Abstract
Members of the bacterial genus Streptococcus are responsible for causing a wide variety of infections in humans. Many Streptococci use quorum-sensing systems to regulate several physiological properties, including the ability to incorporate foreign DNA, tolerate acid, form biofilms, and become virulent. These quorum-sensing systems are primarily made of small soluble signal peptides that are detected by neighboring cells via a histidine kinase/response regulator pair.
Streptococcal infections
Members of the genus Streptococcus (i.e., streptococci) are ubiquitous parasites of humans. Some are part of the indigenous microflora that are involved in opportunistic infections such as dental caries and others are exogenous pathogens that cause infections ranging from mild respiratory or skin diseases to life-threatening conditions such as pneumonia, septic shock, and necrotizing fasciitis. Contemporary research has found that many species of streptococci and other Gram positive bacteria have evolved similar peptide pheromone quorum-sensing systems that probably help them adapt to and survive host-imposed fluctuations in their local environment and coincidently regulate the expression of virulence factors that promote their pathogenicity. This review focuses on streptococcal peptide signaling pathways that are population density–dependent and that impact on vital survival and virulence traits.
Biofilms
Biofilms are dense aggregates of surface-adherent microorganisms embedded in an exopolysaccharide matrix. The study of bacteria residing in biofilms as an interactive community rather than free-living planktonic cells has recently gained a great deal of attention. This has arisen, in part, because of the estimate by the Centers for Disease Control and Prevention that 65% of human bacterial infections involve biofilms. Many species of streptococci are known to form biofilms; however, the relationship between the pathogenic state and the biofilm mode of growth has been most clearly established with the oral streptococci, which are known to initiate dental caries when the bacteria are living in the biofilm environment of dental plaque.
Dental plaque
The human oral cavity is a complex ecosystem that supports an extremely diverse microflora consisting of about 500 species of microorganisms (1). Numerous physical and nutritional interactions between oral bacteria contribute to a complex biofilm community (Figure 1) (2). Streptococci, including Streptococcus mutans, are ubiquitous in the oral microbiota of humans. S. mutans is considered to be a principal etiological agent of dental caries, where it can cause dissolution of tooth enamel by acid end-products resulting from carbohydrate metabolism. The tooth surface is an indispensable natural habitat for S. mutans (3). S. mutans’ dental biofilm tropism most likely reflects its evolution of glucan synthesis and binding functions as well as its relative aciduricity. Its competitiveness for this ecological niche may also relate to cell density–dependent regulation of its acid tolerance response (ATR), natural genetic competence, and bacteriocin activity. Since S. mutans has evolved to depend on a biofilm lifestyle for survival and persistence in the oral cavity combined with its role as an opportunistic pathogen, it has become the best-studied example of a biofilm-forming, disease-causing Streptococcus (4). Biofilm-like populations of pathogenic streptococci may also reach higher densities in confined areas like heart valves, prosthetic devices, sinuses, tonsillar crypts, terminal respiratory passages, and in infectious skin lesions.
Figure 1.
