Abstract
Juvenile Batten disease is an autosomal recessive pediatric neurodegenerative disorder caused by mutations in the CLN3 gene. The CLN3 protein primarily resides in the lysosomal membrane, but its function is unknown. We demonstrate that CLN3 interacts with SBDS, the protein mutated in Shwachman–Bodian–Diamond syndrome patients. We demonstrate that this protein–protein interaction is conserved between Btn1p and Sdo1p, the respective yeast Saccharomyces cerevisiae orthologs of CLN3 and SBDS. It was previously shown that deletion of BTN1 results in alterations in vacuolar pH and vacuolar (H+)-ATPase (V-ATPase)-dependent H+ transport and ATP hydrolysis. Here, we report that an SDO1 deletion strain has decreased vacuolar pH and V-ATPase-dependent H+ transport and ATP hydrolysis. These alterations result from decreased V-ATPase subunit expression. Overexpression of BTN1 or the presence of ionophore carbonyl cyanide m-chlorophenil hydrazone (CCCP) causes decreased growth in yeast lacking SDO1. In fact, in normal cells, overexpression of BTN1 mirrors the effect of CCCP, with both resulting in increased vacuolar pH due to alterations in the coupling of V-ATPase-dependent H+ transport and ATP hydrolysis. Thus, we propose that Sdo1p and SBDS work to regulate Btn1p and CLN3, respectively. This report highlights a novel mechanism for controlling vacuole/lysosome homeostasis by the ribosome maturation pathway that may contribute to the cellular abnormalities associated with juvenile Batten disease and Shwachman–Bodian–Diamond syndrome.
INTRODUCTION
Juvenile neuronal ceroid lipofuscinosis (JNCL), or Batten disease, is a fatal pediatric neurodegenerative disease, characterized pathologically by the premature accumulation of hydrophobic autofluorescent storage material within patient cells. JNCL is an autosomal recessive disorder caused by mutations in the CLN3 gene (1). Although the CLN3 protein has been reported to be in several subcellular locations, it is primarily thought to be an endosomal and lysosomal resident protein (2–4). Despite many studies, the precise function of CLN3 is unknown.
Both Schizosaccharomyces pombe and Saccharomyces cerevisiae yeast models have been developed to study the primordial cellular function of CLN3. In both yeast models, human CLN3 complements deletion of the orthologous gene, BTN1, indicating a trans-kingdom conservation of the protein function, and yeast Btn1p resides in the vacuole, the analogous organelle to the lysosome (5–9). Like CLN3, Btn1p has been implicated in several cellular processes. In S. pombe, Btn1p is implicated in cell-cycle progression, vacuolar and endocytic pH regulation and Golgi homeostasis and protein sorting (8,10,11). In the S. cerevisiae model, Btn1p has been linked to vacuolar arginine transport, ion homeostasis, nitric oxide synthesis and vacuolar pH regulation and vacuolar (H+)-ATPase (V-ATPase) coupling (6,12–16). However, these observations are based on studies characterizing cellular effects resulting from the loss of Btn1p. Thus, it is unclear whether the observed phenomena are direct consequences resulting from the absence of Btn1p, or whether they are the cells’ attempt to compensate for the lack of Btn1p.
In order to gain insights into CLN3 function, a yeast two-hybrid screen was performed to identify proteins that interact with CLN3. CLN3 has five or six transmembrane regions, with the predicted lumenal and cytosolic portions of the protein being used in this screen (17,18). We subsequently identified an interaction between the C-terminus of CLN3 and Shwachman–Bodian–Diamond syndrome protein (SBDS). Mutations in SBDS are associated with the inherited pediatric disorder Shwachman–Bodian–Diamond syndrome, which is characterized by bone marrow and pancreatic dysfunction, mild mental retardation and hematologic abnormalities with an increased risk of leukemia (19–23). Depletion of SBDS via small hairpin RNA interference in cells resulted in altered expression of genes involved in brain development, bone morphogenesis, blood cell proliferation and cell adhesion (24).
