Abstract
Objective: The novel estrogen receptor-α (ER-α) variant ER-α36 is reported to be functional in the estrogen signaling pathway and is related to tamoxifen resistance in breast cancer. However, ER-α36 tends to be a favorable factor for survival in patients without tamoxifen therapy. To investigate the mechanisms behind this paradox, we determined the differences between the transcriptional profiles of ER-α36 and full-length ER-α (ER-α66) in breast cancers and matched normal tissues. Methods: We analyzed ER-α36 and ER-α66 messenger RNA (mRNA) levels in 74 pairs of breast cancers and matched normal tissues using a real-time quantitative polymerase chain reaction (PCR) assay, and correlated the results with their clinicopathological characteristics. Results: Breast cancers expressed lower ER-α36 mRNA levels than matched normal tissues regardless of their ER-α66 expression status. Down-regulation of ER-α36 mRNA was correlated with local progression, lymph node metastasis, and advanced cancer stage. The level of ER-α66 mRNA was lower in ER-α negative breast cancers compared with matched normal tissues. No differences in ER-α66 mRNA levels were observed during cancer progression. Conclusion: Down-regulation of ER-α36 is associated with carcinogenesis and progression of breast cancer.
Keywords: Breast cancer, Estrogen receptor, Estrogen receptor-α36 (ER-α36), ER-α66
1. Introduction
Breast cancer is the most commonly diagnosed cancer type in women and ranks the second in female cancer deaths. About 194 280 new occurrences and 40 610 deaths are expected each year in the United States (Jemal et al., 2009). While surgery, chemotherapy, and radiotherapy have become established modalities for breast cancer treatment, endocrine therapy, such as tamoxifen and aromatase inhibitors, can provide further survival benefits. Among all molecular factors involved in breast cancer management, the expression status of the 66 kDa estrogen receptor-α (ER-α, here termed ER-α66) has been widely accepted as a prognostic marker and a predictor for endocrine therapy response (Dunnwald et al., 2007; Fisher et al., 1988; Goldhirsch et al., 2005). In general, ER-α66 is considered to be the receptor responsible for the proliferative effect of estrogens in breast cancer cells (Ström et al., 2004); breast cancer patients with tumors positive for ER-α66 respond favorably to antiestrogens compared to women with ER-α66 negative disease.
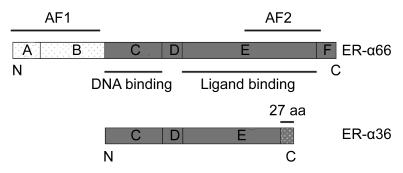
A single ER-α66 was considered to be responsible for all ER-α related estrogen biological actions in breast cancer until the discovery of its splice variants. These variants have been identified in both breast cancer cell lines (Flouriot et al., 2000) and breast cancer tissues (Kumar et al., 2006). The novel 36 kDa variant (here termed ER-α36) which was first identified and cloned in 2005 (Wang et al., 2005), lacks both transcriptional activation function domains (AF-1 and AF-2) but retains the DNA-binding domain, partial dimerization, and ligand-binding domains campared to the full-length ER-α66 (Fig. 1). It was found that ER-α36 may influence ER-α66 mediated transcriptional activity (Wang et al., 2006). In vivo studies of ER-α36 have shown some interesting results. Our earlier studies showed a decreased ER-α36 mRNA level in colorectal cancers compared to their matched normal tissues (Jiang et al., 2008). Not surprisingly, as the breast is one of the main target organs for estrogen, our previous study confirmed ER-α36 expression in breast cancer by immunohistochemical staining (Lee et al., 2008). However, a recent study has demonstrated that women with ER-α66–positive breast cancers that also express high levels of ER-α36 are less likely to benefit from tamoxifen treatment (Shi et al., 2009).
Fig. 1.
