Abstract
The fine-scale structure of the majority of copy number variation (CNV) regions remains unknown. The killer immunoglobulin receptor (KIR) gene complex exhibits significant CNV. The evolutionary plasticity of the KIRs and their broad biomedical relevance makes it important to understand how these immune receptors evolve. In this paper, we describe haplotype re-arrangement creating novel loci at the KIR complex. We completely sequenced, after fosmid cloning, two rare contracted haplotypes. Evidence of frequent hybrid KIR genes in samples from many populations suggested that re-arrangements may be frequent and selectively advantageous. We propose mechanisms for formation of novel hybrid KIR genes, facilitated by protrusive non-B DNA structures at transposon recombination sites. The heightened propensity to generate novel hybrid KIR receptors may provide a proactive evolutionary measure, to militate against pathogen evasion or subversion. We propose that CNV in KIR is an evolutionary strategy, which KIR typing for disease association must take into account.
INTRODUCTION
Genome structural variants, such as copy number variation (CNV), are a significant component of human genetic variation and important genetic determinants of phenotypic variation. A CNV has been defined as a segment of DNA >1 kb and present at variable copy number (1). To date, >20 000 such variants have been identified in the human genome (2).
CNVs frequently involve genes that influence our response to environmental stimuli, including immune response (3). Consequently, there is considerable potential for CNVs to play a significant role in susceptibility to infection and they could, to some extent, explain the variable penetrance of inherited complex polygenic disorders such as autoimmunity. This premise is supported by recent correlations of CNVs with inter-individual variation in immune defence and disease resistance/susceptibility among humans and simians (4–6).
In spite of the biomedical relevance of CNVs, the fine-scale structure of the majority of CNV regions remains unknown with only a fraction resolved at the sequence level. CNV regions involving short-range segmental duplications of DNA with near-identical sequence (niCNV), often representing multi-allelic and highly polymorphic systems, have proven particularly difficult to characterize and present a challenge to high-throughput analysis. Importantly, a significant proportion of CNV regions exhibit higher complexity in possessing hybrid genes, smaller internal CNVs or different breakpoint sites between individuals (7). Current CNV discovery methods (e.g. microarray-based comparative genomic hybridization and fosmid paired-end sequence comparison) provide limited resolution of the underlying genetic organization of such regions.
Since the power to detect a correlation between CNV and phenotype is dependent on accurate determination of the true allelic state of each CNV in each individual, prospective disease association studies must consider how to optimally assess this uncharacterized complexity. To this end, it is important to know whether CNV sites are generally uniquely formed by distinct mechanisms or are commonly created by the same mechanism from independent events. The former can allow such variants to be indirectly analysed by tagging markers without the need for direct identification. Such assessment will influence the design of future platforms for genome-wide association studies.
Mechanistic processes involved in CNV formations, such as non-allelic homologous recombination (NAHR), are being defined (8), but recombination processes in segmental duplications have been less studied. For example, it is unclear to what extent non-homology-based mutational mechanisms operate in these regions (9). Insights into recombination processes can be obtained by studying patterns of genetic variation. However, genotyping of SNPs in segmental duplications is problematic because of the high sequence similarity between segments. Consequently, reliable population genetic data in segmental duplications are presently limited.
The killer immunoglobulin-like receptor (KIR) complex on chromosome 19 is an immune gene family exhibiting substantial segmental or niCNVs. The complex offers a unique opportunity to gain insight into the processes operating in multicopy gene families and segmental CNV regions because over the past 15 years KIRs have been extensively studied in terms of gene structures and haplotype content due to increasing awareness of their broad medical relevance (http://www.ebi.ac.uk/ipd/kir/). We are using a focused sequencing-based approach to study the patterns of variation and the underlying recombination processes in the KIR complex to better understand its relationship with disease and evolutionary history (http://www.sanger.ac.uk/HGP/Chr19/LRC/).
KIR molecules operate in both adaptive and innate immunity by modulating the development and activity of natural killer (NK) and T cell subsets through differential interaction with specific major histocompatibility complex (MHC) class I molecules on target cells. KIR engagement either leads to an activation or inhibition signal to the effector cell depending on the particular structure of the KIR, in doing so regulating cytotoxic activity and cytokine secretion. More than 30 different KIR haplotypes exist based solely on gene content (10). Beyond haplotype diversity, the KIR show significant allelic content with over 50 different alleles described for some genes (http://www.ebi.ac.uk/ipd/kir/). The combined allelic and polygenic diversity produces an extreme degree of heterogeneity among individuals (11,12). Such a high level of diversity probably reflects strong pressure from pathogens on the human NK/T cell immune response, which may account for the evidence of balancing selection (13). Epistatic interactions between KIR and MHC class I strongly influence pathogenesis of some human infections such as HIV/AIDS, cancers, autoimmune diseases, pregnancy disorders, as well as outcome of haematopoietic stem-cell transplantation (14).
The KIR receptor family differs markedly among species and even between primates (15) suggesting that KIR haplotypes evolve rapidly in ways that cannot be accounted for solely by divergence in MHC class I molecules. The evolutionary plasticity of the KIRs and their biomedical relevance makes it important to understand the dynamics of how these receptors evolve.
KIRs are categorized according to whether they have two or three extracellular immunoglobulibin-like domains (2D or 3D, respectively), and whether they posses a short (S) or long (L) cytoplasmic domain. The long-tailed KIRs carry one or two immunoglobulin immunoreceptor tyrosine-based inhibitory motifs (ITIM) that can induce inhibitory signals to the cell. Conversely, the short-tailed KIRs lack ITIMs and possess a charged residue in the transmembrane (TM) region that mediates an association with DAP12, which contains an immunoreceptor tyrosine-based activating motif (ITAM) that can induce cell activation.
The human KIR gene family currently consists of 15 genes (KIR2DL1–4, KIR2DL5A, KIR2DL5B, KIR2DS1–5, KIR3DL1–3 and KIR3DS1) and two pseudogenes (KIR2DP1 and KIR3DP1) encoded within a ∼150 kb region. Each gene spans between 10–16 kb. In general, KIR haplotypes contain between 7 and 15 genes (12). The KIR genes themselves display a high level of sequence similarity, generally being 80–90% identical, and allelic variants of a single KIR gene tend to differ by 2% or less. They are tightly arranged with ∼2 kb of sequence separating each gene. Contributing to the organization of the region is the presence of the framework genes (e.g. KIR3DL3, KIR3DP1, KIR2DL4, KIR3DL1/S1 and KIR3DL2) that feature in their distinctive positions on nearly all haplotypes. Only three human KIR haplotypes have been fully sequenced (16,17).
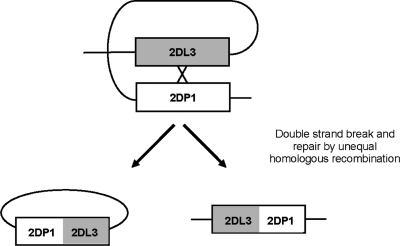
We have previously proposed that the arrangement of KIR genes in close head-to-tail orientation and their high sequence similarity facilitates gene gain and loss by unidirectional alignment and sequential meiotic NAHR (18). Consistent with this, we have identified, by segregation analysis, unusual KIR haplotypes possessing aberrant gene content in families of European origin. These rare KIR haplotypes can help expound the rapid evolution and the genomic and physiological regulation of this gene family. We completely sequenced two unusual, extremely contracted KIR haplotypes using a fosmid cloning strategy. The data provide insight into CNV and hybrid gene formation, as well as special mutational processes that shape the KIR complex.
RESULTS
Identification of a minimal KIR haplotype
Segregation analysis of KIR genes and alleles in a panel of CEPH families from Utah identified an unusual arrangement that did not conform to the general characteristics of KIR haplotypes (19,20). The unusual haplotype was detected at a frequency of 0.6%, in these samples, and appeared to contain only three KIR genes. Our working hypothesis was that the haplotype was created by an NAHR resulting in contraction of the KIR gene complex.