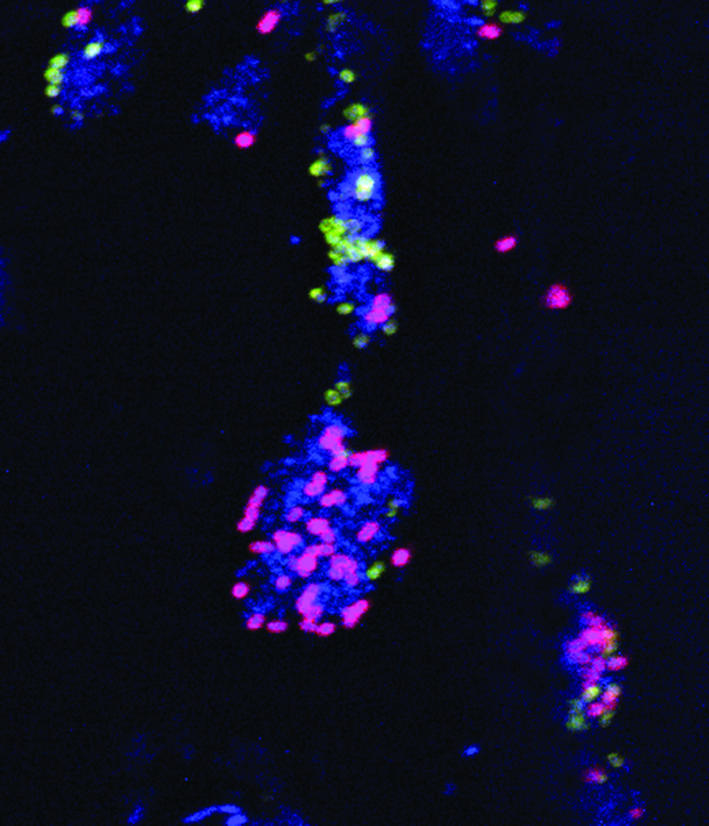
Eight-hour-old dental plaque was visualized using confocal scanning laser microscopy. Antibodies against Streptococcus gordonii DL1 and anti-receptor polysaccharides (RPS), commonly found on Streptococcus oralis, were used for indirect immunofluorescence along with the stain Syto 59. At least two staining types are seen within the aggregates: antibody-reactive (anti-DL1 [green], anti-RPS [purple]) cells found in direct association with antibody-unreactive (blue Syto 59–stained) cells. Single colonies containing all three staining types (anti-DL1, anti-RPS, and antibody unreactive) were frequently seen. Reproduced with permission from The Journal of Bacteriology (2).
Quorum sensing in streptococci
Many bacteria, including streptococci, are known to regulate diverse physiological processes through a mechanism called quorum sensing. In Gram positive bacteria, quorum-sensing systems generally consist of three components, a signal peptide and a two-component regulatory system (also called two-component signal transduction system or TCSTS) that has a membrane-bound histidine kinase sensor and an intracellular response regulator (5). Quorum sensing in Gram positive bacteria has been found to regulate a number of physiological activities, including competence development in Streptococccus gordonii, S. pneumoniae, and S. mutans (6), sporulation in Bacillus subtilus (7), antibiotic biosynthesis in Lactococcus lactis (8), and induction of virulence factors in Staphylococcus aureus (8).
Many of the genes involved in competence induction and the transformation process of streptococci, which is very similar to that described for the mitis group of Streptococcus (including S. pneumoniae), have been identified. Induction of genetic competence in these streptococci is mediated by quorum sensing, which depends on a competence stimulating peptide (CSP) signaling system illustrated in Figure 2.
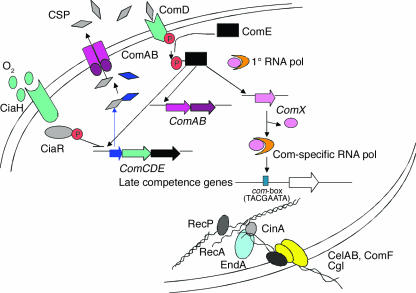
Figure 2.
In Streptococcus pneumoniae, the induction of genetic competence (and potentially virulence) is regulated by a CSP–mediated quorum-sensing system (5, 9). Quorum sensing involves the expression of early gene products encoded by two genetic loci, comAB and comCDE. Genes in the operon, comAB, encode an ATP-binding cassette transporter (ComA) and an accessory protein to ComA (ComB). These secretory proteins are involved in the processing and export of the CSP. The loci, comCDE, respectively encode the precursor to the CSP, a histidine kinase that acts as a CSP receptor, and a response regulator that activates both comAB and comCDE operons. A second two-component regulatory system, CiaH-CiaR, affects the development of competence by negatively regulating comCDE expression. Quorum-sensing signals initiate competence through activity of ComX, a global transcription modulator, which was shown to act as an alternate sigma factor (40). This sigma factor initiates the transcription of competence-specific operons involved in DNA uptake and recombination by recognizing a com-box (also referred to as cin-box) consensus sequence (TACGAATA) in their promoter regions (9, 41, 42). Several of these late competence-specific operons include cilA (ssb2), cilB (dal, like dprA in Haemophilus influenzae), cilC (like comC in Bacillus subtilis), cilD (or cglABCDE), cilE (or celAB), coi, cinA-recA, cfl (like comF in B. subtilis), and dpnA of the DpnII restriction system. pol, polymerase; P, phosphate.