Schematic representation of ER-α variants showing the A–F domains, the function of each domain and the last 27 amino acids (aa) of ER-α36
Surprisingly, positive ER-α36 expression in cancer samples seemed to be a favorable factor for disease-free survival (DFS) and disease-specific survival (DSS) in both ER-α66–positive and ER-α66–negative breast cancer patients who received chemotherapy alone (Shi et al., 2009). Whether this phenomenon is related to the involvement of ER-α36 during carcinogenesis and progression of breast cancer and its potential interactions with ER-α66 remained to be fully elucidated. To address this issue, we performed real-time quantitative polymerase chain reaction (PCR) to compare the mRNA levels of ER-α36 and ER-α66 in breast cancers and their matched normal tissues and correlated the findings with the clinicopathological characteristics of these breast cancer patients.
2. Patients and methods
2.1. Patients and breast tissue samples
A total of 74 consecutive female breast cancer patients who underwent surgical procedures in the First Affiliated Hospital of Zhejiang University from 2006 to 2007 were included in this study. None of these patients received chemotherapy or endocrine therapy before surgery. The median age of all included patients was 52 years (range 25–80 years). Breast cancer samples and their matched normal tissues were collected immediately after surgery with informed consent and stored at −80 °C until used. Pathological diagnosis along with immunohistochemical staining [ER, progesterone receptor (PR), human epidermal growth factor receptor 2 (Her-2)] was performed using sections from the same specimens used for RNA extraction. All matched normal tissues were collected from a region distant from the edge of cancers to avoid contamination with cancer cells.
2.2. RNA extractions and cDNA synthesis
Total RNA was extracted from frozen tissues using TRIzol reagent according to the protocol provided by the manufacturer (Invitrogen, Carlsbad, CA, USA). Total RNA was reverse-transcribed into single-strand complementary DNA (cDNA) using moloney-murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI, USA). Briefly, the RNA was denatured by heating for 5 min at 70 °C, followed by rapid cooling on ice, and then used for reverse transcription (2 μg of total RNA, 25 U of RNase inhibitor, 0.5 mmol/L of each dNTP, 1.5 μmol/L reverse primer and 200 U of M-MLV reverse transcriptase in a total volume of 25 μl). For reverse transcription, tubes were incubated at 42 °C for 60 min.
2.3. Real-time quantitative PCR
TaqMan real-time quantitative PCR was performed using the Line-Gene K Sequence Detection System (Bioer Technology, Hangzhou, China). Specific primer pairs and probes are listed in Table 1. To measure the messenger RNA (mRNA) levels of ER-α36, we used primers directly targeting its 3′ untranslated region, so that the ER-α36 product amplified was not from the full-length ER-α66 product. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an endogenous control for normalization. The real-time quantitative PCR reaction was carried out in triplicate for each sample. Briefly, 25 μl of reaction mixture containing 1 μl of cDNA template, 1 μl each of sense and anti-sense primers, 0.75 μl of 5′ FAM- and 3′ TAMARA-labeled oligonucleotide probe, 2 μl of dNTP mixture, 5 μl of 5× reaction buffer, and 0.125 μl of Taq DNA polymerase was amplified as follows: denaturation at 95 °C for 10 min and 40 cycles at 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. Quantitative analysis was performed using the comparative threshold cycle (C
T) method (Livak and Schmittgen, 2001). The expression level was determined as the ratio between ER-α66 or ER-α36 mRNA and the reference GAPDH mRNA to correct for the variation in the amounts of total RNA, which may directly represent the relative levels of the target transcripts. For analysis of relationships to clinicopathological characteristics, the amounts of ER-α66 and ER-α36 in the cancer samples were normalized to their corresponding normal tissues and were expressed as 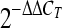 according to the comparative C
T method.
according to the comparative C
T method.
Table 1.