Like CLN3, SBDS is expressed in many tissues, including the brain, and both CLN3 and SBDS have been proposed to function in multiple cellular processes (25). However, most evidence supports a role for SBDS in ribosomal maturation (24,26,27). Specifically, Sdo1p, the yeast homolog to SBDS, was characterized to work in tandem with Efl1p and the shuttling factor Tif6p in the export of the ribosome from the nucleus (26). We have verified that the interaction between SBDS and CLN3 is conserved in S. cerevisiae. Several phenotypes in an SDO1 deletion strain suggest that the absence of SDO1 causes aberrant regulation of Btn1p. Our data support the concept that portions of the ribosome maturation pathway surveys vacuolar function, presumably as a means to adjust protein levels for optimal cellular homeostasis.
RESULTS
CLN3 and SBDS interact
In order to identify CLN3 protein–protein interactions, the C-terminus of CLN3 was screened against a human fetal brain library using Stratagene's Cytotrap yeast two-hybrid method (Supplementary Material, Fig. S1). In this system, the yeast cdc25h strain is temperature sensitive at 37°C due to a mutation in the hSos homolog CDC25, which is required for the activation of the Ras pathway for proper cell-cycle progression. An interaction between CLN3, which was tagged with hSOS, and a library protein, which was myristylated, caused hSos to be translocated to the membrane because of myristylation of the library gene product. As a result, hSos activated the Ras pathway, allowing the cdc25h strain to grow at 37°C. Both genes were under a galactose-inducible promoter, which allowed for expression in the presence of galactose and the absence of glucose.
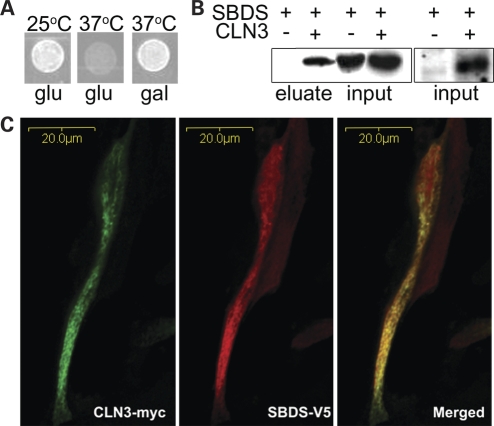
A protein–protein interaction between CLN3 and SBDS was revealed in this screen when the yeast was able to grow at the non-permissible temperature in the presence of galactose (Fig. 1A). The library DNA was identified to encode the N-terminus of SBDS (Supplementary Material, Fig. S2). This interaction was verified by co-immunoprecipitation and co-localization (Fig. 1B and C). CLN3 and SBDS were co-expressed in NIH/3T3 cells with C-terminal c-myc and V5 tags, respectively. CLN3 was immunoprecipitated from the cell lysate using an anti-myc antibody. The resulting eluate was immunoblotted for V5, which showed that SBDS immunopurified with CLN3 (Fig. 1B). Using the same expression system, CLN3 and SBDS were co-expressed in NIH/3T3 cells grown on coated coverglass and were incubated with anti-myc and anti-V5 antibodies and corresponding Alexa fluorescent secondary antibodies. Cells visualized using confocal microscopy show that CLN3 and SBDS partially co-localize (Fig. 1C).
Figure 1.
Human CLN3 and SBDS interact. (A) Cytotrap yeast two-hybrid results showing the interaction-dependent suppression of Cdc25H temperature sensitivity at 37°C upon the galactose-dependent induction of CLN3 and SBDS. (B) Immunoblot using mouse anti-V5 to detect SBDS. SBDS is detectable in the eluate only when CLN3 is present. (C) Confocal microscopy of mouse NIH/3T3 cells co-expressing CLN3-MYC and SBDS-V5. CLN3-MYC and SBDS-V5 partially co-localize.
CLN3–SBDS interaction is conserved in yeast
The yeast S. cerevisiae has orthologs to both CLN3 and SBDS. Human CLN3 is 39% identical and 59% similar in amino acid sequence to yeast Btn1p, and CLN3 complements yeast lacking Btn1p, btn1-Δ (6,7,28). Similarly, human SBDS is 40% identical and 61% similar to yeast Sdo1p.