Evidence for the existence of other CSP-modulated pathways
It has been recently recognized that the quorum-sensing signal in S. pneumoniae initiates competence through the activity of a global transcription modulator, ComX, which acts as an alternate sigma factor during the development of genetic competence (9). Transcription of the comX gene is regulated by ComE, the response regulator of the quorum-sensing system (10). The ComX sigma factor then initiates the transcription of competence-specific operons involved in DNA uptake and recombination by recognizing a com-box (also referred to as cin-box) consensus sequence (TACGAATA) in their promoter regions (9). S. mutans and S. pyogenes also have comX genes present in their genomes (9, 11), and inactivation of one copy of the S. mutans comX renders the cell transformation deficient (12). The presence of the conserved com-box consensus sequence in the promoter region of S. mutans late-competence genes such as comFA, celA, and cglA supports the hypothesis that transcriptional regulation of these genes is also mediated by the ComX sigma factor (13). There is much accumulating evidence that many genes not involved in competence are under the control of the CSP-ComX system. This includes the observations that many “other” S. pneumoniae genes contain the com-box sequence and are activated by CSP (14, 15). These genes likely encode products that aid in the cell’s adaptation to a high cell density, and it is likely that they (or their homologs in other streptococci) contribute to the biofilm phenotype.
LuxS quorum sensing
Recently, the luxS gene has been identified in streptococci (16, 17). The LuxS protein is required for the biosynthesis of the type 2 autoinducer, AI-2, which is involved in quorum sensing in a wide range of bacterial species. Mutation of luxS in S. mutans caused a defect in biofilm formation, while disruption of this gene in S. pneumoniae resulted in reduced virulence in mouse infections. It has been suggested that the AI-2 pathway is a very good target for chemotherapeutic control of bacterial virulence.
Quorum sensing in S. mutans biofilms
Horizontal gene transfer through genetic transformation has been observed in many natural ecosystems, and recent studies suggest that growth of bacteria in biofilms may facilitate horizontal gene transfer among bacterial species via either transformation or conjugation (18, 19). Natural genetic transformation has been extensively studied in streptococci, but these studies relied exclusively on bacteria grown in fluid cultures, where they would become transiently competent after reaching a critical cell density. Since biofilms are more representative of bacterial growth in natural environments and S. mutans is an organism that relies on a biofilm lifestyle, we set forth to investigate the ability of this bacterium to transport and integrate exogenous DNA when living in its biofilm state. To facilitate assays for genetic transformation of biofilm-grown cells, we utilized a chemostat-based continuous flow biofilm system, which allows observation of physiological activities of a bacterial population under controlled growth conditions (20). Using this system, we have demonstrated that growth rates, culture pH, and biofilm age are several important factors that influence competence development of S. mutans growing in biofilms.
The most fascinating finding using this system was that S. mutans cells growing in biofilms were able to incorporate foreign DNA much more efficiently than their free-living counterparts. The transformation frequencies of biofilm-grown cells of S. mutans strains tested were about 10- to 600-fold higher than those of the planktonic cells (20). To our best knowledge, this is the first report to provide direct evidence that biofilm-grown bacteria can be efficiently induced to become genetically competent for transformation. In addition, S. mutans grown in a biofilm appeared to maintain a subpopulation of cells that were constantly competent for taking up DNA from the environment. This static state of competence is in contrast to that observed in fluid where most streptococci, including S. mutans, enter a transient physiological state that usually lasts for only 15–30 minutes during their growth cycle. The evidence from our study and another recent study that demonstrated genetic exchange in S. pneumoniae biofilms (21) clearly suggests that biofilm-growth mode in transformable streptococci appears to favor the induction and maintenance of genetic competence. It also appears that the biofilm environment provides conditions for optimal function of streptococcal cell-cell peptide signaling systems to activate genetic competence and facilitate genetic exchange.
