science and Technology Research and Development Project Fund (No. 2008K09-02), China
References
- 1.Allen M, Saldeen T, Pettersson U, Gyllensten U. Genetic typing of HLA class II genes in Swedish populations: application to forensic analysis. Journal of Forensic Science. 1993;38(3):554–570. [PubMed] [Google Scholar]
- 2.Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández-Viña MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Human Immunology. 2001;62(9):1009–1030. doi: 10.1016/S0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 3.Cao K, Moormann AM, Lyke KE, Masaberg C, Sumba OP, Doumbo OK, Koech D, Lancaster A, Nelson M, Meyer D, et al. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens. 2004;63(4):293–325. doi: 10.1111/j.0001-2815.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Hong W, Shao H, Fu Y, Liu X, Chen D, Xu A. Allelic distribution of HLA class I genes in the Tibetan ethnic population of China. International Journal of Immunogenetics. 2006;33(6):439–445. doi: 10.1111/j.1744-313X.2006.00637.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Ren X, Liu Y, Hu Q, Hong W, Xu A. Human leukocyte antigen class I polymorphism in Miao, Bouyei, and Shui ethnic minorities of Guizhou, China. Human Immunology. 2007;68(11):928–933. doi: 10.1016/j.humimm.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Chen WM, Huang HL, Liu ZH, Tao H, Pan DJ, Lin JH, Xu AL. Analysis of HLA-DQB1 polymorphism by PCR-SSO in Yichu of Yunnan Province. Yi Chuan Xue Bao. 2001;28(2):107–114. (in Chinese) [PubMed] [Google Scholar]
- 7.China, National Bureau of Statistics. Chinese Statistical Yearbook 2000. Beijing, China: China Statistics Press; 2001. pp. 415–416. (in Chinese) [Google Scholar]
- 8.Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, Wu JY, Chen YT. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428(6982):486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 9.Danzer M, Polin H, Pröll J, Hofer K, Fae I, Fischer GF, Gabriel C. High-throughput sequence-based typing strategy for HLA-DRB1 based on real-time polymerase chain reaction. Human Immunology. 2007;68(11):915–917. doi: 10.1016/j.humimm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Du R, Xiao C, Cavalli-sforza LL. Genetic distances between Chinese populations calculated on gene frequencies of 38 loci. Science in China Series C, Life Sciences. 1997;40(6):613–621. doi: 10.1007/BF02882691. (in Chinese) [DOI] [PubMed] [Google Scholar]
- 11.Dunne C, Crowley J, Hagan R, Rooney G, Lawlor E. HLA-A, B, Cw, DRB1, DQB1 and DPB1 alleles and haplotypes in the genetically homogenous Irish population. International Journal of Immunogenetics. 2008;35(4-5):295–302. doi: 10.1111/j.1744-313X.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- 12.Erlich HA, Opelz G, Hansen J. HLA DNA typing and transplantation. Immunity. 2001;14(4):347–356. doi: 10.1016/S1074-7613(01)00115-7. [DOI] [PubMed] [Google Scholar]
- 13.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 14.Guo DL, Dong JZ. Summarization of Chinese Nationalities (Zhonghua Mingzu Zhishi Tonglan) Kunming, China: Kunming Education Publishing House; 2000. pp. 86–94. (in Chinese) [Google Scholar]
- 15.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48(2):361–372. doi: 10.2307/2532296. [DOI] [PubMed] [Google Scholar]
- 16.Hoa BK, Hang NT, Kashiwase K, Ohashi J, Lien LT, Horie T, Shojima J, Hijikata M, Sakurada S, Satake M, et al. HLA-A, -B, -C, -DRB1 and -DQB1 alleles and haplotypes in the Kinh population in Vietnam. Tissue Antigens. 2008;71(2):127–134. doi: 10.1111/j.1399-0039.2007.00982.x. [DOI] [PubMed] [Google Scholar]
