Abstract
Telomere shortening has been linked to rare human disorders that present with bone marrow failure including Fanconi anemia (FA). FANCC is one of the most commonly mutated FA genes in FA patients and the FANCC subtype tends to have a relatively early onset of bone marrow failure and hematologic malignancies. Here, we studied the role of Fancc in telomere length regulation in mice. Deletion of Fancc (Fancc−/−) did not affect telomerase activity, telomere length or telomeric end-capping in a mouse strain possessing intrinsically long telomeres. However, ablation of Fancc did exacerbate telomere attrition when murine bone marrow cells experienced high cell turnover after serial transplantation. When Fancc−/− mice were crossed into a telomerase reverse transcriptase heterozygous or null background (Tert+/− or Tert−/−) with short telomeres, Fancc deficiency led to an increase in the incidence of telomere sister chromatid exchange. In contrast, these phenotypes were not observed in Tert mutant mice with long telomeres. Our data indicate that Fancc deficiency accelerates telomere shortening during high turnover of hematopoietic cells and promotes telomere recombination initiated by short telomeres.
INTRODUCTION
Telomeres are specialized structures consisting of tandem repeats, TTAGGG in human and mouse, together with telomere-associated proteins to form caps at the ends of linear chromosomes (1,2). Telomeres prevent the recognition of chromosome termini as broken DNA ends and are critical to maintaining genomic stability. Telomere dysfunction, resulting from loss of telomere repeats or loss of protection by telomere-associated proteins, can trigger DNA damage responses, cell apoptosis, cell proliferation defects or genome instability. Telomere dysfunction has also been linked to bone marrow failure syndromes and tumor formation (3,4). Telomerase is essential in telomere length maintenance by replenishing telomere loss due to incomplete DNA replication (1). In mice, deficiency in either telomerase core component, telomerase RNA (Terc) or telomerase reverse transcriptase (Tert) leads to progressive telomere shortening (5–8), which is accompanied by cell proliferation defects and apoptosis in highly proliferating organs including the bone marrow (9–11). Furthermore, Terc or Tert heterozygous mice bred for increasing generations also exhibit progressive telomere shortening and loss of tissue renewal capacity (8,12,13). In humans, mutations in the telomerase components are associated with accelerated telomere shortening and the development of bone marrow failure syndromes, such as dyskeratosis congenita, acquired aplastic anemia and idiopathic pulmonary fibrosis (14). Previous reports show that peripheral blood cells derived from Fanconi anemia (FA) patients have shorter telomeres compared with age-matched healthy donors (15–19), but it is unclear whether telomere attrition in hematopoietic cells from FA patients contributes to the pathogenesis of bone marrow failure in FA.
Telomere shortening is considered a biological clock counting down cellular replicative senescence (20). Cells may overcome this barrier and become immortalized by maintaining telomere length through activation of telomerase and homologous recombination (HR)-mediated pathways (20,21). In human and murine telomerase-deficient cells, short telomeres can initiate HR between telomere sister chromatids, or telomere sister chromatid exchange (T-SCE), by which telomere length is maintained (22–26). Furthermore, short telomeres are also capable of initiating telomere recombination in the presence of telomerase (27). Although it is not entirely clear what molecules regulate telomere recombination, a loss of function in the pathways controlling telomere length maintenance, telomere capping or telomere chromatin can affect telomere recombination. For example, inactivation of Werner (Wrn) protein promotes T-SCEs in spontaneously immortalized telomerase-null mouse embryonic fibroblasts (28). Alterations in telomere capping or epigenetic modifications due to disruption of murine telomere capping proteins (e.g. Pot1 or Trf2 in combination with Ku70) or histone methyltransferases (e.g. Suv4-20h or Suv39h) can also contribute to elevation of T-SCEs (29–32).
FA is an autosomal recessive disorder characterized by cancer susceptibility, bone marrow failure and cellular sensitivity to DNA inter-strand cross-linking agents. To date, 13 FA proteins (FANCA, B, C, D1, D2, E, F, G, I, J, L, M and N) have been identified. Increasing evidence demonstrates that FA proteins play an important role in genome integrity via DNA replication-dependent repair (33,34). Several FA proteins form the FA nuclear core complex, which is required for the monoubiquitination of FANCD2 and FANCI and the localization of FA proteins to chromatin, possibly at the sites of DNA repair. FANCD2 and FANCI function as signal transducers and DNA-processing molecules in a DNA damage response network consisting of ATR, BRCA1 and a RecQ helicase, BLM. Abrogation in any of FA core components disrupts the monoubiquitination of FANCD2 and FANCI. FA proteins may respond to endogenous DNA damage, such as DNA inter-strand cross-links or oxidative DNA damage (35). Whether the FA pathway can respond to dysfunctional telomeres is yet to be determined.
FANCC is one of the most commonly mutated genes in FA patients and is conserved among vertebrates (36). Its encoded protein, FANCC, is 63 kDa in size with no discernable motifs or domains (33,34). FANCC is a member of the FA core complex where it interacts with FANCE and FANCF, and it also associates with the Bloom syndrome protein complex in a supercomplex called BRAFT (33,34,37). In addition to participating in monoubiquitination of FANCD2 and FANCI as part of the core complex, FANCC is also involved in HR pathways (38–40). These observations, together with prior reports showing telomere shortening in FA patients, led us to investigate whether FANCC regulates telomere length and telomere recombination in vivo. In this study, we employed Fancc-deficient mouse models in the strain background with long (C57BL/6 strain) or short telomeres (telomerase reverse transcriptase mutant strain after successive breedings) and wild-type recipient mice that had undergone serial transplantations with Fancc−/− bone marrow cells. Using these genetically modified murine models, we demonstrate that Fancc plays a role in stress-induced telomere attrition and short telomere-initiated recombination in primary murine hematopoietic cells.