Sequences of primers and probes used to detect ER-α variants
| Primer | Probe | Sequence |
| ER-α36 | Forward | 5′-CCAAGAATGTTCAACCACAACCT-3′ |
| Reverse | 5′-GCACGGTTCATTAACATCTTTCTG-3′ | |
| Probe | 5′ FAM-TATTTATGTTCCAGTCCCACCTGAGTAGCAAAGTGAACAC-TAMARA 3′ | |
| ER-α66 | Forward | 5′-AAGAAAGAACAACATCAGCAGTAAAGTC-3′ |
| Reverse | 5′-GGGCTATGGCTTGGTTAAACAT-3′ | |
| Probe | 5′ FAM-TTTCTTTTCGCCATTGCCTAGCTTGCCGT-TAMARA 3′ | |
| GAPDH | Forward | 5′-CTTAGCACCCCTGGCCAAG-3′ |
| Reverse | 5′-GATGTTCTGGAGAGCCCCG-3′ | |
| Probe | 5′ FAM-CATGCCATCACTGCCACCCAGAAGA-TAMARA 3′ |
2.4. Statistical analysis
All data were analyzed using the SPSS 13.0 software package (SPSS Inc., Chicago, IL, USA). The differences in ER-α mRNA levels between breast cancers and their matched normal tissues were tested using the two-tailed Wilcoxon signed-rank test, whereas associations between ER-α mRNA levels and clinicopathological characteristics in breast cancer were tested using the nonparametric Mann-Whitney test. A P value of <0.05 was considered to be statistically significant.
3. Results
3.1. ER-α36 and ER-α66 mRNA levels in breast cancers and their matched normal tissues
A real-time quantitative PCR assay was developed to quantify the mRNA levels of ER-α36 and ER-α66 in breast cancers and their matched normal tissues. Amplified products of target mRNA were detected in all samples included in the study.
Of the 74 cancer samples, 52 were ER-α positive and 22 were ER-α negative as determined by immunohistochemical staining. Relative ER-α36 and ER-α66 mRNA levels are shown in Table 2. A significantly higher level of ER-α66 mRNA was observed in both cancer samples and normal tissues in the ER-α positive group compared with the ER-α negative group. No such difference was found in the levels of ER-α36 mRNA.
Table 2.
Relative ER-α36 and ER-α66 mRNA levels (−ΔC T) in breast cancers and matched normal tissues
| ER-α36 mRNA* |
ER-α66 mRNA* |
|||||
| ER-α positive | ER-α negative | P value | ER-α positive | ER-α negative | P value | |
| Cancer | −12.53 (3.17) | −12.52 (4.12) | 0.745 | −2.24 (5.08) | −8.00 (5.60) | <0.001 |
| Normal | −11.27 (3.93) | −10.43 (6.10) | 0.394 | −3.01 (3.58) | −4.51 (3.15) | 0.030 |
Values of ER-α36 and ER-α66 mRNA levels [median (quartile interval)] were normalized to GAPDH in the same samples (Mann-Whitney test, P<0.05 taken as indicating a significant difference)
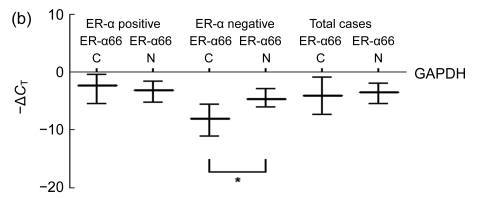
When comparing cancer samples with their matched normal tissues, a lower ER-α36 mRNA level was observed in 39 (75.0%) of the 52 ER-α positive breast cancer samples and in 17 (77.3%) of the 22 ER-α negative breast cancer samples. Quantitative analysis (Fig. 2a) showed that the ER-α36 mRNA levels in both ER-α positive and negative breast cancers were significantly lower than those in their matched normal tissues (P<0.001 and P=0.002, respectively), and when considering all cases together (P<0.001).
Fig. 2.
Median values (represented by horizontal lines in the middle) and quartile intervals (represented by horizontal lines on the top/bottom) of relative ER-α36 and ER-α66 mRNA levels in breast cancers (C) and matched normal tissues (N) after being normalized to GAPDH in the same samples (Wilcoxon signed-ranks test, * P<0.05)
(a) ER-α36 mRNA levels in breast cancers regardless of ER-α status were decreased compared with matched normal tissues; (b) ER-α66 mRNA levels in ER-α negative breast cancers were decreased compared with matched normal tissues
The ER-α66 mRNA levels in ER-α negative breast cancers were significantly lower than those in their matched normal tissues (P=0.001). However, no significant differences between the levels of ER-α66 mRNA in breast cancers and their matched normal tissues were observed in the ER-α positive group or when considering all cases together (Fig. 2b).