- 17.Hong W, Chen S, Shao H, Fu Y, Hu Z, Xu A. HLA class I polymorphism in Mongolian and Hui ethnic groups from Northern China. Human Immunology. 2007;68(5):439–448. doi: 10.1016/j.humimm.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Hu SP, Luan JA, Li B, Chen JX, Cai KL, Huang LQ, Xu XY. Genetic link between Chaoshan and other Chinese Han populations: evidence from HLA-A and HLA-B allele frequency distribution. American Journal of Physical Anthropology. 2007;132(1):140–150. doi: 10.1002/ajpa.20460. [DOI] [PubMed] [Google Scholar]
- 19.Hung SI, Chung WH, Jee SH, Chen WC, Chang YT, Lee WR, Hu SL, Wu MT, Chen GS, Wong TW, et al. Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmaco-genetics and Genomics. 2006;16(4):297–306. doi: 10.1097/01.fpc.0000199500.46842.4a. [DOI] [PubMed] [Google Scholar]
- 20.Jiang B, Li Y, Wu H, He XM, Li CT, Li L, Tang R, Xie Y, Mao YM. Application of HLA-DRB1 genotyping by oligonucleotide micro-array technology in forensic medicine. Forensic Science International. 2006;162(1-3):66–73. doi: 10.1016/j.forsciint.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 21.Kurz B, Steiert I, Heuchert G, Müller CA. New high resolution typing strategy for HLA-A locus alleles based on dye terminator sequencing of haplotypic group-specific PCR-amplicons of exon 2 and exon 3. Tissue Antigens. 1999;53(1):81–96. doi: 10.1034/j.1399-0039.1999.530109.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee HC, Ladd C, Bourke MT, Pagliaro E, Tirnady F. DNA typing in forensic science. I. Theory and background. The American Journal of Forensic Medicine and Pathology. 1994;15(4):269–282. doi: 10.1097/00000433-199412000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens. 2005;65(5):437–447. doi: 10.1111/j.1399-0039.2005.00386.x. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Li CT, Liu Y. Research on the application feasibility of HLA-DRB1 genotyping for forensic identification by oligonucleotide chip. Fa Yi Xue Za Zhi. 2004;20(2):81–84. 81. (in Chinese) [PubMed] [Google Scholar]
- 25.Li Y, Yan H, Xue WJ, Tian PX, Ding XM, Pan XM, Feng XS, Tian XH, Xiang HL, Hou J. Allograft rejection-related gene expression in the endothelial cells of renal transplantation recipients after cytomegalovirus infection. Journal of Zhejiang University-SCIENCE B. 2009;10(11):820–828. doi: 10.1631/jzus.B0920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, Chu CC, Chen PJ, Su YW, Lim KH, Tsai ZU, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Medical Genetics. 2003;4(1):9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo YM, Patel P, Mehal WZ, Fleming KA, Bell JI, Wainscoat JS. Analysis of complex genetic systems by ARMS-SSCP: application to HLA genotyping. Nucleic Acids Research. 1992;20(5):1005–1009. doi: 10.1093/nar/20.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonjou C, Thomas L, Borot N, Ledger N, de Toma C, LeLouet H, Graf E, Schumacher M, Hovnanian A, Mockenhaupt M, et al. A marker for Stevens-Johnson syndrome …: ethnicity matters. The Pharmacogenomics Journal. 2006;6(4):265–268. doi: 10.1038/sj.tpj.6500356. [DOI] [PubMed] [Google Scholar]
- 29.Lu XZ, Hong KX, Qin GM, Li CX, Zhu JH, Shao YM. DNA typing of HLA-A locus in Chinese Yi ethnic group by PCR-SSP. Immunological Journal. 2003;19(6):2. (in Chinese) [PubMed] [Google Scholar]
- 30.Lu XZ, Hong KX, Qin GM, Chen JP, Ruan YH, Li CH, Zhu JH, Shao YM. Genotyping of HLA-Cw locus in Chinese Yi ethnic group by PCR-SSP. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2003;17(1):62–65. (in Chinese) [PubMed] [Google Scholar]