RESULTS
Fancc deficiency does not compromise telomeres in a mouse strain with intrinsically long telomeres
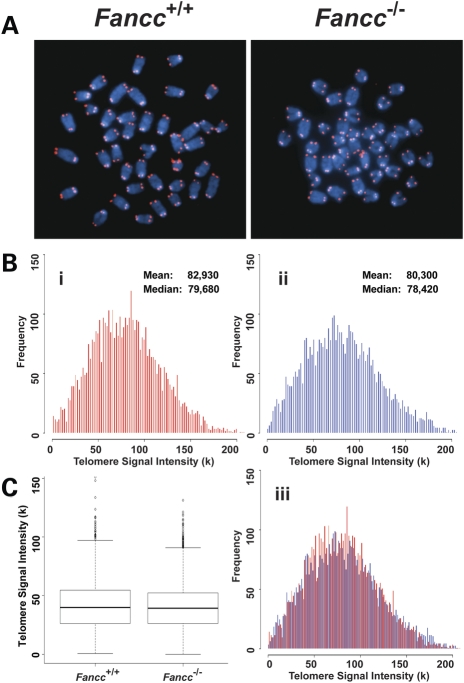
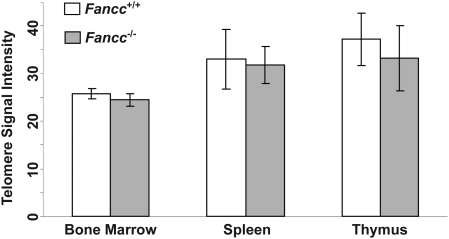
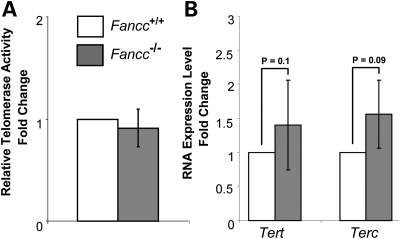
FA patients were reported to harbor short telomeres (15–19). To investigate whether FANCC plays a direct role in telomere length maintenance in vivo, we examined telomere length of wild-type and Fancc−/− mice in the C57BL/6 genetic background, known to have several-fold longer telomeres than humans (41). Hematopoietic cells were isolated from 2- to 4-month-old mice. The mean and median telomere signal intensity as well as the distribution of individual telomere signal intensities were similar between wild-type and Fancc−/− mouse bone marrow cells by Q-FISH analysis (Fig. 1). In addition, the average telomere signal intensity was comparable between wild-type and Fancc−/− bone marrow, spleen and thymus cells via Flow-FISH measurements (Fig. 2). No significant differences in telomerase activity and the expression of the telomerase core components, Tert and Terc, were observed between wild-type and Fancc−/− mouse bone marrow cells (Fig. 3).
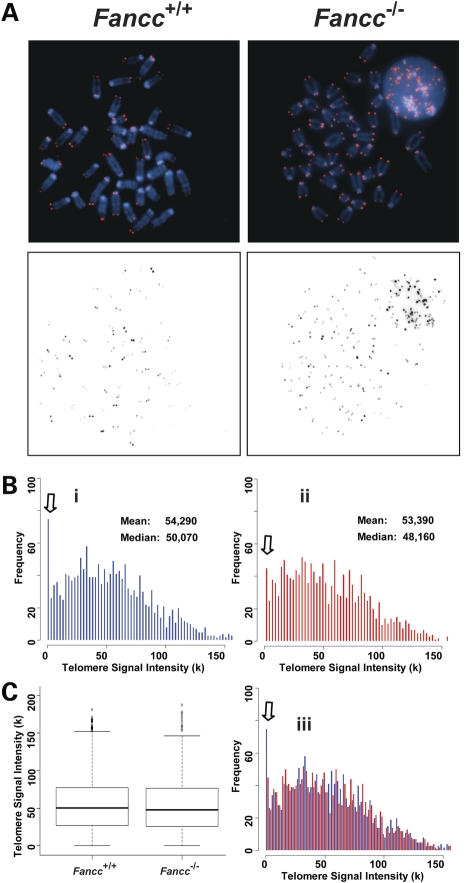
Figure 1.
Telomere lengths are comparable between wild-type and Fancc−/− bone marrow cells derived from C57BL/6 mice. Q-FISH analysis of bone marrow cells derived from wild-type and Fancc−/− mice (n = 6). (A) Representative metaphase spreads of wild-type (Bi) and Fancc−/− (Bii) bone marrow cells showing DAPI staining (blue) and telomere fluorescence signals (red). There was no significant difference in the mean telomere signal intensities and distribution of telomere signal intensities between two genotypes in overlapping histogram (Biii) and box-plot (C).
Figure 2.
Average telomere lengths in wild-type and Fancc−/− hematopoietic cells derived from C57BL/6 mice. Flow-FISH analysis of average telomere signal intensity of bone marrow cells, splenocytes and thymocytes derived from wild-type and Fancc−/− mice. Error bars represent the standard error from different mice of each genotype (n = 5).
Figure 3.
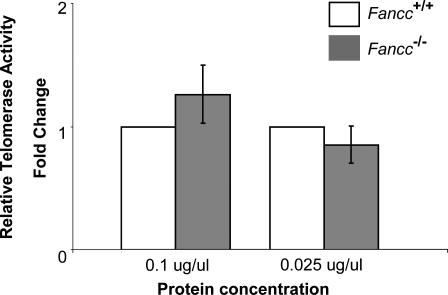
Telomerase activity and Tert and Terc RNA levels in wild-type and Fancc−/− mouse bone marrow cells derived from C57BL/6 mice. qT-PCR analysis indicates a comparable telomerase activity (A), and Tert and Terc RNA levels (B) in wild-type and Fancc−/− mouse bone marrow cells (n = 6). Error bars represent the standard error obtained from different mice of each genotype. P-values are not significant between wild-type and Fancc−/− mouse bone marrow cells.