3.2. Association of ER-α36 and ER-α66 mRNA levels with clinicopathological characteristics in breast cancers
Associations between ER-α36 and ER-α66 mRNA levels in breast cancer samples and patient age, menopausal status, tumor size, lymph node metastasis, and tumor stage were further analyzed (Table 3). The ER-α36 mRNA level in cancers over 2 cm was significantly lower compared with cancers less than 2 cm (P=0.014). Similarly, cancers with lymph node metastasis showed significantly decreased ER-α36 mRNA levels compared to those without lymph nodes involvement (P=0.023). The ER-α36 mRNA level was also decreased in advanced diseases compared to early stage cancers (P=0.031). However, the ER-α66 mRNA levels in breast cancers did not show any significant correlation with age, menopausal status, tumor size, lymph node metastasis, and tumor stage.
Table 3.
Association of ER-α mRNA levels with clinicopathological characteristics in breast cancers
| Characteristics | N | ER-α36 |
ER-α66 |
||
| mRNA level | P | mRNA level | P | ||
| Age | |||||
| ≥45 years | 61 | 0.39 (0.81) | 0.696 | 0.82 (3.36) | 0.098 |
| <45 years | 13 | 0.34 (1.69) | 0.09 (4.19) | ||
| Menopausal status | |||||
| Postmenopausal | 46 | 0.30 (0.84) | 0.275 | 0.64 (3.37) | 0.439 |
| Premenopausal | 28 | 0.43 (1.20) | 0.58 (2.88) | ||
| Tumor size | |||||
| >2 cm | 37 | 0.20 (0.54) | 0.014 | 0.37 (1.78) | 0.089 |
| ≤2 cm | 37 | 0.46 (1.23) | 0.99 (4.30) | ||
| Lymph node metastasis | |||||
| Positive | 36 | 0.19 (0.52) | 0.023 | 0.50 (3.87) | 0.607 |
| Negative | 38 | 0.49 (1.61) | 0.83 (2.95) | ||
| Tumor stage | |||||
| III+IV | 12 | 0.02 (0.60) | 0.031 | 0.37 (5.01) | 0.747 |
| I+II | 62 | 0.41 (1.03) | 0.76 (3.38) | ||
The values of ER-α36 and ER-α66 mRNA [median (quartile interval)] are the relative levels in cancers versus matched normal samples (Mann-Whitney test, P<0.05 taken as indicating a significant difference)
4. Discussion
Estrogen controls the physiology of the female reproductive system and the mammary glands and is considered to be a mitogen in the genesis and progression of breast cancers. These effects result mainly from the binding of estrogen to its specific receptors, ERs. The human ER-α66, which belongs to the nuclear receptor family of transcription factors, contains six conserved domains, A–F (Ponglikitmongkol et al., 1988) (Fig. 1). Transcriptional activation is mediated by two activation function (AF) domains. The ligand-independent AF-1 is located at the N-terminal of the receptor (domain A/B) and the ligand-dependent AF-2 resides in the C-terminal (domain E/F) (Kumar et al., 1987; Kong et al., 2003). Previous studies have reported the presence of several alternatively spliced ER-α66 mRNAs in both normal and malignant breast tissues (Poola et al., 2000; Gotteland et al., 1995; Pfeffer et al., 1995; Leygue et al., 1996). However, few translated protein products of those variants have been found naturally in breast cancers.
The novel variant ER-α36, in contrast to the full-length ER-α66, lacks both AF-1 and AF-2 but has a unique 27 amino acid domain at the C-terminal (Wang et al., 2005). This structural change may significantly influence the response of ER-α36 to estrogens and antiestrogens compared to ER-α66. Patients with positive ER-α36 expression are less likely to benefit from tamoxifen treatment because of the tamoxifen mediated activation of the mitogen-activated protein kinase (MAPK) pathway in these patients. However, positive ER-α36 expression in cancer samples tends to be a favorable factor of DFS and DSS in breast cancer patients who do not receive tamoxifen therapy (Shi et al., 2009). The underlying mechanisms of this paradox are still unclear.