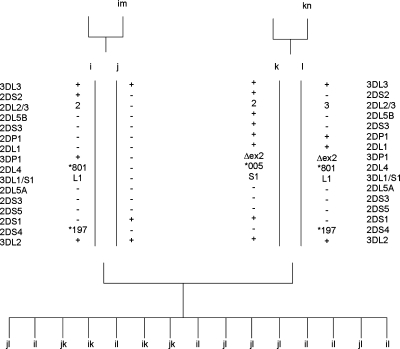
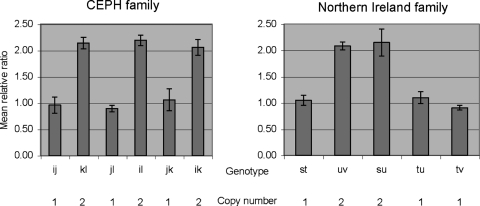
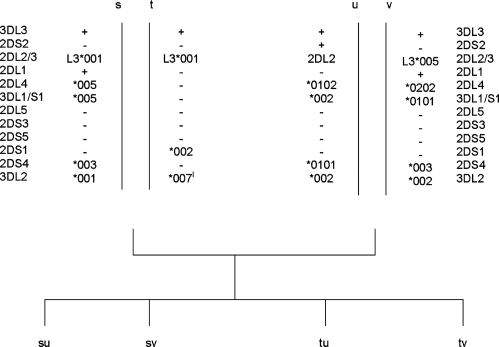
The contracted haplotype was identified in a three-generation CEPH family from Utah. Segregation analysis of KIR genes/alleles in the family indicated that only three KIR genes, KIR3DL3, KIR2DS1 and KIR3DL2, segregate on one haplotype (designated j) and the framework KIR genes KIR3DP1, KIR2DL4 and KIR3DL1/S1 were absent (Fig. 1). Other haplotypes i, k, l, m and n, segregating in the family, had gene compositions consistent with known KIR haplotypes. Using real-time PCR to measure gene dosage, we confirmed that individuals with the j haplotype had only one copy of KIR2DL4, and those without the j haplotype had two copies of this gene (Fig. 2).
Figure 1.
Segregation of contracted haplotypes in a CEPH family. KIR haplotypes were determined by segregation analysis in all members of a three-generation family. Allele designations correspond to HGNC nomenclature (http://www.genenames.org/genefamily/kir.php).
Figure 2.
Gene copy number determination of KIR2DL4 at the genomic level using quantitative real-time multiplex PCR. The results of duplicate experiments are expressed as the mean relative ratio of KIR2DL4 to a reference gene with an SD. Individuals carrying the j or t haplotype possess one copy of KIR2DL4.
The minimal KIR haplotype possesses four KIR genes including two hybrid genes
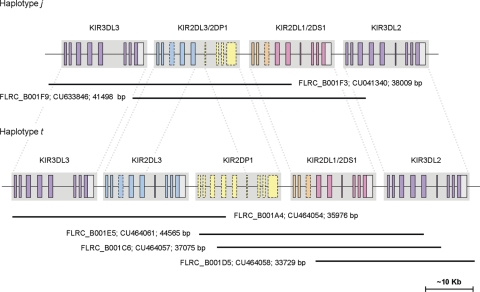
The j haplotype was sequenced after cloning in fosmids to determine the precise gene composition (see Materials and Methods). The sequence confirmed that j comprises a minimal KIR haplotype in which multiple KIR genes have been deleted and novel KIR genes have arisen. The haplotype contains just four complete KIR genes encoded within ∼60 kb comprising two novel hybrid genes sandwiched between two framework KIR genes, KIR3DL3 and KIR3DL2 (Fig. 3). The KIR2DL4 and KIR3DL1/S1 framework genes are completely deleted from the haplotype. Both novel genes display intact open reading frames and typical KIR exon structures. The first novel KIR, termed KIR2DL3/2DP1, is identical to KIR2DL3 (OMIM604938; accession no. L41268) in the 5′ region, but is identical to the KIR2DP1 pseudogene (accession no. AC011501) from exon 6 to the final exon and is predicted to encode a functional molecule. The second novel KIR (termed KIR2DL1/2DS1) is identical to KIR2DL1 (OMIM604936; accession no. L41267) in the 5′ region and to KIR2DS1 (OMIM604952; accession no. AF022046) from exon 4 to the final exon. The clone sequences encompassing the complete haplotype and both the novel KIR sequences have been submitted to GenBank and are available to view in tile path form with complete annotation in Chromoview (http://www.sanger.ac.uk/cgi-bin/humpub/chromoview) and VEGA (http://vega.sanger.ac.uk/info/data/LRC_Homo_sapiens.html). The genomic sequences of the novel KIR genes indicate that the contracted KIR haplotype was formed by two deletion events, fusing KIR2DL3 with KIR2DP1, and KIR2DL1 with KIR2DS1, which derived KIR2DL3/2DP1 and KIR2DL1/2DS1, respectively. To verify our supposition of double-deletion formation and to further characterize the recombination processes that derived this haplotype, we sought to identify a haplotype with one of the hybrid genes but not the other.
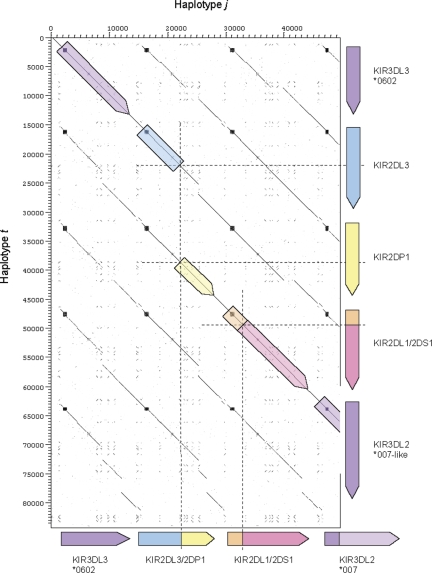
Figure 3.
Gene annotation of the sequenced portions of the j and t haplotypes. Locations of fosmid clone insert sequences are shown below each haplotype.
A putative ancestral precursor of the minimal KIR haplotype
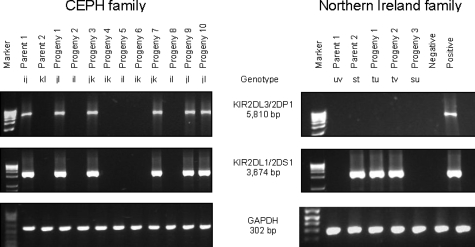
Using long-range PCRs designed to detect the novel genes (KIR2DL3/2DP1 and KIR2DL1/2DS1), we identified a family in which KIR2DL1/2DS1, but not KIR2DL3/2DP1, segregated on one haplotype (Fig. 4). This family, of Caucasian descent, originated from Northern Ireland. Segregation analysis of KIR genes/alleles in the family indicated that KIR3DL3, KIR2DL3, KIR2DS1 and KIR3DL2 segregated on the t haplotype and KIR3DP1, KIR2DL4 and KIR3DL1/S1 were absent from this haplotype (Fig. 5) (21). Haplotypes s, u and v segregating in the family have a gene composition consistent with known KIR haplotypes. Using real-time PCR, we confirmed that individuals carrying a t haplotype had one copy of KIR2DL4, and those without the t haplotype had two copies of this gene (Fig. 2).
Figure 4.
Segregation of KIR2DL3/2DP1 and KIR2DL1/2DS1 in the CEPH and the Caucasian family from Northern Ireland, as analysed by gene-specific LR-PCR. Results from the corresponding GAPDH genomic PCR are shown.
Figure 5.
Segregation of contracted haplotypes in a Caucasian family from Northern Ireland. lHaplotype sequencing subsequently showed that the t haplotype possesses a novel KIR3DL2 allele. This allele was provisionally labelled *007-like because it only differs from allele *007 by a single non-synonymous nucleotide substitution in exon 5; an adenosine to guanosine corresponding to a substitution of a glutamic acid to a glycine residue in the D2 domain of the predicted translated product.