Accumulating evidence suggest that telomere integrity depends not only on telomere length but also on the proper capping of chromosome ends by telomere-associated proteins and telomere special structures (3,4,42). Therefore, telomeres with normal length do not necessarily reflect that they are functionally capped. To explore whether FANCC plays a direct role in telomeric end-capping in vivo, we examined Fancc−/− bone marrow cells for evidence of telomeric end-capping defects, i.e. chromosome end-to-end fusion and telomere signal-free end (SFE). Fancc−/− bone marrow cells did not exhibit chromosome end-to-end fusions and SFEs (Fig. 1 and Table 1). Furthermore, Fancc−/− bone marrow cells did not display spontaneous chromosomal abnormalities, e.g. chromosome breakages and fragments (Table 1). Together, these results suggest that FANCC does not play a direct role in regulating telomere length, telomerase activity and telomeric end-capping, when telomeres are long and functional.
Table 1.
Frequencies of chromosomal and telomeric abnormalities in bone marrow cells derived from wild-type and Fancc−/− mice in C57BL/6 genetic background and G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− mice
| Cell types | Aneuploidya | Chr. fragments and breaksb | SFEsb |
|---|---|---|---|
| Wild type | 7.9 ± 1.8% | 0% | 0% |
| Fancc−/− | 16 ± 4.3% | 0% | 0% |
| P-value | 0.1842 | NA | NA |
| G2 Tert−/−Fancc+/+ | 21.6 ± 1.7% | 0.42 ± 0.21% | 4.33 ± 1.22% |
| G2 Tert−/−Fancc−/− | 7.2 ± 4.9% | 0.63 ± 0.29% | 0.73 ± 0.32% |
| P-value | 0.07328 | 0.3948 | 0.0024 |
SFE, telomere signal-free end. The data were obtained from wild-type and Fancc−/− mice (n = 6) as well as G2 Tert−/− Fancc+/+ and G2 Tert−/− Fancc−/− mice (n = 6), and more than 50 bone marrow cells from each mouse were scored. P-values were calculated by comparing wild-type with Fancc−/− or G2 Tert+/− Fancc+/+ with Tert+/− Fancc−/− bone marrow cells.
aPercentage of abnormal cells.
bPercentage of abnormal events per chromosome.
Inactivation of Fancc accelerates telomere attrition in serially transplanted bone marrow cells
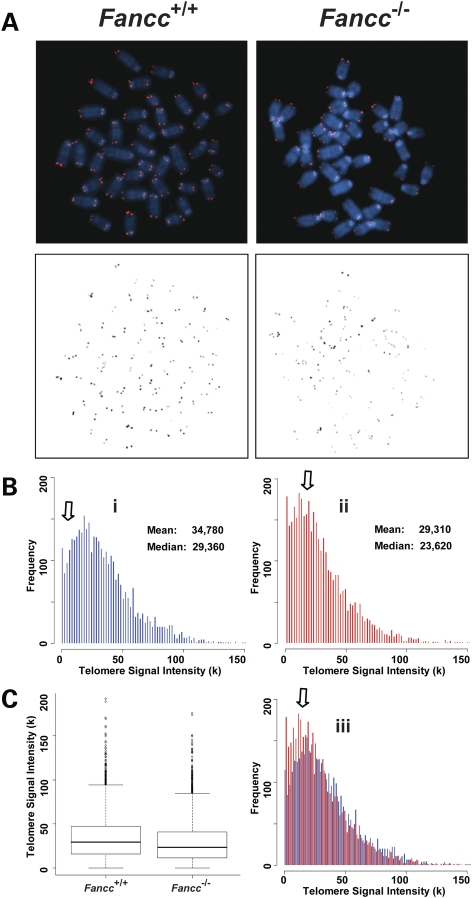
In both humans and mice, telomere dysfunction leads to cell proliferation defects and apoptosis in highly proliferating organs, especially in the bone marrow (9–11,14). Fancc−/− mice do not have obvious bone marrow abnormalities (43). Thus, it is not surprising that the mutant mice do not have any detectable telomere defects. On the other hand, Fancc−/− bone marrow cells display decreased hematopoietic stem cell repopulating ability after primary and secondary transplantations (44,45). It is unclear whether telomere length is altered in Fancc−/− hematopoietic cells during serial bone marrow transplantation and consequently contributes to the decreased repopulating ability of these cells. Utilizing Q-FISH analysis, we examined telomere length of wild-type and Fancc−/− bone marrow cells, which had previously undergone serial transplantation in lethally irradiated secondary recipient mice. Wild-type bone marrow cells from secondary transplant recipients had a decrease in mean and median telomere signal intensity compared with wild-type bone marrow cells from untransplanted mice (mean telomere signal intensity was 34 780 and 82 930, after and before transplantation, respectively) (Figs 1 and 4). Interestingly, Fancc−/− bone marrow cells from secondary transplant recipients had an additional reduction in telomere signal intensity compared with transplanted wild-type cells (Fig. 4). The appearance of chromosome ends with greatly reduced or no detectable telomere signals was increased in Fancc−/− hematopoietic cells from secondary transplant recipients (Table 2). To investigate whether altered telomerase activity during serial transplantation accelerated telomere attrition in Fancc−/− bone marrow cells, we next examined telomerase activity. Deletion of Fancc had no effect on telomerase activity in serially transplanted bone marrow cells (Fig. 5). Furthermore, spontaneous chromosomal abnormalities, including aneuploidy and chromosome breakages, were not significantly different between wild-type and Fancc−/− hematopoietic cells (Table 2). Collectively, these data indicate that Fancc deficiency accelerates telomere attrition during the hematologic stress of serial bone marrow transplantation.
Figure 4.