In the present study, we performed real-time quantitative PCR procedures on samples from 74 paired breast cancers and normal breast tissues to quantify precisely the mRNA levels of ER-α66 and ER-α36. While our results showed that all cancers and normal tissues coexpress both ER-α mRNAs, cancer samples showed a decreased mRNA level of ER-α36 compared with their matched normal tissues regardless of their ER-α status. Moreover, our further analysis of the correlation between the level of ER-α36 mRNA in breast cancer and several clinicopathological characteristics showed a parallel finding. The results showed that relatively large tumors (>2 cm), tumors with lymph nodes metastases, or advanced stage (stage III+IV) tumors had lower ER-α36 mRNA levels than those found in tissues with less advanced disease. These findings suggest that down-regulation of ER-α36 is associated with carcinogenesis and progression of breast cancer, which may partially explain the fact that relatively high ER-α36 expression tends to be a favorable factor for survival in patients without tamoxifen therapy. Based on previous studies, alternatively spliced variants of ER-α66 could regulate the mRNA level of the full-length counterpart present in cells as well as the expression of functional proteins (Erenburg et al., 1997; Fuqua et al., 1992; Garcia Pedrero et al., 2003). However, it was reported that the specific novel variant ER-α36 lacks intrinsic transcriptional activity. Interestingly, when co-transfecting with ER-α66 in ER negative HEK293 cells, ER-α36 can strongly inhibit estrogen-dependent and -independent transactivation activities by AF-1 and AF-2 domains of ER-α66 (Wang et al., 2006). Given that breast cells typically express ER-α66 as confirmed by our study, it is reasonable to speculate that ER-α36 functions in a dominant negative pattern in the breast estrogen transactivation pathway, and may inhibit ER-α66 mediated transcriptional activation of estrogen-responsive genes. Therefore, decreased ER-α36 expression may lead to abnormal transcriptional activation and account in part for carcinogenesis and the progression of breast cancer.
Several studies have shown that the absolute and relative mRNA levels of ER-α66 and its variants change during breast cancer carcinogenesis and progression, but in a pattern dependent on ethnicity (Koduri et al., 2000; Anandappa et al., 2000; Poola and Speirs, 2001). A profile of unchanged ER-α66 mRNA level between breast cancers and matched normal tissues in immunohistochemically ER-α positive cancers and reduced ER-α66 mRNA level in cancer samples compared with matched normal tissues in ER-α negative cancers was reported in African-American women, but not in Caucasian women (Poola et al., 2002). Similar results were found in a study of Taiwanese women: an unchanged ER-α66 mRNA level was observed during breast malignant progression (Hsiao et al., 2006). Our study adds a quantitative transcriptional profile of ER-α66 mRNA for Chinese women, which shows a close resemblance to results from African-American and Taiwanese women. Moreover, our study is the first to show the quantitative profile of the mRNA levels of the novel variant ER-α36 in carcinogenesis and progression of breast cancer.
5. Conclusion
Our study showed that the full-length ER-α66 and novel functional variant ER-α36 mRNAs are coexpressed in breast cancers and normal breast tissues. By applying real-time quantitative PCR techniques, our study revealed the quantitative transcriptional profile of the novel functional variant ER-α36 and showed that its mRNA level is decreased in breast cancers compared with matched normal tissues. Furthermore, down-regulation of the level of ER-α36 mRNA was observed in locally advanced or lymphatic metastases disease. All these findings suggest that ER-α36 may account in part for carcinogenesis and progression of breast cancer. Modulating ER-α36 activity may serve as a potential therapeutic strategy for breast cancers. However, because of the unavailability of commercial ER-α36 antibodies, further conclusions are not possible at this time. Here we provide evidence only of the mRNA levels of ER-α and its novel variant, ER-α36. The influence of stroma cells and lymphocytes should also be taken into consideration. Further methodological improvement, including the application of micro-dissection, may refine the results.