Quorum sensing in biofilm formation
The discovery that the S. mutans quorum-sensing system functions optimally in growing biofilms led us to investigate other roles of this system in biofilm formation and biofilm physiology. Dental plaque is a complex biofilm community that harbors the most diverse resident microflora associated with humans. Bacteria in dental biofilms, including S. mutans, are frequently exposed to various stresses, such as extreme nutrient shortage or excess, low pH, high osmolarity, oxidation, and consumption of antimicrobial agents or antibiotics by the host. Formation of a biofilm is considered an important mechanism used by a bacterium for adaptation to this environment (22). Although adaptation to environmental stress by genetic transformation is believed to occur very infrequently, such a rare event can be highly significant if the transforming DNA, such as an antibiotic resistance gene or a virulence factor, provides a selective advantage to the recipient cells (23). In addition to providing the community with an abundant extracellular gene pool, the biofilm environment facilitates the bacteria with a localized neighborhood where cell-cell signaling mechanisms likely abound.
Recent studies using genetic dissection of biofilm development have revealed that the formation of biofilms involves multiple, convergent signaling pathways and a genetic program for the transition from planktonic growth state to the biofilm mode of growth (24). In Gram negative bacteria such as Pseudomonas aeruginosa, cell-cell signaling through quorum sensing has been found to play an important role in biofilm differentiation (25). The first evidence to suggest that quorum-sensing systems might influence the structure of Gram positive biofilms came from a recent study of S. gordonii where a biofilm-defective mutant was found to have a transposon insertion in the comD gene encoding the histidine kinase sensor protein of the TCSTS required for genetic competence (26). This implied that biofilm formation by S. gordonii involved cell-cell communication through quorum sensing. To test if the S. mutans ComCDE quorum-sensing system was involved in biofilm formation, we examined the ability of S. mutans mutants defective in various components of the system to form biofilms. We found that inactivation of any one of the genes encoding the components of the quorum-sensing signaling system results in the formation of an abnormal biofilm (Figure 3) (12). Particularly, the comC mutant (unable to produce or secrete the signal peptide) formed a biofilm that lacked the wild-type architecture, whereas the comD and comE mutants defective in sensing and responding to the signal peptide formed biofilms with a reduced biomass. The architectural change in the comC mutant biofilms may be associated with a defect in cell separation with mutations in this gene resulting in the formation of large aggregates or “weblike” biofilms that were easily removed from the surface. The observation that the mutants unable to produce or secrete the CSP formed biofilms that differed from those formed by the mutant defective in the comD or comE genes suggested that S. mutans has multiple CSP receptors. Although we have clearly demonstrated that the ComCDE quorum-sensing system is directly connected to the ability of S. mutans to form biofilms, the molecular and biochemical mechanisms involved in expression of the “wild-type biofilm phenotype” remain to be investigated.
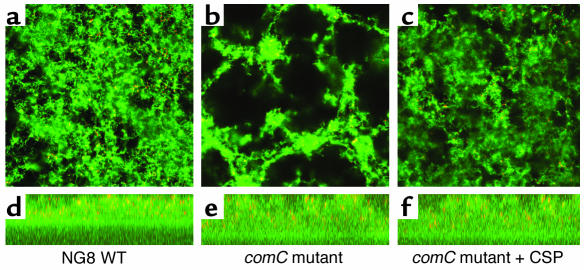
Figure 3.
Scanning confocal laser microscope images of 16 h S. mutans biofilms. Panels a–c show the X-Y planes (top view), while the panels d–f show the X-Z planes (side view). The panels illustrate (a, d) a normal biofilm formed by the parent strain NG8 and (b, e) an aberrant biofilm formed by the comC– mutant defective in making a CSP. The wild-type appearance of the biofilm is restored by addition of synthetic CSP (c, f). Reproduced with permission from The Journal of Bacteriology (12).