Accelerated telomere attrition is observed in Fancc−/− bone marrow cells after two serial bone marrow transplantations. Q-FISH analysis of wild-type and Fancc−/− bone marrow cells after two serial bone marrow transplantations (n = 4). (A) Representative metaphase spreads of wild-type and Fancc−/− bone marrow cells showing DAPI staining (upper panel, blue) and telomere fluorescence signals (upper panel, red; lower panel, black). There was a decrease in telomere signal intensities in Fancc−/− cells (Bii) in comparison to wild-type cells (Bi), shown here as shift of the dynamic range in overlapping histogram (Biii) and box-plot (C).
Table 2.
Frequencies of chromosomal abnormalities in wild-type and Fancc−/− bone marrow cells after two series of transplantations
| Cell types | Aneuploidya | Fragment and breaksb | SFEsb |
|---|---|---|---|
| Wild type | 17 ± 6% | 0.77 ± 0.32% | 2.7 ± 1% |
| Fancc−/− | 20.7 ± 4% | 0.48 ± 0.27% | 4.2 ± 0.8% |
| P-value | 0.7034 | 0.3619 | 0.03123 |
The data were obtained from wild-type and Fancc−/− mice (n = 4), and more than 50 bone marrow cells from each mouse were scored.
aPercentage of abnormal cells.
bPercentage of abnormal events per chromosome.
Figure 5.
Telomerase activity in wild-type and Fancc−/− bone marrow cells after two serial bone marrow transplantations. qT-PCR analysis indicates a comparable telomerase activity in serially transplanted wild-type and Fancc−/− mouse bone marrow cells (n = 4). Error bars represent the standard error from different mice of each genotype.
Fancc deficiency promotes short telomere-initiated T-SCE in late generation Tert mutant mice
Although these data suggest that FANCC does not directly control long telomeres, it is unclear whether FANCC regulates short telomeres. A body of evidence suggests that short telomeres can initiate recombinogenic events, including T-SCEs (22–27), even in the presence of telomerase (27). However, the molecular mechanism of short telomere-initiated telomere recombination is not well known. Given that FANCC is involved in HR pathways, we questioned whether FANCC may be involved in HR to resolve endogenous DNA damage, such as critically shortened telomeres. To test this hypothesis, we introduced Fancc+/− into Tert+/− mice (Supplementary Material, Fig. S1). After successive breedings (Supplementary Material, Fig. S1), bone marrow cells from Fancc+/+ and Fancc−/− (HG5 in Tert+/− background and G2 in Tert−/− background) exhibited decrease in overall telomere length and increases in the appearance of chromosome ends with greatly reduced, or non-existent, telomere signals as detected by telomere restriction fragment and Q-FISH analysis (Supplementary Material, Fig. S1, Fig. 6 and Table 1). These mutant mice with short telomeres allowed us to investigate the impact of Fancc deficiency on short telomere-initiated T-SCEs in the presence or absence of telomerase.
Figure 6.
Critically short telomeres are detectable in G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− mutant mice. Q-FISH analysis of bone marrow cells derived from G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− mice (n = 6). Representative metaphase spreads of G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− mouse bone marrow cells showing DAPI staining (upper panel, blue) and telomere fluorescence signals (upper panel, red; lower panel, black). SFEs are detectable in G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− mice (see arrows).
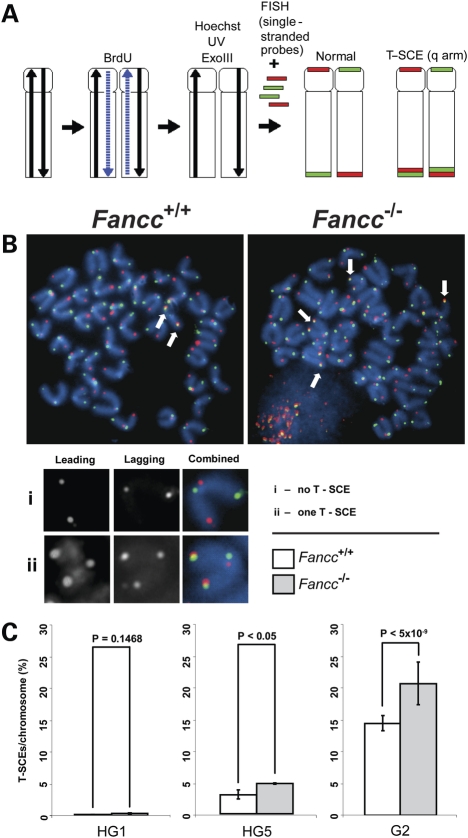
We examined the frequencies of T-SCEs in heterozygous generation 1 (HG1) and HG5 Tert+/− Fancc+/+ and Tert+/− Fancc−/− bone marrow cells via CO-FISH analysis. In HG1 mice with long telomeres (Supplementary Material, Fig. S2), T-SCEs were nearly undetectable in both Tert+/− Fancc+/+ and Tert+/− Fancc−/− mice (Fig. 7). In HG5 mice with short telomeres (Supplementary Material, Fig. S2), higher frequencies of T-SCEs were observed in Tert+/− Fancc−/−, compared with Tert+/− Fancc+/+ (average 5.5% T-SCEs/chromosome in Tert+/− Fancc−/− and 3.5% T-SCEs/chromosome in Tert+/− Fancc+/+) (Fig. 7). T-SCEs further increased in G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− bone marrow cells harboring critically short telomeres (see SFEs in Fig. 6 and Table 1), and the latter displayed more T-SCE events (average 24% T-SCEs/chromosome in Tert−/− Fancc−/− and 13% T-SCEs/chromosome in Tert−/− Fancc+/+) (Fig. 7). Thus, deletion of Fancc leads to elevated T-SCEs in late generation telomerase mutant mice with short telomeres. These observations suggest that short telomeres in late generation telomerase mutant mice become prone to T-SCEs and that inactivation of Fancc promotes T-SCEs.
Figure 7.