The t haplotype was sequenced in its entirety after cloning in fosmids, and eliminating clones from the partner haplotype of the heterozygous cells, as described earlier. The clone sequences comprising the complete t haplotype have been submitted to GenBank and the fully annotated tile paths are available to view in ChromoView and VEGA. Sequencing confirmed that the t haplotype carries one of the hybrid genes (KIR2DL1/2DS1), but not the other (KIR2DL3/2DP1). In total, the t haplotype comprised five KIR genes, KIR3DL3, KIR2DL3, the KIR2DP1 pseudogene, the novel KIR2DL1/2DS1 gene and KIR3DL2 (Fig. 3). The KIR2DL4 and KIR3DL1/S1 framework genes are again deleted on the haplotype, as they are on the j haplotype as described earlier.
The minimal haplotype could have been derived from the Northern Ireland haplotype by intra-chromosomal recombination
The j and t haplotypes share the same alleles of KIR3DL3 (*0602) as well as the novel KIR2DL1/2DS1 gene sequence. In fact, pairwise alignment of the j and t haplotype sequences demonstrated that, apart from a ∼16.7 kb deletion on the j haplotype between exon 5 of KIR2DL3 and exon 6 of KIR2DP1 and a single nucleotide difference in intron 4 of KIR2DL1/S1, the j sequence is identical to the t haplotype over its entire ∼49.5 kb length (Figs 3 and 6). The extended sequence identity between the two haplotypes on both sides of the KIR2DL3–KIR2DP1 deletion indicates that the j and t haplotypes are related by descent and that intra-chromosomal (intra-chromatid or inter-chromatid) recombination between an ancestral KIR2DL3 gene and an ancestral KIR2DP1 gene (KIR2DL3 and KIR2DP1 map adjacent to each other in the centromeric half of the KIR complex) on the same haplotype created the hybrid gene KIR2DL3/2DP1 (Fig. 7). This contention implies that the t haplotype, or a closely related sequence, is the ancestral precursor of the j haplotype. NAHR between two homozygous haplotypes is another possible scenario, although the t haplotype was not represented in the full Utah CEPH panel of 47 families, indicating the rare nature of this haplotype and the unlikely possibility of NAHR. To examine further the ancestral relationship of the j and t haplotypes, we sequenced the highly polymorphic LILRB3 locus, located ∼500 kb centromeric of the KIRs, in both the Utah and Irish families. Thirty-seven LILRB3 polymorphisms within three exons and adjoining intron sequence were tracked. Segregation analysis of their alleles in the two families showed that the j and t haplotypes carry an identical coding haplotype of LILRB3 and differ at only two polymorphic sites (intronic) out of the 37 identified (Table 1), corroborating the close ancestral relationship between the j and t two haplotypes.
Figure 6.
Dot matrix analysis of the j and t KIR haplotype sequences. The plot shows the four KIR genes of the j haplotype on the x-axis and the 5 KIR genes of the t haplotype on the y-axis. Regions of similarity are identified as a concentration of dots forming diagonal lines. Minisatellites can be visualized as boxes. The deletion breakpoints on both haplotypes are indicated by dashed lines. The intergenic regions and introns of the KIR loci are well conserved.
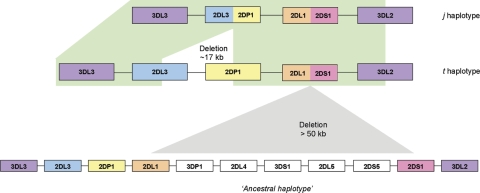
Figure 7.
The presumed order of genomic deletions in the formation of the j haplotype. First, a >50 kb deletion fused the KIR2DL1 and KIR2DS1 genes creating the KIR2DL1/S1 hybrid gene. Subsequently, a smaller ∼17 kb deletion fused the KIR2DL3 and KIR2DP1 genes to form the KIR2DL3/2DP1 gene. A representative KIR ‘B’ haplotype is shown below the deletion haplotypes as an example of one potential ancestral haplotype involved in the derivation of haplotype t. The represented haplotype was the most frequent observed in a panel of 85 Caucasoid individuals with a frequency of 0.124 (41). Apart from the ∼17 kb deletion and a single nucleotide, the j and t haplotype sequences are identical, raising the possibility of intra-chromosomal recombination in the formation of the j haplotype, and implicating the t haplotype as the single precursor of the j haplotype.
Table 1.
LILRB3 haplotypes
| Position | Location | Haplotypes |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| i | j | k | l | s | t | u | v | ||
| +1105 | Exon 4 | G | G | G | A | A | G | A | A |
| +1659 | Intron 4 | C | C | C | C | T | C | C | C |
| +1802 | Exon 5 | C | C | C | C | C | C | G | G |
| +1807 | C | C | A | A | A | C | A | C | |
| +1809 | C | C | T | T | T | C | T | C | |
| +1830 | C | T | C | T | C | T | T | T | |
| +1869 | C | C | C | C | C | C | T | C | |
| +1879 | A | A | A | A | A | A | G | A | |
| +1890 | G | G | G | G | G | G | C | G | |
| +1891 | G | G | G | G | G | G | C | G | |
| +1947 | Intron 5 | C | C | C | T | C | T | C | T |
| +2027 | + | + | − | + | + | + | + | + | |
| +2036 | C | C | G | C | C | C | C | C | |
| +2055 | G | A | G | A | G | A | G | A | |
| +2214 | Exon 6 | G | G | A | G | G | G | G | G |
| +2235 | T | T | C | T | T | T | T | T | |
| +2238 | T | T | G | T | T | T | T | T | |
| +2241 | C | C | T | C | C | C | C | C | |
| +2243 | G | G | T | G | G | G | G | G | |
| +2391 | C | T | C | T | C | T | C | T | |
| +2392 | G | A | G | A | G | A | G | A | |
| +2406 | T | C | C | C | C | C | T | C | |
| +2418 | C | T | C | T | C | T | C | T | |
| +2419 | A | T | T | T | A | T | A | T | |
| +2438 | G | A | A | A | G | A | G | A | |
| +2442 | G | A | A | A | G | A | G | A | |
| +2497 | Intron 6 | A | A | C | A | A | A | A | A |
| +2736 | G | T | T | G | G | T | G | T | |
| +2843 | Intron 7 | G | G | G | G | G | A | G | G |
| +2854 | C | G | C | G | C | G | C | G | |
| +2951 | + | + | − | + | − | + | − | + | |
| +3303 | G | A | G | A | G | A | G | A | |
| +3354 | G | A | A | A | A | A | G | A | |
| +3440 | C | C | C | T | C | C | C | C | |
| +3494 | T | C | T | C | T | C | T | C | |
| +3539 | G | A | A | A | A | A | G | A | |
| +3731 | G | C | C | C | C | C | G | C | |
The KIR genes that constitute the t haplotype, including the two ancestral genes forming the KIR2DL1/2DS1 hybrid (3DL3, 2DL3, 2DP1, 2DL1, 2DS1 and 3DL2), have been observed on various KIR haplotypes (some of which contain additional genes) and in the same order as on the t haplotype. From family studies, the frequencies of these haplotypes range from <1 to 6.7% (19,22). KIR2DL1/2DS1 could, therefore, have been derived from an intra-chromosomal recombination between an ancestral KIR2DL1 gene (KIR2DL1 maps to the centromeric half of the KIR gene complex) and an ancestral KIR2DS1 gene (KIR2DS1 maps to the telomeric half of the KIR gene complex). Alternatively, misalignment of KIR genes on two parental homologous chromosomes during synapsis may have resulted in NAHR between ancestral KIR2DL1 and KIR2DS1 genes on different haplotypes and consequential formation of KIR2DL1/2DS1. Whether KIR2DL1/2DS1 was formed by intra-chromosomal recombination or by NAHR, the resulting progeny haplotype containing the novel hybrid gene, KIR2DL1/2DS1, would theoretically not include the framework KIR genes, KIR3DP1, KIR2DL4, KIR3DL1/S1, or KIR2DL5A, KIR2DS3 and KIR2DS5, as observed on both the j and t haplotypes.