CSP modulation of the ATR
The pH levels in dental biofilms are highly variable and frequently shift from above pH 7.0 in the resting pH state to as low as pH 3.0 during the ingestion of dietary carbohydrates by the host. Thus, pH exerts a significant ecological pressure on S. mutans, and its ability to tolerate and grow in low pH environments is crucial to its survival and eventual dominance in dental plaque, leading to caries (27). Considerable evidence has shown that S. mutans has evolved a number of sophisticated mechanisms to survive these pH changes including induction of an ATR in which exposure of S. mutans cells to a mild or moderately acidic pH (5.0–6.0) results in enhanced survival of a significant proportion of the cell population in a lower pH of 3.0–3.5 (28). This ATR involves a number of de novo proteins that appear to be important for adaptation to an acidic environment (29). Although many of the molecular mechanisms of the ATR in S. mutans remain unclear, this “signal pH” that results in synthesis of protective proteins appears to be important for induction of the ATR.
It is widely accepted that bacteria living in biofilms are more resistant to mechanical, physical, and chemical stresses. Since S. mutans normally resides in high cell density biofilms, the ability to withstand acid in this physiological state is likely an important adaptive response. We therefore addressed the question of whether acid adaptation involved cell density–dependent events or cell-cell signaling in biofilms. Changes in external pH can significantly influence many physiological parameters, such as energy coupling, ion transport, proton movement, and export of metabolic products, thereby triggering numerous secondary signals. During growth at pH 5.0, Escherichia coli can signal stress tolerance to other unadapted cells by secreting a proteinlike molecule, termed extracellular induction component (EIC) (30). Although the signal molecule remains unidentified, induction of stress (including acid) adaptation in E. coli presumably involves cell-cell communication. Since S. mutans normally encounters acid while living in dense biofilm communities, we proposed that quorum sensing at high cell densities of S. mutans might facilitate its survival against low pH challenges. Testing this hypothesis has led us to find that the ATR interfaces with the density-dependent signaling pathway that also initiates genetic competence. We have demonstrated that mutants defective in the comC, D, or E genes have a diminished log-phase ATR and the neutralized culture filtrates prepared from acid-adapted wild-type cells also induce a partial log-phase ATR in cells that have never encountered the signal pH (31). S. mutans grown at high cell density established adaptation to the signal pH more rapidly than cells grown at low density. Similarly, S. mutans cells grown in a high cell-density biofilm were more resistant to the killing pH than planktonic-phase cells. In fact, S. mutans cells grown in biofilms not only survived better than the planktonic cells but also were capable of growth at the lower pH following a glucose pulse. Based on the evidence obtained from this work, we propose that S. mutans, upon exposure to low pH in a growing culture, releases extracellular signal molecules, one of which is the CSP, to enhance induction of acid adaptation in the population. It is likely that optimal induction of acid adaptation in a population of S. mutans requires a coordinated activity through mechanisms involving both low pH induction and cell density–dependent intercellular signals.
Biofilms likely provide bacterial cells with a unique environment to fully express their adaptive survival mechanisms. Because of three-dimensional structures, high cell density, and diffusion barriers, bacterial cells at different locations within a biofilm may not sense the same degree of extracellular stress simultaneously. The cells that first sense a pH stress may rapidly process the information and pass their “secondary signal” to the other members of the population through cell-cell signaling systems to initiate a coordinated protective response against potentially lethal forces, like acid. Unlike some planktonic cells that need to reach a critical concentration of signal molecules and cell density, biofilms can allow signal molecules to accumulate rapidly in the local environment to initiate coordinated activities far more quickly (25). In addition, physiological states of bacterial cells living in a biofilm, in terms of growth rate, growth phase, or metabolic activities, are heterogeneous; this allows the cells to respond to stress in different ways. Apparently, biofilm populations have several advantages over their free-living counterparts since the cells have more time, a sufficient concentration of signal molecules, and high population density to adapt to stress relative to planktonic cells. The high cell density biofilms may provide a unique environment for induction of acid adaptation via quorum sensing in S. mutans. It is likely that the S. mutans quorum-sensing signaling pathway is significant for the ATR to intersect with the regulatory networks initiating genetic competence as well as a switch to the “biofilm phenotype.”