Fancc deletion increases the frequencies of T-SCE in late generation Tert mutant mice. CO-FISH analysis of mouse bone marrow cells with indicated genotypes. (A) A schematic representation of CO-FISH procedure. In brief, newly synthesized strands are removed, leaving parental strands to be detected by fluorescent-labeled telomeric C-rich or G-rich probes. A chromosome with more than two telomere signals is considered to be positive for T-SCE. (B) Representative metaphase spreads of G2 Tert−/− Fancc+/+ and Tert−/− Fancc−/− mouse bone marrow cells showing DAPI staining (blue), leading strand telomere fluorescence signals (red) and lagging strand telomere fluorescence signals (green). Arrows indicate T-SCEs. (C) The frequencies of T-SCEs in HG1, HG5 and G2 Fancc+/+ and Fancc−/− mice. Error bars represent standard errors from different mice of each genotype (n = 3).
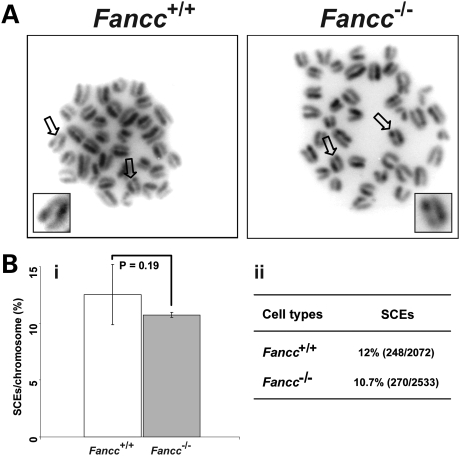
Inactivation of FANCC in chicken DT40 cells results in elevated genome SCEs (40). We hypothesized that inactivation of Fancc in mice would cause spontaneous genome SCEs that in turn contribute to T-SCE events. We examined the frequencies of genome SCEs in HG5 Tert+/− Fancc+/+ and Tert+/− Fancc−/− mouse bone marrow cells, and our data showed that the frequencies of genome SCEs in these mice were comparable (average 12% SCEs/chromosome in Tert+/− Fancc+/+ and 10.7% SCEs/chromosome in Tert+/− Fancc−/−) (Fig. 8). Thus, deletion of Fancc does not affect the rate of spontaneous genome SCEs in primary hematopoietic cells derived from late generation Tert mutant mice.
Figure 8.
The frequencies of genome SCEs are comparable between HG5 Tert+/− Fancc+/+ and Tert+/− Fancc−/− mouse bone marrow cells. (A) Representative metaphase spreads of bone marrow cells with indicated genotypes, showing genome SCE events. (B) (i) The incidences of genome SCEs are comparable between Tert+/− Fancc+/+ and Tert+/− Fancc−/− bone marrow cells (n = 4). Arrows indicate genome SCE events. (ii) The frequencies are derived from the number of SCE events divided by total number of chromosomes (%).
DISCUSSION
In this study, we examined the role of FANCC in telomere length regulation and telomere recombination in vivo. Although deletion of Fancc did not directly affect telomere length or end-capping in a strain with long telomeres, it led to an increase in the incidence of T-SCEs in late generation telomerase mutant mice with short telomeres. These genetic data support the notion that FANCC does not directly regulate long telomeres, but does regulate short telomere-initiated telomere recombination. Thus, ablation of FANCC function may promote telomere recombination. To our knowledge, this is the first study to demonstrate a molecular event that regulates short telomere-initiated telomere recombination in non-transformed murine tissues.
Fancc−/− mice did not show telomere attrition or telomere defects in the C57BL/6 genetic background that has exceedingly longer telomeres than humans. A similar observation was reported in a mouse model deficient in Fancg (46). Interestingly, in an experimental system that dramatically increases the hematopoietic stem and progenitor cell turnover (i.e. serial bone marrow transplantation), Fancc−/− bone marrow cells showed elevated telomere shortening compared with wild-type bone marrow cells, even though telomerase activity was comparable in these cell types. Since Fancc−/− hematopoietic stem and progenitor cells exhibit increased cycling compared with wild-type cells (47,48), it is possible that higher turnover of Fancc−/− bone marrow cells may accelerate telomere shortening, which may, in turn, contribute to decreased Fancc−/− hematopoietic stem cell repopulation ability (44). Thus, telomere shortening may be an indirect consequence of Fancc deficiency.
Late generation Tert−/− mice displayed short telomeres and increased T-SCE events. When Fancc was deleted, the incidence of T-SCEs was exacerbated. These observations suggest that inactivation of FANCC promotes short telomere-initiated T-SCEs. It is possible that Fancc inhibits short telomere-initiated telomere recombination in primary murine hematopoietic cells, but inactivation of Fancc leads to loss of this suppressive mechanism. The exact mechanism of how FANCC suppresses telomere recombination is unclear. FANCC does not directly bind to DNA (49), and it may thus regulate proteins or pathways that are involved in telomere recombination. As a component of the FA core complex, FANCC regulates the monoubiquitination of FANCD2. A previous report demonstrates that the modified FANCD2 localizes to telomeres in an immortalized cell line that maintains telomeres through HR-based mechanisms; however, depletion of FANCD2 causes increased short telomeres and decreased T-SCEs in this line (50). These telomere phenotypes are in contrast to the observations in HG5 Tert+/− Fancc−/− and G2 Tert−/− Fancc−/− mice. A possible explanation for these apparent inconsistencies could be that FA proteins may not only serve as checkpoint proteins that prevent primary cells from the engagement of illegitimate telomere recombination, but also regulate the maintenance of telomere recombination to keep the shortest telomeres intact in immortalized cells. Alternatively, these discrepant findings may be because the former study employed an immortalized cell line that had already escaped checkpoints for the illegitimate telomere recombination, whereas our studies utilized primary hematopoietic cells with intact checkpoints for illegitimate telomere recombination. FANCC also associates with the Bloom syndrome protein complex (37). It has been shown that the frequencies of spontaneous SCEs in fancc and blm double-mutant chicken DT40 cells are similar to those in blm single mutant (40). These studies suggest a functional linkage between FANCC and BLM in a common pathway to suppress SCE. Deletion of Fancc leads to reduced levels of Blm in mice (A.N. Suhasini and R.M. Brosh, personal communication). Decreased Blm levels may thus serve a role in promoting telomere recombination in Fancc and Tert double-mutant mice. Although ablation of another member of RecQ helicase proteins, Wrn, also results in elevated T-SCEs in late generation telomerase-null mice (28), the Wrn and telomerase double null cells display elevated incidence of critically short telomeres (28). In contrast, Fancc deficiency reduces the incidence of critically short telomeres in late generation telomerase-null mice (Table 1). These observations do not support the involvement of Wrn in Fancc-regulated telomere recombination in mice. However, deletion of fancc in chicken DT40 cells can lead to elevated spontaneous SCEs that depend upon a key HR protein, the RAD51 paralog, XRCC3 (40). In addition, ablation of murine telomere capping proteins (e.g. Pot1 or Trf2 in combination with Ku70) and histone methyltransferases (e.g. Suv4-20h or Suv39h) can cause elevated T-SCEs (29–32). It is unknown whether FANCC regulates XRCC3, telomere-binding proteins or histone methyltransferases in telomere recombination.