The KIR haplotype restructuring is associated with Alu-mediated recombination
The minimum recombination interval in which the recombination must have taken place for KIR2DL3/2DP1 and KIR2DL1/2DS1, as delimited by gene-specific nucleotides in exons, are 3261 and 1866 bp, respectively. For hybrid KIR genes, precise definition of the limits between the regions derived from particular KIR can be restricted by the high sequence similarity of these genes and the limited number of genomic (intron) sequences presently available. However, using the complete genomic sequences of the j haplotype and its hypothetical precursor, the t haplotype, we were able to map accurately the breakpoints involved in the derivation of the short haplotypes, to facilitate identification of elements associated with the generation of de novo KIR genes and haplotypes. We located an interval of 93 bp within intron 5 where the KIR2DL3 and KIR2DP1 sequences were interrupted to form the resulting KIR2DL3/2DP1 gene (Fig. 8). The 93 bp interval comprises part of an Alu short interspersed nuclear element (SINE) in reverse orientation. Alu elements are frequent contributors to unequal crossovers and have been implicated in a variety of chromosomal rearrangements (23,24). Notably, the Alu sequence that is represented in the interval is precisely the portion of Alu that has previously been associated with increased recombination (Supplementary Material, Fig. S1) (23,25). Different explanations have been posited for the heightened recombination of the Alu component. These explanations are supported by the fact that the recombination hotspot is sited near to the L1 endonuclease cleavage site of the Alu element, that it contains the prokaryotic chi sequence, and that it tends to have high GC content (23,25).
Figure 8.
The KIR2DL3/2DP1 gene is a product of recombination between KIR2DL3 (CU464054) and KIR2DP1 (CU464061). Nucleotide positions that differ between CU464054 and CU464061 are shown.
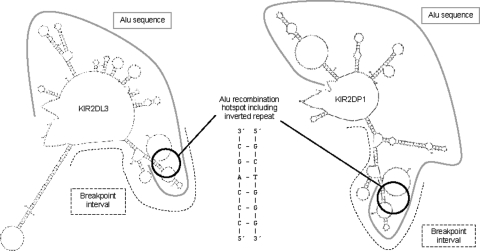
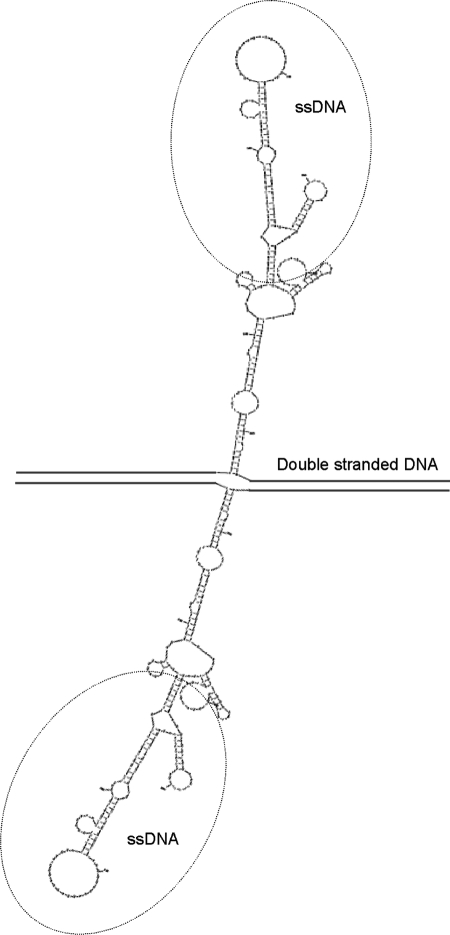
To further investigate the promiscuous nature of the Alu recombination hotspot, the sequences surrounding the breakpoints involved in formation of the KIR2DL3/2DP1 gene were analysed by M-Fold to determine potential single-stranded DNA secondary or non-B DNA structures. Interestingly, the part of the Alu associated with heightened recombination was capable of adopting a cruciform conformation comprising multiple hairpin loops (Fig. 9). Such structures can be accessible substrates for the MR(X)N nuclease complex or other nucleases, thereby making them susceptible to strand breaks. We observed that the sequence of the Alu recombination hotspot subsumes centrally an inverted repeat within the Alu (CCCAGC—22 nucleotides—GCTGGG), which is located near the pinnacle of the cruciform and contributes to the non-B structure formation. Alignment of complete Alu sequences showed that the 22-nucleotide hotspot sequence is conserved among all Alu subclasses (Supplementary Material, Fig. S2). The 22-nucleotide hotspot can adopt a hairpin structure incited by the inverted repeat TCCCA—6 nucleotides—TGGGA (Supplementary Material, Fig. S3). An AT-rich site of ∼90 nucleotides is located within the Alu break site in KIR2DL3. This sequence can adopt an extended hairpin and could also have participated in the deletion at this site therefore. Figure 10 shows a schematic representation of one way the KIR2DL3/2DP1 hybrid could have been formed by an intra-chromatid recombination through strand break repair.
Figure 9.
Potential ssDNA secondary structures formed at the KIR2DL3 (left) and KIR2DP1 (right) breakpoint regions ssDNA. These structures can serve as recognition signals to induce strand breaks that cause genomic rearrangements by recombination-repair. The dashed line indicates the breakpoint intervals. The solid grey line depicts the full ∼282 bp Alu sequence. The black circle contains the core 22 nucleotides of the Alu recombination hotspot (5′-TGTAATCCCAGCACTTTGGGAG-3′).
Figure 10.
Proposed mechanism behind the formation of the KIR2DL3/2DP1 gene; deletion of ∼17 kb by homologous unequal intra-chromosomal recombination (Alu-mediated). A DNA break is introduced at the recombination hotspot site of an Alu repeat within a KIR gene. A second break occurs 17 kb away that contains sequence homologous to the first break site. The two homologous sequences serve as a substrate for double-strand break repair, which leads to deletion of the intervening sequences between the break sites.
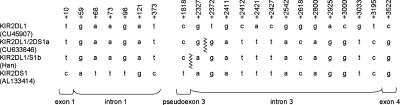
The recombination sites within KIR2DL1 and KIR2DS1 involved in the formation of the hybrid KIR2DL1/2DS1 were located to a 44 bp interval within intron 3 of each gene (Fig. 11). The breakpoints lie within a mammalian long terminal repeat (LTR)-transposon 1 (MLT1) element, an ancient member of the highly repetitive mammalian apparent LTR retrotransposon (MaLR) superfamily of repeats (26) that include the transposon-like human element (THE-1) repeats. Interestingly, THE-1 sequences have been associated with recombination hotspots and genome instability in humans (27). Analysis by M-fold predicted that the breakpoint regions within the MLT1 sequence are susceptible of forming long protruding multi-loop structures (Fig. 12), which could, as described earlier, have catalysed the formation of the deletion.
Figure 11.
The KIR2DL1/2DS1 gene is a product of recombination between KIR2DL1 (CU45907) and KIR2DS1 (AL133414). Nucleotide positions that differ between CU45907 and AL133414 are shown. KIR2DL1/2DS1a (j, t haplotypes, Ukrainian and African American) and KIR2DL1/2DS1b (Han Chinese).
Figure 12.
Potential ssDNA conformation formed from the MLT1 sequence at the KIR2DS1 breakpoint region. The breakpoint interval sequences are circled. Such protrusive non-B DNA structures may catalyse KIR rearrangements.
A degenerate 13-mer hotspot-promoting motif associated with THE-1 elements and other repeat elements has recently been identified (28). This motif occurs 16 times on the t KIR haplotype, once every ∼5 kb on average and with typically three motifs within each KIR gene. Four instances are within AluS elements, one is in a long terminal repeat (LTR33A) and another is in a long interspersed nuclear element (LIPA3). The remaining 10 are in non-repeat DNA. The motifs within KIR are sometimes closely paired (<410 bp) in reverse orientation, potentially stimulating single-strand DNA folding. The motif is present less than 170 bp from the KIR2DL1/S1 breaksite and ∼4 kb from the KIR2DL3/2DP1 breaksite. Interestingly, the motif overlaps the previously described 22-nucleotide Alu hotspot sequence, potentially implicating DNA secondary structure formation and strand break in hotspot activity of the motif, depending on its genetic background (Supplementary Material, Figs S2 and S3).