- 31.Mack SJ, Tu B, Lazaro A, Yang R, Lancaster AK, Cao K, Ng J, Hurley CK. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish eastern European Americans from the general European American population. Tissue Antigens. 2009;73(1):17–32. doi: 10.1111/j.1399-0039.2008.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Man CB, Kwan P, Baum L, Yu E, Lau KM, Cheng AS, Ng MH. Association between HLA-B*1502 allele and antiepileptic drug-induced cutaneous reactions in Han Chinese. Epilepsia. 2007;48(5):1015–1018. doi: 10.1111/j.1528-1167.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- 33.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Mach B, Mayr WR, et al. Nomenclature for factors of the HLA system, 2002. European Journal of Immunogenetics. 2002;29(6):463–515. doi: 10.1046/j.1365-2370.2002.00359.x. [DOI] [PubMed] [Google Scholar]
- 34.Mayr WR. The detection of HLA antigens in bloodstains. Zeitschrift für Rechtsmedizin Journal of Legal Medicine. 1988;101(4):209–217. doi: 10.1007/BF00200226. [DOI] [PubMed] [Google Scholar]
- 35.Middleton D, Hawkins BR, Williams F, Meenagh A, Moscoso J, Zamora J, Arnaiz-Villena A. HLA class I allele distribution of a Hong Kong Chinese population based on high-resolution PCR-SSOP typing. Tissue Antigens. 2004;63(6):555–561. doi: 10.1111/j.0001-2815.2004.00234.x. [DOI] [PubMed] [Google Scholar]
- 36.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods in Enzymology. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 38.Ogata S, Shi L, Matsushita M, Yu L, Huang XQ, Shi L, Sun H, Ohashi J, Muramatsu M, Tokunaga K, et al. Polymorphisms of human leucocyte antigen genes in Maonan people in China. Tissue Antigens. 2007;69(2):154–160. doi: 10.1111/j.1399-0039.2006.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersdorf EW, Smith AG, Haase AM, Martin PJ, Hansen JA. Polymorphism of HLA-DRw52-associated DRB1 genes as defined by sequence-specific oligonucleotide probe hybridization and sequencing. Tissue Antigens. 1991;38(4):169–177. doi: 10.1111/j.1399-0039.1991.tb01891.x. [DOI] [PubMed] [Google Scholar]
- 40.Pozzi S, Longo A, Ferrara GB. HLA-B locus sequence-based typing. Tissue Antigens. 1999;53(3):275–281. doi: 10.1034/j.1399-0039.1999.530308.x. [DOI] [PubMed] [Google Scholar]
- 41.Saito S, Ota S, Yamada E, Inoko H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens. 2000;56(6):522–529. doi: 10.1034/j.1399-0039.2000.560606.x. [DOI] [PubMed] [Google Scholar]
- 42.Santamaria P, Lindstrom AL, Boyce-Jacino MT, Myster SH, Barbosa JJ, Faras AJ, Rich SS. HLA class I sequence-based typing. Human Immunology. 1993;37(1):39–50. doi: 10.1016/0198-8859(93)90141-M. [DOI] [PubMed] [Google Scholar]
- 43.Scheltinga SA, Johnston-Dow LA, White CB, van der Zwan AW, Bakema JE, Rozemuller EH, van den Tweel JG, Kronick MN, Tilanus MG. A generic sequencing based typing approach for the identification of HLA-A diversity. Human Immunology. 1997;57(2):120–128. doi: 10.1016/S0198-8859(97)00204-8. [DOI] [PubMed] [Google Scholar]
- 44.Shen CM, Zhu BF, Liu ML, Li SB. Genetic polymorphisms at HLA-A, -B, and -DRB1 loci in Han population of Xi’an city in China. Croatian Medical Journal. 2008;49(4):476–482. doi: 10.3325/cmj.2008.4.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi L, Ogata S, Yu JK, Ohashi J, Yu L, Shi L, Sun H, Lin K, Huang XQ, Matsushita M, et al. Distribution of HLA alleles and haplotypes in Jinuo and Wa populations in Southwest China. Human Immunology. 2008;69(1):58–65. doi: 10.1016/j.humimm.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Shi M, Bai R, Wan L, Yu X, Chang L. Population genetics for Y-chromosomal STRs haplotypes of Chinese Tujia ethnic group. Forensic Science International Genetics. 2008;2(4):e65–e68. doi: 10.1016/j.fsigen.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Single RM, Meyer D, Hollenbach JA, Nelson MP, Noble JA, Erlich HA, Thomson G. Haplotype frequency estimation in patient populations: the effect of departures from Hardy-Weinberg proportions and collapsing over a locus in the HLA region. Genetic Epidemiology. 2002;22(2):186–195. doi: 10.1002/gepi.0163. [DOI] [PubMed] [Google Scholar]