FA patients without functional FA proteins are predisposed to bone marrow failure and malignancies (33,34). FANCC may therefore control cell viability and immortalization of primary cells, including bone marrow cells, by safeguarding genome stability. Telomere dysfunction triggers cell apoptosis and genomic instability preferentially in highly proliferating organs, e.g. bone marrow. In rare events, cells may overcome this barrier and become immortalized by activating the pathway involved in maintaining telomere length (21). Increasing evidence suggests that short telomeres initiate telomere recombination likely due to a dysfunction in the highly regulated mechanisms controlling HR. We have shown that inactivation of FANCC facilitates short telomere-initiated telomere recombination. In addition, Fancc deficiency can accelerate telomere shortening during high hematopoietic cell turnover. It is possible that FANCC may function to control cell viability and immortalization of primary cells by influencing telomere attrition and telomere recombination.
MATERIALS AND METHODS
Mice
Fancc and Tert knockout mice (Fancc−/−and Tert−/−) in C57BL/6 genetic background were produced as described previously (7,8,43,44). Mouse breeding strategy is illustrated in Supplementary Material, Figure S1. In brief, Tert and Fancc heterozygous mice (Tert+/− Fancc+/−) were generated by interbreeding Tert+/− and Fancc+/− mice and were named as HG1 mice. HG1 mice from separate mating events were mated to obtain HG2 mice, which were crossed again until generation 5 (HG5). Tert−/− Fancc+/− mice were generated from HG5 Tert+/− Fancc+/− breeders and were named as G1 mice. G1 mice from separate mating events were mated to obtain G2. All animal experiments were carried out according to the ‘Guide for the Care and Use of Laboratory Animals’ (National Academy Press, USA, 1996) and were approved by the NIA IACUC.
Serial mouse bone marrow transplantation
Bone marrow was flushed from tibias and femurs of experimental mice and low-density mononuclear cells were prepared by density centrifugation (ficoll-hypaque density 1.119, Sigma, St Louis, MO, USA). Primary and secondary transplants were conducted as described previously with slight modification (44,45). Briefly, low-density mononuclear bone marrow cells (2 × 106 cells) were resuspended in 200 μl IMDM supplemented with 20% fetal bovine serum (Biowhittaker, Walkersville, MD, USA). Cells were transplanted into congenic lethally irradiated B6.SJL-PtrcaPep3b/BoyJ recipient mice obtained from the Stem Cell Transplant Mouse Core in the Indiana University Cancer Center. Bone marrow from primary recipients was harvested 12 months after transplantation and prepared for secondary transplantation. Secondary transplants were conducted exactly the same as primary transplants. Four months after secondary transplantation, bone marrow was harvested for telomere studies.
Telomere length measurements
Flow-FISH
The average telomere fluorescence in splenocytes, thymocytes and bone marrow cells was scored for each mouse and the data were pooled from the indicated number of mice in each genotype and measured according to the previously published protocol with minor modifications (51). A telomere-specific FITC-conjugated (CCCTAA)3 PNA probe (0.3 μg/ml, Panagene) was used.
Q-FISH
Mice were injected with 100 μl of 0.5% colchicine intraperitoneally for ∼30 min before being scarified. Bone marrow cells were then collected by flushing 1 ml of phosphate-buffered saline from femurs. Collected bone marrow cells were immediately incubated in 0.075 m KCl for 15 min in 37°C, followed by fixation in ice-cold (3:1) methanol and glacial acetic acid. Metaphase spreads were then hybridized with a Cy3-labeled PNA (CCCTAA)3 probe (0.3 μg/ml, Panagene) and counterstained with 4,6-diamidino-2-phenylindole as described previously (52). Images were captured using Cytovision™ software (Applied Imaging Corp.) on a fluorescence microscope (Axio2; Carl Zeiss, Germany), followed by quantification of telomere fluorescence signals using the TFL-Telo software (a kind gift from P. Lansdorp, Vancouver, BC, USA). For histograms and box-plots, data from different mice of each genotype were pooled and scored and R statistical package (http://www.r-project.org) along with R.utils package and Biobase package (http://www.brajul.com/R) was used. The frequencies of telomeres within a given range of telomere signal intensities were plotted against the telomere signal intensity using arbitrary units. Metaphases from different mice of each genotype were scored for chromosomal and telomeric abnormalities (i.e. frequencies of cells with more or less than 40 chromosomes, frequencies of chromosomal fragmentations and breakages, and frequencies of chromosome ends with no detectable telomere signals) as described previously (26,53).