The KIR haplotype restructuring uses pseudogene sequence and switches KIR promoters
KIR2DP1 is considered to be a silent gene or pseudogene (29) due to a single base-pair deletion in exon 4 that creates a premature stop codon in exon 5 and is predicted to cause nonsense-mediated decay of the transcript. Conversely, exons 1–5 of the KIR2DP1 recombinant, KIR2DL3/2DP1, share complete sequence identity to the KIR2DL3 gene and do not contain any non-sense mutations. Exons 6–9 of KIR2DP1 are homologous to the corresponding exons of other type I KIR2D and have uninterrupted reading frames. All exons have legitimate splice junctions, although intron 7 is non-canonical, and KIR2DL3/2DP1 is preceded by a promoter sequence that is functional in the KIR2DL3 gene (30). This suggests that, unlike KIR2DP1, the recombinant KIR2DL3/2DP1 could be transcribed.
Since KIR2DL3/2DP1 and KIR2DL3 share identical sequences over their extracellular domains, KIR2DL3/2DP1 is likely to bind the same ligands as KIR2DL3, such as HLA-C group 1 molecules encoding amino acid serine (Ser) at residue 77 and asparagine (Asn) at residue 80 of the MHC-C molecule. However, differences in the primary structures of the intracellular cytoplasmic portions suggest that they could be functionally distinct. Although KIR2DL3/2DP1 shows the typical feature of an inhibitory KIR by not possessing a charged residue in the transmembrane region, it is unusual in character among inhibitory KIR in possessing a single intracellular ITIM (V/I/L/SxYxxL/V/I/S) (Supplementary Material, Fig. S4). The single ITIM present is distinct from that of other KIR in possessing a valine at the fourth position (VTYVQL); created by a dinucelotide substitution within the respective codon. The same substitution is found in one allele of KIR2DL1 (*009) but not in any other KIR sequences.
To assess the relationship between the intracellular region of KIR2DP1 that is incorporated into KIR2DL3/2DP1 with the intracellular region of other KIR, a phylogenetic tree (Supplementary Material, Fig. S5) was built from a nucleotide sequence (exons 6–8) alignment using the neighbour-joining method. The tree demonstrated clear relationships between KIR2DP1 and other known type I KIR2DL genes. KIR2DP1 grouped with KIR2DL1, KIR2DL2 and KIR2DL3.
NK cell RNA from a donor with the KIR2DL3/2DP1 gene was unavailable to directly test endogenous expression. However, expression of KIR2DL3/2DP1 transcription was detected in the B lymphoblastoid CEPH cell line from which the gene was identified after two rounds of KIR2DL3/2DP1-specific RT–PCR. Sequencing over the breakpoint region confirmed that the transcript originated from the recombinant gene.
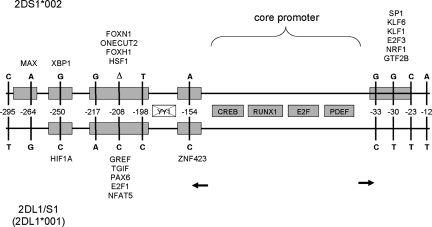
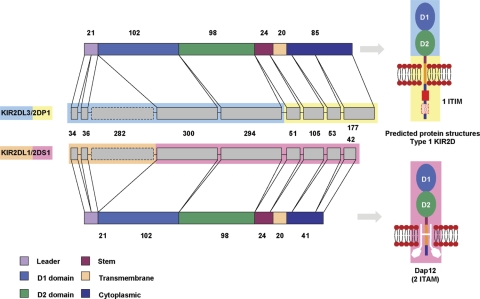
Transcription of the KIR2DL1/2DS1 hybrid was confirmed by RT–PCR analysis using RNA of purified peripheral NK cells isolated from a donor carrying the novel gene (Supplementary Material, Fig. S6). The full-length cDNA of KIR2DL1/2DS1 (AM999888) contains an open reading frame of 915 bp that codes for a polypeptide of 304 amino acids. The switching of promoters through KIR2DL1/S1 hybrid formation (Fig. 13), where the KIR2DS1 promoter has been converted to KIR2DL1 promoter sequence could alter transcriptional expression of the KIR2DS1 hybrid gene relative to the wild-type KIR2DS1 (30) (Supplementary Material, Fig. S7).The exon structures and predicted receptor structures of KIR2DL1/2DS1 and KIR2DL3/2DP1 are shown in Figure 14.
Figure 13.
Schematic of the 300 bp KIR bi-directional core promoter region of KIR2DL1/S1 (upper) with respect to KIR2DS1 (lower). The positions of known and predicted TFBSs are indicated by boxes. Shaded grey boxes represent core promoter TFBSs. Coloured boxes represent significant TFBS matches corresponding to nucleotide polymorphisms, with their identity (HUGO gene symbol nomenclature) given against the gene in which the TFBS is present. For example, the Sp1 site is present in KIR2DS1 but not in KIR2DL1/2DS1. Transcription initiation sites for forward and reverse promoters of the bidirectional promoter are shown by the rightward and leftward arrows, respectively (Supplementary Material Fig. S7). The vertical lines indicate the positions of polymorphic nucleotides. Numbering indicates the positions of the polymorphic residues relative to the translation initiation codon of the KIR2DL1/S1 gene, where the base A of the ATG codon is denoted nucleotide +1. The nucleotide present at each variable position is shown for both genes. The YY1 site is present neither in KIR2DS1 nor KIR2DL1/S1.
Figure 14.
Hybrid KIR gene organization and predicted protein structures. The coding regions of the exons are represented as grey boxes; their size in base pairs is shown. Pseudoexons are indicated with a dashed line. The way in which the exons code for each protein domain/region is shown. The main structural characteristics of KIR proteins are shown where the domains and regions are represented as boxes of different colours according to the key. The approximate length (amino acids) of each domain or region is shown next to their corresponding box. Predicted KIR protein structures are displayed adjacent to their respective gene. The structural characteristics of two immunoglobulin-like domain KIR proteins are shown. The intact ITIM site of KIR2DL3/2DP1 is shown as a red box. The disrupted ITIM is represented by a dashed red box. The KIR2DL1/S1 is presented with the associated adaptor molecule, DAP12.
A different KIR2DL1/2DS1 hybrid gene identified in a Chinese population
Since we found the KIR2DL1/2DS1 hybrid locus in two geographically separated, though both Caucasian, families we wanted to determine whether it was present in other populations. Based on KIR typing information and using the long-range PCR assays as described earlier, we identified additional carriers of a KIR2DL1/S1 hybrid gene. The first example was of African American descent, identified in a reference DNA panel from the International Cell Exchange (http://www.hla.ucla.edu/cellDNA/Cell/history.htm). The second was an infertile psoriatic arthritis patient of Ukrainian origin. This individual is homozygous for the t KIR haplotype. Sequencing established that the KIR2DL1/2DS1 recombination break interval in these subjects was identically sited to that of the j and t haplotypes (Fig. 11).
To assess a possible wider global distribution of the hybrid genes, using the long-range PCRs designed to detect the novel genes (KIR2DL3/2DP1 and KIR2DL1/2DS1), we screened 1214 unrelated individuals from 52 geographically distinct worldwide populations. A KIR2DL1/2DS1 gene was identified in Han Chinese with a carrier frequency of 1.1%. However, sequence analysis revealed that the deletion break site that formed the Han Chinese KIR2DL1/2DS1 gene, although within intron3/pseudoexon 3, was located slightly differently to that of the Caucasian KIR2DL1/2DS1 gene (Fig. 11), signifying that a deletion event giving rise to a KIR2DL1/2DS1 composite gene has occurred independently at least twice. Using real-time PCR to measure gene dose, we confirmed that the KIR2DL4 locus had been deleted in the formation of the Han Chinese KIR2DL1/2DS1 gene (data not shown).