The ComC signal peptide may activate more than one signal transduction pathway
Our previous study revealed that an S. mutans comC mutant unable to produce CSP formed a biofilm that differed from that formed by the mutant defective in the comD or comE genes suggested that there might be a second receptor that also responded to the CSP but activated a different pathway in order to invoke the phenotype (12, 20). Based on the available information, we have proposed a “two-receptor” cell-cell signaling model to illustrate how the quorum-sensing system in S. mutans functions to regulate genetic competence, biofilm formation, and the ATR (12). The principle of this model is that the signal peptide (CSP) encoded by comC can simultaneously interact with two cognate receptors, one encoded by comD and another encoded by an unknown gene. These receptors likely transfer the input signal through two different pathways. Although the genes encoding the components involved in the second transduction pathway remain unknown, the work in our lab has recently characterized another TCSTS, named HK/RR11, which is also demonstrated to involve biofilm formation and acid resistance in S. mutans (32).
One of the defects observed in HK/RR11 mutant biofilms was the development of a spongelike architecture composed of cells in very long chains, a feature that we previously observed with the biofilm formed by the comC mutant unable to produce CSP. Since there is no putative substrate, signal or function assigned for the HK/RR11 transduction system, we suspected that the TCSTS encoded by hk/rr11 may activate a second pathway to respond to the CSP. To test this hypothesis, we added CSP to the HK/RR11 mutant cultures to assess the effect on chain formation by the hk11 and rr11 mutant biofilm cells. The results revealed that addition of CSP to the mutant cultures had no observable impact on the length of cell chains comprising the mutant biofilms. This result was consistent with HK11 acting as a CSP receptor but provided no direct evidence to conclusively assign a role to HK11 as a CSP receptor. A study for characterizing the relationship between the comC-encoded signal peptide and the HK/RR11 signal transduction pathway is now underway.
Evaluation of CSP analogs to act as inhibitors of biofilm formation
Analogs of quorum-sensing peptides can competitively inhibit the activity of the peptide-mediated phenotype. It has been demonstrated that analogous signal peptides from Staphylococcus epidermidis can inhibit the agr signaling system of Staphylococcus aureus, thereby modulating S. aureus virulence (33). Since peptide signaling–sensing systems are very similar among Gram positive bacteria (including the S. mutans CSP system) analogs of the S. mutans CSP may be able to interfere with the quorum-sensing process, leading to inhibition of induction of the “biofilm phenotype.”
Mucosal pathogens
Quorum sensing among mucosal pathogens may not require high cell density. In contrast to the very dense dental plaques colonized by S. mutans and several other oral streptococci, pathogenic streptococci that infect mucosal tissues colonize environments where bacterial microcolonies are usually less dense due to bathing effects of secretions and desquamation of epithelium. Yet, several of these streptococcal and other Gram positive species have evolved peptide pheromone signaling pathways that have autoregulating functions analogous to the quorum-sensing systems that we described above for S. mutans. The finding that initiation of genetic competence in S. pneumoniae is sensitive to relatively low levels of CSP at about 107 bacteria/ml (34) suggests that pathogenic streptococci of mucosal surfaces may respond to gradients of cell densities that occur frequently during natural infections, raising the question of whether quorum sensing affects pathways by which pathogenic streptococci grow to predominance over normally protective indigenous species. It has been recently established that mutants of S. pneumoniae defective in the quorum-sensing competence induction pathway have diminished virulence in a murine model relative to the parent strain (35). Differential fluorescence induction also showed that several Com genes were induced during infection in mice (36). Clearly, the role of quorum sensing during infection by S. pneumoniae warrants further examination.