Telomere restriction fragment analysis
The analysis was carried out as described by Hemann and Greider (54). Approximately 1 × 106 mouse bone marrow cells were embedded in agarose plugs. DNA was digested with DnpII (BioLabs) and electrophoresed through 1% w/v pulsed-field grade agarose (Bio-Rad, CA, USA) in 1× Tris–acetate–EDTA buffer. Electrophoresis was carried out in a CHEF DR-III pulsed-field apparatus (Bio-Rad) at 14°C with 3 V/cm and a switch time of 10 s for 48 h. The gel was denatured, dried, probed with 32P-labeled (AATCCC)4 probe and visualized by autoradiography.
Measurement of T-SCE and genome SCE
Chromosome orientation FISH (CO-FISH) was used to measure the frequency of T-SCE (22). The measurement for genome SCEs was carried out as described previously (26). Briefly, bone marrow cells were flushed from femurs and tibias and cultured with Iscove's modified Dulbecco's medium (IBCO-BRL, Gaithersburg, MD, USA) supplemented with 20% fetal calf serum (Hyclone, Logan, UT, USA) in the presence of interleukin 6 (200 U/ml) and stem cell factor (100 ng/ml; Peprotech, Rocky Hill, NJ, USA) (44). Bone marrow cells were subcultured in medium containing a 3:1 ratio of BrdU/BrdC (Sigma) at a final concentration of 1 × 10−5 m and collected around 12 or 24 h for detecting T-SCE or SCE, respectively. Colcemid (0.1 μg/ml) was added 2 h before harvest. Metaphase spreads were then stained with Hoechst 33258, exposed to UV light and digested with exonuclease III to remove newly synthesized DNA strands. Hybridization and wash conditions were identical to those described for telomere FISH (52). A chromosome with more than two telomeric DNA signals by FITC-labeled (CCCTAA)3 or Cy3-labeled (TTAGGG)3 PNA probes (0.3 μg/ml, Panagene) was scored as T-SCE-positive. A SCE was scored each time a color switch between dark or light sister chromatids occurred. The frequencies of T-SCEs and SCEs were obtained from different mice of each genotype.
Analysis of telomerase activity and Tert and Terc levels
Telomerase activity was measured by using Biomax Telomerase detection kit (Biomax, Inc., MD, USA) according to the manufacturer's recommendations. Briefly, freshly isolated bone marrow cells were lysed, and the cell extracts were then added to a pre-mix for quantitative telomerase activity in a real-time PCR. For detecting Tert and Terc levels, total RNA was extracted from mouse bone marrow using RNeasy kit (Qiagene) and then reverse transcribed with random hexamers by the SuperScript III first-strand synthesis system for RT–PCR (Invitrogen Life Technologies). The PCR included cDNA reaction, 2× Sybr master mix (Bio-Rad), and a set of primers for mouse Tert, Terc or Beta-Actin (Supplementary Material, Fig. S3). The levels of Tert and Terc were normalized to that of Beta-Actin. MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) was used to conduct the reaction where each sample was done in triplicates and performed according to the manufacturer's instructions. Tert-deficient mouse bone marrow cell extracts or cDNA were used as a control. Relative telomerase activity was expressed as log of CT value. Relative Tert and Terc RNA levels were normalized to Beta-Actin and expressed using the comparative CT method according to the manufacturer's instructions.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by the Intramural Research Program of the NIA, National Institutes of Health (Y.L.), U.S. Public Health Services Grants R01 HL077175 (L.H.), P30 CA82709 (L.H.), T32 DK07519 (M.M.) and the Riley Children's Foundation (L.H.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Weidong Wang, Vilhelm Bohr, Robert Brosh, Michael Seidman and Richard Hodes for critical discussion and comments on the manuscript, and Dr. Shengyuan Luo and his team for genotyping animals.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Greider C.W. Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harb. Symp. Quant. Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- 3.d'Adda di Fagagna F., Teo S.H., Jackson S.P. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 5.Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 6.Yuan X., Ishibashi S., Hatakeyama S., Saito M., Nakayama J., Nikaido R., Haruyama T., Watanabe Y., Iwata H., Iida M., et al. Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice. Genes Cells. 1999;4:563–572. doi: 10.1046/j.1365-2443.1999.00284.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Snow B.E., Hande M.P., Yeung D., Erdmann N.J., Wakeham A., Itie A., Siderovski D.P., Lansdorp P.M., Robinson M.O., et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 8.Erdmann N., Liu Y., Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6080–6085. doi: 10.1073/pnas.0401580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H.W., Blasco M.A., Gottlieb G.J., Horner J.W., 2nd, Greider C.W., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 10.Rudolph K.L., Chang S., Lee H.W., Blasco M., Gottlieb G.J., Greider C., DePinho R.A. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 11.Herrera E., Samper E., Martin-Caballero J., Flores J.M., Lee H.W., Blasco M.A. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hathcock K.S., Hemann M.T., Opperman K.K., Strong M.A., Greider C.W., Hodes R.J. Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3591–3596. doi: 10.1073/pnas.012549799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao L.Y., Armanios M., Strong M.A., Karim B., Feldser D.M., Huso D., Greider C.W. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005;123:1121–1131. doi: 10.1016/j.cell.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Savage S.A., Alter B.P. The role of telomere biology in bone marrow failure and other disorders. Mech. Ageing Dev. 2008;129:35–47. doi: 10.1016/j.mad.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball S.E., Gibson F.M., Rizzo S., Tooze J.A., Marsh J.C., Gordon-Smith E.C. Progressive telomere shortening in aplastic anemia. Blood. 1998;91:3582–3592. [PubMed] [Google Scholar]
- 16.Leteurtre F., Li X., Guardiola P., Le Roux G., Sergere J.C., Richard P., Carosella E.D., Gluckman E. Accelerated telomere shortening and telomerase activation in Fanconi's anaemia. Br. J. Haematol. 1999;105:883–893. doi: 10.1046/j.1365-2141.1999.01445.x. [DOI] [PubMed] [Google Scholar]