The Ukrainian KIR2DL1/S1 carrier was homozygous for the KIR3DL2*007 allele which is present on the j haplotype, but was negative for the KIR3DL3*00602 (as determined by SSP-PCR on the genomic DNA targeting a nucleotide that distinguishes the *006 allele) which is present on both the j and t haplotypes. Neither KIR3DL2*007 nor KIR3DL3*0602 alleles were present in the African American or Han Chinese KIR2DL1/2DS1 carriers, suggesting that these deletion haplotypes have diverged from the t haplotype by recombination. Alternatively, they may have formed by independent recombination events as proposed above for the Han Chinese haplotype based on their differently located break sites. So, despite the KIR3DL2*007 and KIR3DL3*0602 link to the j and t haplotypes, they do not appear to act as strict markers for all KIR2DL1/2DS1 carrying haplotypes. Allele frequencies of KIR3DL2*007 and KIR3DL3*00602 in worldwide populations are given in Supplementary Material, Figure S8.
Long-range PCR identified further variant genes in African, Japanese and Maya populations at carrier frequencies ranging from 3.2 to 25% (Supplementary Material, Fig. S9). However, sequence analysis showed that all of these cases appear to comprise short-tract gene conversions or mutations of KIR2DP1 or KIR2DS1. These composite genes are, therefore, unrelated and distinct from the KIR2DL3/2DP1 and KIR2DL1/S1 genes described earlier.
DISCUSSION
Formation of hybrid KIR genes
It is generally accepted that gene fusion events, which are common in tumours, are deleterious (31). We propose that the head-to-tail arrangement of closely related KIR is a substrate for promiscuous generation of novel hybrid genes, providing a flexible evolutionary adaptive strategy. Gene gain–loss has been noted before as a feature of the plastic KIR complex (16). The heightened propensity to generate novel hybrid KIR receptors may provide, we suggest, a further proactive evolutionary measure, to militate against pathogen evasion or subversion (32).
Repeat elements in the LRC may facilitate sequence exchange
KIRs exhibit dense clustering of particular repeat elements within their introns, including MaLR, MLT1D, Alu, L2 and long-interspersed nuclear elements (LINEs) (16). These families of repeats, including LINES (33), have all recently been associated with genomic deletions or recombination. The LILR, a multi-gene family adjacent to KIR, are less densely arranged, contain fewer repeats of this type within genes and exhibit higher stability in terms of CNV compared with KIR (16). Interestingly, chromosome 19, on which the KIRs are housed, has the highest repeat density, and notably Alu density, of all human chromosomes along with the highest gene density and an unusual enrichment of immune genes (34–37).
The precise localization of the KIR2DL3/2DP1 breakpoint within an Alu sequence recombination hotspot suggests that repetitive elements incorporated within the KIR genes facilitate their evolution. The breakpoints of the KIR3DL1/2v hybrid gene reported recently (38) also occur within Alu sequence and can adopt stem-loop structure (Supplementary Material, Fig. S10). The recombination sites within KIR2DL1 and KIR2DS1 involved in the formation of the hybrid KIR2DL1/2DS1 lie within a MaLR repeat and near an AT-rich site, both of which have been identified as structural risk factors for chromosome breakage. We conjecture that the plasticity of the KIR complex in terms of its hyper-mutability is intrinsic and relates to inherent sequence elements associated with chromosome fragility among closely arranged homologous KIR genes. These elements could catalyse the formation of chromosomal rearrangements by being prone to forming non-B conformations. These structures serve as recognition signals to induce chromosomal double-strand breaks that cause genomic rearrangements by recombination-repair. Another possibility is that the epigenetic architecture of these regions may facilitate rearrangements. In other words, the combined effect of DNA breakage-prone elements in addition to the arrangement of KIR genes in close head-to-tail orientation facilitates the rapid diversification of gene CNV in the cluster.
Evolutionary relationship between the j and t haplotypes
Our data are compatible with intra-chromosomal recombination in the formation of the hybrid genes. The near complete sequence identity between the j and t haplotypes points to a common ancestry in relatively recent human history. However, it is problematic to use conventional mutation rate estimates to calculate when the j and t haplotypes diverged from a single common ancestor because a significant proportion of a KIR haplotype is coding sequence and much of the non-coding DNA may be functional. In addition, the single nucleotide difference between the two haplotypes in intron 4 of KIR2DL1/2DS1 could have arisen by a localized conversion event with a different KIR gene, rather than by mutation. This aside, using a range of plausible mutation rates (see Materials and Methods), it can be estimated that the time to the most common recent ancestor of the two sequences that have accumulated only 1 SNP within 49 458 kb (the length of the region represented in both haplotypes) is 9620–15 553 years.
Novel hybrid KIR form recurrently
The sharing of hybrid genes among different populations implies that these specific genomic variants predated the dispersal of modern humans (out of Africa), recurred independently in different populations or were due to admixture between populations. The differently sited chromosome breakages in the KIR2DL1/S1 gene between individuals are consistent with independent formation. The germline rates of de novo meiotic deletions and duplications within the KIR locus could be assessed by sperm-based assays of meiosis (39).
We previously reported a hybrid KIR gene, KIR2DL5A/3DP1, which was identified on an extended haplotype (18) and proposed that this gene was formed by an unequal crossover event. The reciprocal gene, KIR3DP1/2DL5A (KIR2DL5B; AF217486), has also been identified (40), providing further evidence for recurrent recombination within KIR. However, reciprocal haplotypes to the ones described herein have not been observed in any family studies published to date (11,19,20,40–42). Neither are the reciprocal hybrids (KIR2DS1/2DL1 or KIR2DP1/2DL3) found within in the current mRNA sequences from NCBI reference sequence database (refseq_mrna). It might be predicted that deletion haplotypes occur more commonly because they can result from both intra-chromosomal and NAHR, whereas extended haplotypes can only be formed by NAHR. On the other hand, current typing methods may miss the extended haplotypes.
Hybrid genes need to be considered when KIR typing samples
The detection of novel KIR haplotypes and loci in different non-Caucasian populations highlights the importance of considering diverse worldwide populations for full characterization of KIR haplotype variation. It also prompts caution in KIR analysis of populations of different ethnic origins. Current KIR typing designs are predominantly based on Caucasian KIR sequences and haplotype structures. Inaccurate genotypes may result because present methods are blind to the existence of hybrid genes and localized genomic re-arrangements in different ethnic populations. Further resolution of structural variation associated with KIR genetic diversities in human populations promises to provide further insights into complex disease and quantitative genetic traits.
The power to identify a relationship between DNA variation and phenotype is limited by the sensitivity with which the genetic variation is measured in each individual. The structural complexity and high diversity of the KIR region needs special attention. A recent study highlights the importance of distinguishing between alleles of KIR genes in disease studies (43). Both copy number and allelic/isotype are important in CNV analysis. If sites are commonly regenerating, it makes them less amenable to indirect interrogation by surrogate markers. Some CNV may be in LD with flanking SNPs/STRs and may effectively detect associations in certain haplotypes and populations but CNVs that have occurred multiple times independently will not be as readily detectable through SNP-based association studies. Additionally, epistasic interactions between co-evolving CNV gene families may conceal potential disease effects.
KIR genes vary both in copy number and allelic sequence
One way by which CNVs diversify human phenotypes is by varying transcript levels through gene dosage. This effect has also been shown for the KIR loci (30,44). However, an additional contribution is apparent; paralogous KIRs subtly diversify the family repertoire such that each KIR gene potentially encodes a receptor with variable specificity for HLA class I. The duplication of KIR genes is not discretely modular; the breakpoints of duplication are not identical and have various boundaries among duplicated modules. As described in this paper, allelic diversity is generated as a result of NAHR, in addition to homologous recombination events. The ‘patchwork’ shuffling of KIR sequences by recombination appears to allow distribution of the structural or regulatory attributes of existing KIR in the creation of new genes, as well as being involved in relegation of genes to pseudogenes and vice versa. In addition, diversification by mutation and recombination continues between genes post-duplication (38). In other words, the functional effects for KIR do not simply relate to only copy number. They also influence allotypes in terms of ligand affinity or expression capacity. The same scenario may apply to other niCNV genes encoding receptor/ligand molecules.