Bacteriocins
Bacteriocins are antimicrobial peptides that are generated by some bacteria and target others that are sensitive. The signaling networks that regulate bacteriocin production and transport as well as immunity to bacteriocins involve peptide pheromone sensing pathways that are very similar to those involved in genetic competence (8). Operons encoding bacteriocin peptide precursors, cognate peptide processing transporters, and two-component regulatory sensors and response regulators are known to be under the control of cell density. For example, the virulence-related blp and com regulons of S. pneumoniae have many analogous features including highly similar signal peptides and peptide secretion systems (15). Moreover, activation of the competence and bacteriocin peptide pheromone response regulators may, in turn, activate downstream pathways that intersect in a common regulatory gene. One such example is the previously mentioned comX, a transcriptional regulator of so-called “late” competence genes (9). The genome of M1 S. pyogenes strain SF370 contains comX and a complete bacteriocin salA1 locus; yet, it lacks the genes comABC that encode the CSP and its secretion apparatus (11). This implies that S. pyogenes comX (and downstream pathways) may be activated in response to environmental signals other than CSPs. S. pyogenes is a pathogen with a host range limited to humans and a unique set of virulence properties not shared by other streptococci. It is possible that it has evolved a sophisticated bacteriocin peptide regulatory system to help its population competitively emerge on mucosal surfaces, yet protect its genome from contamination with foreign DNA by deleting (or never evolving) genes for the cell density–dependent CSP. Recently, Jenkinson’s lab reported that the sensory and immunity pathways for the pheromone antibiotics SalA1 of S. pyogenes and SalA of the indigenous oral species Streptococcus salivarius are conserved, closely related, and cross-sensitive (37). Such novel findings suggest that determinants of population dynamics on bathed mucosal surfaces probably encompass subtle combinations of complex environmental sensing systems that are not limited to cell density, bacteriocin production, and competence stimulation. It is also possible that S. pyogenes utilizes a bacteriocin-like hemolysin as a quorum-sensing molecule. Recently, Enterococcus faecalis was shown to regulate gene expression by such a mechanism involving its cytolysin, a molecule with both antimicrobial and hemolytic activity (38). S. pyogenes has a hemolysin that is genetically very similar to quorum-sensing autoinduction operons found in streptococci (39).
Fluctuations in total bacterial burden and population density also determine the pathogenicity of bacterial microcolonies and biofilms. For example, the ability to suppress competing streptococci through bacteriocin activity may be considered a virulence property if it allows the pathogenic species to emerge sufficiently to damage the host. Evidence is growing that cell density–dependent gene regulation also affects the expression of many other virulence-related proteins of pathogenic streptococci. For example, virulence factor expression and regulation are, in part, determined by growth phase and activation of two-component regulatory sensors of environmental signals, which are characteristics of quorum sensing in Gram positive bacteria (38). Thus, the extracellular concentration of autoregulating peptides appears to serve as one of several environmental conditions that regulate virulence genes through signal transduction pathways that are initiated via two-component regulatory systems. In S. pyogenes, in addition to sagA, there appear to be several coordinated regulatory loci such as mga, csrRS, fasBCA, and rgg that affect the expression of genes for its numerous and diverse virulence factors.
Such a pattern of global regulation is reminiscent of the induction of virulence genes via quorum-sensing pathways in biofilms of Pseudomonas aeruginosa. Therefore, it is likely that the genomes of streptococci have evolved density-dependent regulons to control expression of downstream genes that affect bacterial survival in response to changing environmental conditions on mucosal and tooth surfaces, and the selective survival of the pathogenic streptococcal species leads to clinical infection. Although the key molecules of the quorum-sensing pathways of pathogenic streptococci such as S. pneumoniae, S. pyogenes, and S. mutans are distinct from those first reported for Gram negative bacteria, they seem to serve an analogous function, to modulate physiologic homeostasis and adaptation to environmental conditions in response to fluctuations in population density. The use of this information to exploit these pathways to control streptococcal infections is now being implemented and in the near future we may see a more selective and targeted approach to controlling persistent biofilm-dwelling bacteria.
Acknowledgments
Special thanks to Paul Kolenbrander and Rob Palmer from the National Institute of Dental and Craniofacial Research (NIDCR) for providing Figure 1. Work conducted by the authors was generously funded by the Canadian Institutes of Health Research, the NIDCR, and the Canada Foundation for Innovation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: acid tolerance response (ATR); two-component signal transduction system (TCSTS); competence-stimulating peptide (CSP); extracellular induction component (EIC).