6. Acknowledgement
We thank Dr. Zhao-yi WANG of the Department of Medical Microbiology and Immunology, Creighton University Medical School, USA, for his advice and for critically reading the manuscript.
Footnotes
Project supported by the National Basic Research Program (973) of China (No. 2009CB521704), the National Natural Science Foundation of China (No. 30772510), the Ministry of Health of China (No. WKJ2006-2-008), the Department of Science and Technology of Zhejiang Province (No. 2007C24011), the Natural Science Foundation of Zhejiang Province (No. R206060), China
References
- 1.Anandappa SY, Sibson R, Platt-Higgins A, Winstanley JH, Rudland PS, Barraclough R. Variant estrogen receptor alpha mRNAs in human breast cancer specimens. Int J Cancer. 2000;88(2):209–216. doi: 10.1002/1097-0215(20001015)88:2<209::AID-IJC10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 2.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9(1):R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erenburg I, Schachter B, Mira y Lopez R, Ossowski L. Loss of an estrogen receptor isoform (ER alpha delta 3) in breast cancer and the consequences of its reexpression: interference with estrogen-stimulated properties of malignant transformation. Mol Endocrinol. 1997;11(13):2004–2015. doi: 10.1210/me.11.13.2004. [DOI] [PubMed] [Google Scholar]
- 4.Fisher B, Redmond C, Fisher ER, Caplan R. Relative worth of estrogen or progesterone receptor and pathologic characteristics of differentiation as indicators of prognosis in node negative breast cancer patients: findings from National Surgical Adjuvant Breast and Bowel Project Protocol B-06. J Clin Oncol. 1988;6(7):1076–1087. doi: 10.1200/JCO.1988.6.7.1076. [DOI] [PubMed] [Google Scholar]
- 5.Flouriot G, Brand H, Denger S, Metivier R, Kos M, Reid G, Sonntag-Buck V, Gannon F. Identification of a new isoform of the human estrogen receptor-alpha (hER-α) that is encoded by distinct transcripts and that is able to repress hER-α activation function 1. EMBO J. 2000;19(17):4688–4700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuqua SA, Fitzgerald SD, Allred DC, Elledge RM, Nawaz Z, McDonnell DP, O′Malley BW, Greene GL, McGuire WL. Inhibition of estrogen receptor action by a naturally occurring variant in human breast tumors. Cancer Res. 1992;52(2):483–486. [PubMed] [Google Scholar]
- 7.Garcia Pedrero JM, Zuazua P, Martinez-Campa C, Lazo PS, Ramos S. The naturally occurring variant of estrogen receptor (ER) ERDeltaE7 suppresses estrogen-dependent transcriptional activation by both wildtype ERalpha and ERbeta. Endocrinology. 2003;144(7):2967–2976. doi: 10.1210/en.2002-0027. [DOI] [PubMed] [Google Scholar]
- 8.Goldhirsch A, Glick JH, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol. 2005;16(10):1569–1583. doi: 10.1093/annonc/mdi326. [DOI] [PubMed] [Google Scholar]
- 9.Gotteland M, Desauty G, Delarue JC, Liu L, May E. Human estrogen receptor mRNA variants in both normal and tumor tissues. Mol Cell Endocrinol. 1995;112(1):1–13. doi: 10.1016/0303-7207(95)03576-S. [DOI] [PubMed] [Google Scholar]
- 10.Hsiao WC, Cho WC, Lin PW, Lin SL, Lee WY, Young KC. Quantitative profile of estrogen receptor variants/isoforms in Taiwanese women with breast cancer. Eur J Surg Oncol. 2006;32(5):492–497. doi: 10.1016/j.ejso.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Teng R, Wang Q, Zhang X, Wang H, Wang Z, Cao J, Teng L. Transcriptional analysis of estrogen receptor alpha variant mRNAs in colorectal cancers and their matched normal colorectal tissues. J Steroid Biochem Mol Biol. 2008;112(1-3):20–24. doi: 10.1016/j.jsbmb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Koduri S, Fuqua SA, Poola I. Alterations in the estrogen receptor alpha mRNA in the breast tumors of African American women. J Cancer Res Clin Oncol. 2000;126(5):291–297. doi: 10.1007/s004320050345. [DOI] [PubMed] [Google Scholar]