The KIR2DL1/S1 hybrid reflects the transient nature of activating KIR in evolution. Activating KIRs appear to be especially short lived and recurrently evolving (45), implying that they are subject to transient shifts in selection pressure over time. This is consistent with the association of activating KIRs with resistance to infection, reproductive success and susceptibility to autoimmunity.
Selective advantages of novel hybrid KIR
KIR, in combination with MHC class I variants, are associated with resistance to infectious and other diseases (10). The KIR2DL3/2DP1 hybrid, in effect, encodes a KIR2DL3-like molecule with an altered cytoplasmic tail. This could provide a change in signalling potential (46), suitable for certain infections. In the case of the KIR2DL1/2DS1 hybrid, the switching of the promoter is predicted to alter transcriptional control by local or more widespread effects on the KIR locus. The persistence of the recombinant genes/haplotypes in populations may hint at selection or re-occurrence since most selectively neutral de novo mutations are lost by chance (random sampling of gametes) within a small number of generations (<15 generations; population size 200) (47).
CNV is an important evolutionary strategy in KIR and other gene families in the immune system
Gene duplications are essentially products of legitimate ‘successful’ expansions that have been fixed in the population. On the other hand, ‘unsuccessful’ and ‘decommissioned’ duplicates remain in the genome as pseudogenes, which can act as substrates for future adaptations of the gene family. Immune gene families seem particularly versatile at rapid diversification. Both the KIR and MHC are regions of high plasticity, high polymorphism and they are polygenic. Pseudogenes are generally considered to be non-functional relics but we have shown that KIR pseudogenes may be put to use. Similarly, it was shown some years ago in analysis of the series of MHC class I bm mutants that pseudogenes have a function as sequence donors for diversification by gene conversion (48).
Interestingly, NAHR events sometimes converge in relatively narrow hotspots across the genome (49). It may be that non-B structures in sequence motifs are responsible for this. Somewhat ironically, it appears therefore that retroviral integrations that occurred in the ancient past have in more recent times been exploited in the human genome to diversify defence genes to counter modern day virus threats.
MATERIALS AND METHODS
KIR genotyping, copy number and haplotype determination
Genomic DNA was genotyped for presence or absence of KIR genes either by using PCR amplification with locus-specific primers (PCR-SSP) (50) or by using polymerase chain reaction-sequence specific oligonucleotide probes (PCR-SSOP) (51). KIR haplotypes were determined by segregation analysis in families (20). Allelic discrimination was performed either by PCR-SSOP (21) or by direct sequencing of KIR genes. Allele nomenclature was derived from IPD-KIR (http://www.ebi.ac.uk/ipd/kir/). KIR2DL4 copy number was determined by quantitative PCR (18). Long-range PCR-SSP assays were designed for detection of the KIR hybrid genes. The KIR2DL3/2DP1 forward and reverse primers, sited in exon 5 and exon 7, were 5′-GGCTCTTTCCGTGACTCTCCA-3′ and 5′-AATCAGAACGTGCAGGTGTCTT-3′, respectively. The KIR2DL1/2DS1 forward and reverse primers, sited in exons 1 and 4, were 5′-CGGCAGCACCATGTCGCTCT-3′ and 5′-GGTCCCTGCCAGGTCTTGCT-3′, respectively. The amplification product sizes for the KIR2DL3/2DP1 and KIR2DL1/2DS1 are 5810 bp and 3674 bp, respectively. An internal control was included in each PCR to validate proper amplifications.
Fosmid preparation, clone selection and insert sequencing
KIR haplotypes were sequenced using EpiFOS fosmid vectors (Epicentre). Deletions associated with the hybrid genes were captured in at least two different fosmids and were confirmed by PCR-SSP in the original genomic DNA. The CEPH DNA was obtained from an EBV-transformed lymphoblast cell line maintained by the Coriell Institute. DNA was mechanically sheared, blunt-end repaired and size selected using pulse-field gel electrophoresis. Bands corresponding to fragments >32 kb and <42 kb were excised. DNA was extracted from the low-melting point agarose and ligated into the pCC1FOS vector. Fosmid clones were prepared using Phage T1-resistant EP1300-T1 Escherichia coli plating strain. Packaged fosmid clones were plated onto LB-chloramphenicol bio-assay plates. Following overnight incubation, colonies were lifted onto Immobilon-Ny + membranes (Millipore Ltd). Membranes were hybridized with a 32P-labelled KIR probe and visualized with X-ray film. Positive clones were picked and fosmid DNA was extracted using FosmidMAX DNA purification. Clones were selected for sequencing after performing PCR-SSP to determine their KIR gene content. Shotgun sequencing and directed finishing of the clone inserts were carried out by the Wellcome Trust Sanger Institute. The generated sequence data were processed by a suite of in house programs (http://www.sanger.ac.uk/Software/sequencing/) prior to assembly. For the finishing phase, the GAP4 program was used to edit and select reactions to eliminate ambiguities and close sequence gaps. Each clone has been finished according to the agreed international finishing standard (http://genome.wustl.edu/gsc/Overview/finrules/hgfinrules.html). All sequences presented in this paper have been submitted to the EMBL/Genbank/DBB database and allocated accession numbers. For purposes of clarity, all fosmid clones are referred to using their accession numbers. Clones were assigned to a particular haplotype by comparing the gene and allelic content of the sequences with the haplotypes determined by segregation analysis. Overlaps between clones were analysed using the bl2seq program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and clones assigned to the same haplotype were all >9 kb in length and showed no discrepancies.
Gene annotation and sequence analysis
The finished genomic sequence was analysed using an automatic Ensembl pipeline (52). Interspersed repeats were identified using RepeatMasker (http://repeatmasker.genome.washington.edu) and simple repeats were detected by Tandem Repeat Finder (http://tandem.bu.edu/trf/trf.html). Sequence with repeats masked was searched against vertebrate cDNAs and ESTs using WU-BLASTN and EST_GENOME, and against a non-redundant SWISS-PROT/TrEMBL database using WU-BLASTX. Genes were annotated according to human annotation workshop (HAWK) guidelines (http://www.sanger.ac.uk/HGP/havana/hawk.shtml). Pairwise sequence alignments were carried out using the dot-matrix program, ‘dotter’ (http://sonnhammer.sbc.su.se/Dotter.html). Promoter sequences were examined for potential transcription factor binding sites using the MatInspector Program (www.genomatix.de). Potential secondary structure formed within a single-strand DNA sequence was determined using the M-fold server (http://mfold.bioinfo.rpi.edu). Structures are predicted from DNA sequences flanking the breakpoints of the deletion and are examples because alternative conformations are possible. Default parameters were used except for the ‘DNA’ setting and 100 ∼ 150 mm [Na+] and 10 ∼ 15 mm [Mg++] for in vivo human conditions. Structures were depicted as two-dimensional stem-loop base parings. Alignment of KIR sequences for phylogentic analysis was performed using Geneious Pro 4.6 software (www.geneious.com). The neighbour-joining method, using a Tamura-Nei genetic distance model, was used to build a phylogenetic diagram from exons 6–8 of KIR genes. The dating of the last shared common ancestor of the j and t haplotypes was calculated as described previously (53).