- 48.Spínola H, Brehm A, Bettencourt B, Middleton D, Bruges-Armas J. HLA class I and II polymorphisms in Azores show different settlements in Oriental and Central islands. Tissue Antigens. 2005;66(3):217–230. doi: 10.1111/j.1399-0039.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 49.Spínola H, Middleton D, Brehm A. HLA genes in Portugal inferred from sequence-based typing: in the crossroad between Europe and Africa. Tissue Antigens. 2005;66(1):26–36. doi: 10.1111/j.1399-0039.2005.00430.x. [DOI] [PubMed] [Google Scholar]
- 50.Takeshima SN, Matsumoto Y, Aida Y. Short communication: establishment of a new polymerase chain reaction-sequence-based typing method for genotyping cattle major histocompatibility complex class II DRB3. Journal of Dairy Science. 2009;92(6):2965–2970. doi: 10.3168/jds.2008-1999. [DOI] [PubMed] [Google Scholar]
- 51.Tereba A. Tools for analysis of population statistics. Profiles DNA. 1999;2:3. [Google Scholar]
- 52.Trachtenberg E, Vinson M, Hayes E, Hsu YM, Houtchens K, Erlich H, Klitz W, Hsia Y, Hollenbach J. HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China. Tissue Antigens. 2007;70(6):455–463. doi: 10.1111/j.1399-0039.2007.00932.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang HF, Cheng YZ, Wang HP, Chen ZM, Lou JY, Jin J. CD19-positive acute myeloblastic leukemia with trisomy 21 as a sole acquired karyotypic abnormality. Journal of Zhejiang University-SCIENCE B. 2009;10(11):833–838. doi: 10.1631/jzus.B0820362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, Massachusetts, United States: Sinauer Associates Inc; 1996. pp. 204–206. [Google Scholar]
- 55.Wen G, Huang Y, Hao P, Qi Q, Li H, Zhou L, Zhou L, Yu L. Analysis of HLA-DRB1, DQB1 allele polymorphism in the Kunming Yi nationality population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2004;21(5):522–523. (in Chinese) [PubMed] [Google Scholar]
- 56.Wen SH, Lai MJ, Yang KL. Human leukocyte antigen-A, -B, and -DRB1 haplotypes of cord blood units in the Tzu Chi Taiwan Cord Blood Bank. Human Immunology. 2008;69(7):430–436. doi: 10.1016/j.humimm.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 57.Xu MY, Hong KX, Ma J, Deng XL, Li J, Peng H, Ruan YH, Qin GM, Zhang YZ, Xing H, et al. Analysis of HLA-B locus gene polymorphism in Sichuan Yi ethnic group and Xinjiang Uygur ethnic group. Yi Chuan. 2006;28(8):913–917. (in Chinese) [PubMed] [Google Scholar]
- 58.Yang G, Deng YJ, Hu SN, Wu DY, Li SB, Zhu J, Zhu BF, Liu Y. HLA-A, -B, and -DRB1 polymorphism defined by sequence-based typing of the Han population in Northern China. Tissue Antigens. 2006;67(2):146–152. doi: 10.1111/j.1399-0039.2006.00529.x. [DOI] [PubMed] [Google Scholar]
- 59.Yang ZL, Guo Y, Gao L, Wang Y, Dong YL, Tang WR, Li KY, Yan W, Xiao CJ. Hypertension survey in Yi ethnic group in Yunnan Province, China. Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25(9):817. (in Chinese) [PubMed] [Google Scholar]
- 60.Yuliwulandari R, Kashiwase K, Nakajima H, Uddin J, Susmiarsih TP, Sofro AS, Tokunaga K. Polymorphisms of HLA genes in western Javanese (Indonesia): close affinities to Southeast Asian populations. Tissue Antigens. 2009;73(1):46–53. doi: 10.1111/j.1399-0039.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 61.Zetterquist H, Olerup O. Identification of the HLA-DRB1*04, -DRB1*07, and -DRB1*09 alleles by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Human Immunology. 1992;34(1):64–74. doi: 10.1016/0198-8859(92)90086-3. [DOI] [PubMed] [Google Scholar]
- 62.Zhu BF, Shen CM, Wu QJ, Deng YJ. Population data of 15 STR loci of Chinese Yi ethnic minority group. Legal Medicine (Tokyo) 2008;10(4):220–224. doi: 10.1016/j.legalmed.2007.12.004. [DOI] [PubMed] [Google Scholar]