- 17.Hanson H., Mathew C.G., Docherty Z., Mackie Ogilvie C. Telomere shortening in Fanconi anaemia demonstrated by a direct FISH approach. Cytogenet. Cell Genet. 2001;93:203–206. doi: 10.1159/000056985. [DOI] [PubMed] [Google Scholar]
- 18.Callen E., Samper E., Ramirez M.J., Creus A., Marcos R., Ortega J.J., Olive T., Badell I., Blasco M.A., Surralles J. Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Hum. Mol. Genet. 2002;11:439–444. doi: 10.1093/hmg/11.4.439. [DOI] [PubMed] [Google Scholar]
- 19.Alter B.P., Baerlocher G.M., Savage S.A., Chanock S.J., Weksler B.B., Willner J.P., Peters J.A., Giri N., Lansdorp P.M. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shay J.W. Telomerase in human development and cancer. J. Cell. Physiol. 1997;173:266–270. doi: 10.1002/(SICI)1097-4652(199711)173:2<266::AID-JCP33>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Cesare A.J., Reddel R.R. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Bailey S.M., Brenneman M.A., Goodwin E.H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bechter O.E., Shay J.W., Wright W.E. The frequency of homologous recombination in human ALT cells. Cell Cycle. 2004;3:547–549. [PubMed] [Google Scholar]
- 24.Londono-Vallejo J.A., Der-Sarkissian H., Cazes L., Bacchetti S., Reddel R.R. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Giannone R.J., Liu Y. Telomere sister chromatid exchange in telomerase deficient murine cells. Cell Cycle. 2005;4:1320–1322. doi: 10.4161/cc.4.10.2075. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Erdmann N., Giannone R.J., Wu J., Gomez M., Liu Y. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10256–10260. doi: 10.1073/pnas.0504635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrish T.A., Greider C.W. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 2009;5:e1000357. doi: 10.1371/journal.pgen.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laud P.R., Multani A.S., Bailey S.M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R.A., Chang S. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Celli G.B., Denchi E.L., de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 30.Wu L., Multani A.S., He H., Cosme-Blanco W., Deng Y., Deng J.M., Bachilo O., Pathak S., Tahara H., Bailey S.M., et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 31.He H., Multani A.S., Cosme-Blanco W., Tahara H., Ma J., Pathak S., Deng Y., Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benetti R., Gonzalo S., Jaco I., Schotta G., Klatt P., Jenuwein T., Blasco M.A. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel K.J., Joenje H. Fanconi anemia and DNA replication repair. DNA Repair. 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 35.Pang Q., Andreassen P.R. Fanconi anemia proteins and endogenous stresses. Mutat. Res. 2009;668:42–53. doi: 10.1016/j.mrfmmm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ameziane N., Errami A., Leveille F., Fontaine C., de Vries Y., van Spaendonk R.M., de Winter J.P., Pals G., Joenje H. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum. Mutat. 2008;29:159–166. doi: 10.1002/humu.20625. [DOI] [PubMed] [Google Scholar]
- 37.Meetei A.R., Sechi S., Wallisch M., Yang D., Young M.K., Joenje H., Hoatlin M.E., Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niedzwiedz W., Mosedale G., Johnson M., Ong C.Y., Pace P., Patel K.J. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 40.Hirano S., Yamamoto K., Ishiai M., Yamazoe M., Seki M., Matsushita N., Ohzeki M., Yamashita Y.M., Arakawa H., Buerstedde J.M., et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemann M.T., Greider C.W. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blasco M.A. Telomere length, stem cells and aging. Nat. Chem. Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 43.Chen M., Tomkins D.J., Auerbach W., McKerlie C., Youssoufian H., Liu L., Gan O., Carreau M., Auerbach A., Groves T., et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat. Genet. 1996;12:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 44.Haneline L.S., Gobbett T.A., Ramani R., Carreau M., Buchwald M., Yoder M.C., Clapp D.W. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94:1–8. [PubMed] [Google Scholar]
- 45.Haneline L.S., Li X., Ciccone S.L., Hong P., Yang Y., Broxmeyer H.E., Lee S.H., Orazi A., Srour E.F., Clapp D.W. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc−/− hematopoietic stem cells and decreases the risk of clonal evolution. Blood. 2003;101:1299–1307. doi: 10.1182/blood-2002-08-2404. [DOI] [PubMed] [Google Scholar]
- 46.Franco S., van de Vrugt H.J., Fernandez P., Aracil M., Arwert F., Blasco M.A. Telomere dynamics in Fancg-deficient mouse and human cells. Blood. 2004;104:3927–3935. doi: 10.1182/blood-2003-10-3626. [DOI] [PubMed] [Google Scholar]
- 47.Haneline L.S., Broxmeyer H.E., Cooper S., Hangoc G., Carreau M., Buchwald M., Clapp D.W. Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from Fac−/− mice. Blood. 1998;91:4092–4098. [PubMed] [Google Scholar]
- 48.Li X., Plett P.A., Yang Y., Hong P., Freie B., Srour E.F., Orschell C.M., Clapp D.W., Haneline L.S. Fanconi anemia type C-deficient hematopoietic stem/progenitor cells exhibit aberrant cell cycle control. Blood. 2003;102:2081–2084. doi: 10.1182/blood-2003-02-0536. [DOI] [PubMed] [Google Scholar]
- 49.Kruyt F.A., Hoshino T., Liu J.M., Joseph P., Jaiswal A.K., Youssoufian H. Abnormal microsomal detoxification implicated in Fanconi anemia group C by interaction of the FAC protein with NADPH cytochrome P450 reductase. Blood. 1998;92:3050–3056. [PubMed] [Google Scholar]
- 50.Fan Q., Zhang F., Barrett B., Ren K., Andreassen P.R. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 2009;37:1740–1754. doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rufer N., Dragowska W., Thornbury G., Roosnek E., Lansdorp P.M. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat. Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 52.Zijlmans J.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez M., Wu J., Schreiber V., Dunlap J., Dantzer F., Wang Y., Liu Y. PARP1 is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemann M.T., Greider C.W. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.