Hybrid gene expression analysis and diversity panel screening
We designed primers to specifically amplify full-length KIR2DL3/2DP1 and KIR2DL1/2DS1 from cDNA derived from the lymphoblastoid CEPH cell line and from MACS sorted peripheral NK cells (www.miltenyibiotec.com). Total RNA extractions were carried out using the RNeasy Mini Kit (Qiagen). cDNAs were made using first-strand cDNA synthesis (Invitrogen) on mRNA using an oligo dT primer. Full-length gene sequences were determined following cloning into TOPO vector (Invitrogen). The KIR2DL3/2DP1 forward and reverse RT–PCR primers were 5′-CTCATGGTCGTCAGCATGGT-3′ and 5′-GAAAACGCAGTGATCCAACTGTA-3′, respectively. The KIR2DL1/2DS1 forward and reverse RT–PCR primers were 5′-GCAGCACCATGTCGCTCT-3′ and 5′-GACTGTGGTGCTCGTGGA-3′, respectively. We screened 1214 unrelated individuals from 52 geographically distinct worldwide populations within the Human Genome Diversity Cell Line Panel (http://www.cephb.fr/en/hgdp/diversity.php) and unrelated individuals from Japanese, Han Chinese and Yoruban populations from the HapMap Panel (http://ccr.coriell.org/).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the MRC (www.mrc.ac.uk). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all staff of the DNA Sequencing Division at the Wellcome Trust Sanger Institute; the Northern Ireland family for participation in this study; Stephan Beck and Mike Quail for help organizing sequencing or subcloning at the Sanger Institute; Harminder Sehra for annotation of the clones; Howard Cann, Nigel Carter, Richard Redon for providing DNA samples; Louise Boyle and Chiwen Chang for assistance with fosmid transfections and NK cell isolations; Jyothi Jayaraman for KIR genotype profiling; Des Jones for LILRB3 primer sequences.
Conflict of Interest statement. The authors declare no conflicts of interest.
REFERENCES
- 1.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 3.Korbel J.O., Kim P.M., Chen X., Urban A.E., Weissman S., Snyder M., Gerstein M.B. The current excitement about copy-number variation: how it relates to gene duplications and protein families. Curr. Opin. Struct. Biol. 2008;18:366–374. doi: 10.1016/j.sbi.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willcocks L.C., Lyons P.A., Clatworthy M.R., Robinson J.I., Yang W., Newland S.A., Plagnol V., McGovern N.N., Condliffe A.M., Chilvers E.R., et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J. Exp. Med. 2008;205:1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y., Chung E.K., Wu Y.L., Savelli S.L., Nagaraja H.N., Zhou B., Hebert M., Jones K.N., Shu Y., Kitzmiller K., et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degenhardt J.D., de Candia P., Chabot A., Schwartz S., Henderson L., Ling B., Hunter M., Jiang Z., Palermo R.E., Katze M., et al. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in Rhesus Macaques (Macaca mulatta) PLoS Genet. 2009;5:e1000346. doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw C.J., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. 2004;13(Spec No. 1):R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 10.Khakoo S.I., Carrington M. KIR and disease: a model system or system of models? Immunol. Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 11.Shilling H.G., Guethlein L.A., Cheng N.W., Gardiner C.M., Rodriguez R., Tyan D., Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J. Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 12.Uhrberg M., Valiante N.M., Shum B.P., Shilling H.G., Lienert-Weidenbach K., Corliss B., Tyan D., Lanier L.L., Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 13.Gendzekhadze K., Norman P.J., Abi-Rached L., Layrisse Z., Parham P. High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics. 2006;58:474–480. doi: 10.1007/s00251-006-0108-3. [DOI] [PubMed] [Google Scholar]
- 14.Carrington M., Martin M.P. The impact of variation at the KIR gene cluster on human disease. Curr. Top. Microbiol. Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 15.Bashirova A.A., Martin M.P., McVicar D.W., Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genomics Hum. Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 16.Wilson M.J., Torkar M., Haude A., Milne S., Jones T., Sheer D., Beck S., Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl Acad. Sci. USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin A.M., Freitas E.M., Witt C.S., Christiansen F.T. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics. 2000;51:268–280. doi: 10.1007/s002510050620. [DOI] [PubMed] [Google Scholar]
- 18.Martin M.P., Bashirova A., Traherne J., Trowsdale J., Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J. Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 19.Martin M.P., Single R.M., Wilson M.J., Trowsdale J., Carrington M. KIR haplotypes defined by segregation analysis in 59 Centre d'Etude Polymorphisme Humain (CEPH) families. Immunogenetics. 2008;60:767–774. doi: 10.1007/s00251-008-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu K.C., Liu X.R., Selvakumar A., Mickelson E., O'Reilly R.J., Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J. Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 21.Middleton D., Meenagh A., Gourraud P.A. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59:145–158. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- 22.Norman P.J., Cook M.A., Carey B.S., Carrington C.V., Verity D.H., Hameed K., Ramdath D.D., Chandanayingyong D., Leppert M., Stephens H.A., et al. SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics. 2004;56:225–237. doi: 10.1007/s00251-004-0674-1. [DOI] [PubMed] [Google Scholar]
- 23.Sen S.K., Han K., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey J.A., Liu G., Eichler E.E. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han K., Lee J., Meyer T.J., Wang J., Sen S.K., Srikanta D., Liang P., Batzer M.A. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 2007;3:1939–1949. doi: 10.1371/journal.pgen.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit A.F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 28.Myers S., Freeman C., Auton A., Donnelly P., McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 29.Vilches C., Rajalingam R., Uhrberg M., Gardiner C.M., Young N.T., Parham P. KIR2DL5, a novel killer-cell receptor with a D0-D2 configuration of Ig-like domains. J. Immunol. 2000;164:5797–5804. doi: 10.4049/jimmunol.164.11.5797. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Pascal V., Martin M.P., Carrington M., Anderson S.K. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kummerfeld S.K., Teichmann S.A. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Hamerman J.A., Ogasawara K., Lanier L.L. NK cells in innate immunity. Curr. Opin. Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Han K., Lee J., Meyer T.J., Remedios P., Goodwin L., Batzer M.A. L1 recombination-associated deletions generate human genomic variation. Proc. Natl Acad. Sci. USA. 2008;105:19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 35.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Kelley J., de Bono B., Trowsdale J. IRIS: a database surveying known human immune system genes. Genomics. 2005;85:503–511. doi: 10.1016/j.ygeno.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Bolzer A., Kreth G., Solovei I., Koehler D., Saracoglu K., Fauth C., Muller S., Eils R., Cremer C., Speicher M.R., et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norman P.J., Abi-Rached L., Gendzekhadze K., Hammond J.A., Moesta A.K., Sharma D., Graef T., McQueen K.L., Guethlein L.A., Carrington C.V., et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner D.J., Miretti M., Rajan D., Fiegler H., Carter N.P., Blayney M.L., Beck S., Hurles M.E. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez-Lozano N., Gardiner C.M., Parham P., Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–319. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- 41.Hsu K.C., Chida S., Geraghty D.E., Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol. Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 42.Uhrberg M., Parham P., Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54:221–229. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 43.Martin M.P., Qi Y., Gao X., Yamada E., Martin J.N., Pereyra F., Colombo S., Brown E.E., Shupert W.L., Phair J., et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pascal V., Yamada E., Martin M.P., Alter G., Altfeld M., Metcalf J.A., Baseler M.W., Adelsberger J.W., Carrington M., Anderson S.K., et al. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J. Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 45.Abi-Rached L., Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J. Exp. Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 47.Kimura M., Ota T. The average number of generations until extinction of an individual mutant gene in a finite population. Genetics. 1969;63:701–709. doi: 10.1093/genetics/63.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nathenson S.G., Geliebter J., Pfaffenbach G.M., Zeff R.A. Murine major histocompatibility complex class-I mutants: molecular analysis and structure-function implications. Annu. Rev. Immunol. 1986;4:471–502. doi: 10.1146/annurev.iy.04.040186.002351. [DOI] [PubMed] [Google Scholar]
- 49.Gu W., Zhang F., Lupski J.R. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M.P., Nelson G., Lee J.H., Pellett F., Gao X., Wade J., Wilson M.J., Trowsdale J., Gladman D., Carrington M. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J. Immunol. 2002;169:2818–2822. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]
- 51.Middleton D., Williams F., Halfpenny I.A. KIR genes. Transpl. Immunol. 2005;14:135–142. doi: 10.1016/j.trim.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Mungall A.J., Palmer S.A., Sims S.K., Edwards C.A., Ashurst J.L., Wilming L., Jones M.C., Horton R., Hunt S.E., Scott C.E., et al. The DNA sequence and analysis of human chromosome 6. Nature. 2003;425:805–811. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- 53.Traherne J.A., Horton R., Roberts A.N., Miretti M.M., Hurles M.E., Stewart C.A., Ashurst J.L., Atrazhev A.M., Coggill P., Palmer S., et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.