- 14.Kong EH, Pike AC, Hubbard RE. Structure and mechanism of the oestrogen receptor. Biochem Soc Trans. 2003;31(Pt 1):56–59. doi: 10.1042/bst0310056. [DOI] [PubMed] [Google Scholar]
- 15.Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51(6):941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 16.Kumar VL, Kumar S, Srivastava A, Kumar V. Observations on the presence of E domain variants of estrogen receptor-alpha in the breast tumors. J Surg Oncol. 2006;94(4):332–337. doi: 10.1002/jso.20588. [DOI] [PubMed] [Google Scholar]
- 17.Lee LM, Cao J, Deng H, Chen P, Gatalica Z, Wang ZY. ER-alpha36, a novel variant of ER-alpha, is expressed in ER-positive and -negative human breast carcinomas. Anticancer Res. 2008;28(1B):479–483. [PMC free article] [PubMed] [Google Scholar]
- 18.Leygue E, Huang A, Murphy LC, Watson PH. Prevalence of estrogen receptor variant messenger RNAs in human breast cancer. Cancer Res. 1996;56(19):4324–4327. [PubMed] [Google Scholar]
-
19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
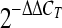 method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - 20.Pfeffer U, Fecarotta E, Vidali G. Coexpression of multiple estrogen receptor variant messenger RNAs in normal and neoplastic breast tissues and MCF-7 cells. Cancer Res. 1995;55(10):2158–2165. [PubMed] [Google Scholar]
- 21.Ponglikitmongkol M, Green S, Chambon P. Genomic organization of the human oestrogen receptor gene. EMBO J. 1988;7(11):3385–3388. doi: 10.1002/j.1460-2075.1988.tb03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poola I, Speirs V. Expression of alternatively spliced estrogen receptor alpha mRNAs is increased in breast cancer tissues. J Steroid Biochem Mol Biol. 2001;78(5):459–469. doi: 10.1016/S0960-0760(01)00118-2. [DOI] [PubMed] [Google Scholar]
- 23.Poola I, Koduri S, Chatra S, Clarke R. Identification of twenty alternatively spliced estrogen receptor alpha mRNAs in breast cancer cell lines and tumors using spliced targeted primer approach. J Steroid Biochem Mol Biol. 2000;72(5):249–258. doi: 10.1016/S0960-0760(00)00033-9. [DOI] [PubMed] [Google Scholar]
- 24.Poola I, Clarke R, DeWitty R, Leffall LD. Functionally active estrogen receptor isoform profiles in the breast tumors of African American women are different from the profiles in breast tumors of Caucasian women. Cancer. 2002;94(3):615–623. doi: 10.1002/cncr.10274. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Dong B, Li Z, Lu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, et al. Expression of ER-{alpha}36, a novel variant of estrogen receptor {alpha}, and resistance to tamoxifen treatment in breast cancer. J Clin Oncol. 2009;27(21):3423–3429. doi: 10.1200/JCO.2008.17.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ström A, Hartman J, Foster JS, et al. Estrogen receptor beta inhibits 17beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci USA. 2004;101(6):1566–1571. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochem Biophys Res Commun. 2005;336(4):1023–1027. doi: 10.1016/j.bbrc.2005.08.226. [DOI] [PubMed] [Google Scholar]
- 28.Wang ZY, Zhang XT, Shen P, et al. A variant of estrogen receptor-alpha, hER-alpha36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc Natl Acad Sci USA. 2006;103(24):9063–9068. doi: 10.1073/pnas.0603339103. [DOI] [PMC free article] [PubMed] [Google Scholar]