References
- 1.Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. U. S. A. 1999; 96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer RJ, Jr, et al. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 2003; 185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlsson J, Soderholm G, Almfeldt I. Prevalence of Streptococcus sanguis and Streptococcus mutans in the mouth of persons wearing full-dentures. Arch. Oral Biol. 1969; 14:243–249. doi: 10.1016/0003-9969(69)90226-x. [DOI] [PubMed] [Google Scholar]
- 4.Burne RA. Oral streptococci . . . products of their environment. J. Dent. Res. 1998; 77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 5.Kleerebezem M, et al. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997; 24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 6.Cvitkovitch DG. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 2001; 12:217–243. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- 7.Lazazzera BA. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 2000; 3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 8.Kleerebezem M, Quadri LE. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides. 2001; 22:1579–1596. doi: 10.1016/s0196-9781(01)00493-4. [DOI] [PubMed] [Google Scholar]
- 9.Lee MS, Morrison DA. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 1999; 181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ween O, Gaustad O, Havarstein LS. Identification of DNA binding sites for ComE, a key regulator of natural competence in. Streptococcus pneumoniae. Mol. Microbiol. 1999; 33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 11.Ferretti JJ, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 2001; 98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YH, et al. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 2002; 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, J.H., et al. 2001. Genetic transformation in Streptococcus mutans: identification of competence genes by functional genomic analysis. Abstracts of the 101st Annual General Meeting of the American Society for Microbiology. Orlando, Florida, USA. (Abstr. I-76)
- 14.Peterson S, et al. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 2000; 182:6192–6202. doi: 10.1128/jb.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Saizieu A, et al. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 2000; 182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merritt J, et al. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 2003; 71:1972–1979. doi: 10.1128/IAI.71.4.1972-1979.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stroeher UH, et al. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 2003; 71:3206–3212. doi: 10.1128/IAI.71.6.3206-3212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen BB, et al. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 1998; 64:2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 1999; 65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YH, et al. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 2001; 183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waite RD, Struthers JK, Dowson CG. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variation. Mol. Microbiol. 2001; 45:1223–1232. doi: 10.1046/j.1365-2958.2001.02674.x. [DOI] [PubMed] [Google Scholar]
- 22.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002; 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowson CG, et al. Horizontal gene transfer and the evolution of resistance and virulence determinants in. Streptococcus. Soc. Appl. Bacteriol. Symp. Ser. 1997; 26:42S–51S. [PubMed] [Google Scholar]
- 24.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998; 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 25.Parsek MR, Greenberg EP. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. U. S. A. 2000; 97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo CY, Corliss DA, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 2000; 182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuramitsu HK. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 1993; 4:159–176. doi: 10.1177/10454411930040020201. [DOI] [PubMed] [Google Scholar]
- 28.Svensater G, et al. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 1997; 12:266–273. doi: 10.1111/j.1399-302x.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 29.Wilkins JC, Homer KA, Beighton D. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 2002; 68:2382–2390. doi: 10.1128/AEM.68.5.2382-2390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowbury RJ, Goodson M. Extracellular sensing and signalling pheromones switch-on thermotolerance and other stress responses in. Escherichia coli. Sci. Prog. 2001; 84:205–233. doi: 10.3184/003685001783238970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YH, et al. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 2001; 183:6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YH, et al. A novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 2002; 184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otto M, et al. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999; 450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 34.Steinmoen H, Knutsen E, Havarstein LS. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. U. S. A. 2002; 99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau GW, et al. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 2001; 40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 36.Bartilson M, et al. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 2001; 39:126–135. doi: 10.1046/j.1365-2958.2001.02218.x. [DOI] [PubMed] [Google Scholar]
- 37.Upton M, et al. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 antibiotic peptides. J. Bacteriol. 2001; 183:3931–3938. doi: 10.1128/JB.183.13.3931-3938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002; 415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 39.Nizet V, et al. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 2000; 68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo P, Morrison DA. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus mutans. J. Bacteriol. 2003; 185:349–358. doi: 10.1128/JB.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova EV, Morrison DA. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 1998; 180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell EA, Choi SY, Masure HR. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 1998; 27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]