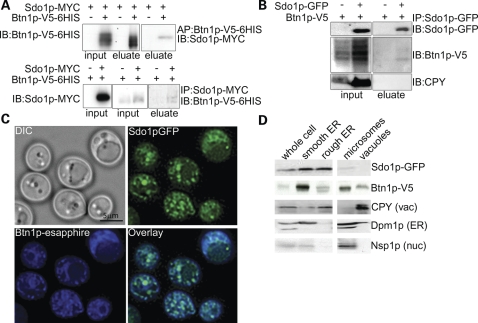
A Btn1p-V5-6HIS construct was used to affinity-purify Btn1p via a cobalt column. The eluate was immunoblotted for Sdo1p-MYC. Sdo1p-MYC was only detectable in eluate containing Btn1p-V5 (Fig. 2A). Next, we immunopurified Sdo1p-MYC and immunoblotted the eluate for Btn1p-V5. Btn1p-V5 was only detectable in eluate containing Sdo1p-MYC (Fig. 2A).
Figure 2.
Yeast Btn1p and Sdo1p interact. (A) Affinity- and immunopurification of Btn1p-V5-6HIS. Sdo1p is detectable only in eluate containing Btn1p-V5-6HIS when Btn1p-V5-6HIS is purified. Btn1p-V5 is detectable only in eluate containing Sdo1p-MYC when Sdo1p-MYC is purified. (B) Co-immunoprecipitation of endogenously expressed Sdo1p and Btn1p from a Golgi and endosome-enriched fractions. Btn1p-V5 was detected only if Sdo1p-GFP was present. (C) Epifluorescent micrographs of endogenously expressed Btn1p-eSapphire and Sdo1p-GFP. Sdo1p and Btn1p co-localize to punctate structures in the cell. (D) Enrichment of organelle fractions by differential centrifugation of cells endogenously expressing Sdo1p-GFP and Btn1p-V5. Sdo1p and Btn1p co-fractionate in the ER and microsome-enriched fractions.
Although the interaction was detected whether Btn1p or Sdo1p was purified, we were concerned that overexpression of the two proteins might lead to a false-positive interaction, especially because of the hydrophobicity of Btn1p. This necessitated confirming the interaction at the endogenous level. Because of the low abundance of Btn1p, endogenous levels of that protein are only detectable upon enrichment of specific cellular fractions. Thus, we performed co-immunoprecipitation experiments by purifying intrachromasomally green fluorescent protein (GFP)-tagged Sdo1p from specific organelle-enriched fractions and by detecting functional Btn1p-V5, which was expressed via its own promoter (Supplementary Material, Fig. S3). Btn1p was only detected in the eluate from the endosomes and Golgi-enriched fraction that contained Sdo1p (Fig. 2B). In addition, soluble carboxypeptidase Y (CPY) and membrane-bound Pep12p, which are not reported to interact with Sdo1p or Btn1p but are found in the endomembrane system, did not co-immunopurify with Sdo1p, further validating the specificity of the Sdo1p–Btn1p interaction (Fig. 2B; Supplementary Material, Fig. S4).
Next, we asked whether the two proteins co-localize, which would indicate that the proteins have the ability to interact at the physiological level. Sdo1p and Btn1p partially co-localized to punctate spots in cells expressing endogenous levels of Sdo1p-GFP and Btn1p-eSapphire (Fig. 2C). Using an overexpression system, the majority of Btn1p was reported to localize to the vacuole (9,29). Here, Btn1p can be seen in punctate spots throughout the cell, in addition to the vacuole. Only a small portion of Btn1p and Sdo1p interacts, and the identity of the punctate spots where the co-localization occurred is unknown. Co-fractionation confirmed that Sdo1p and Btn1p were detectable in common fractions (Fig. 2D). We suspect that the interaction between Btn1p and Sdo1p does not occur at the vacuole, where Btn1p is thought to localize, because Sdo1p was not enriched in the vacuole fraction. Our Sdo1p localization results were consistent with previously published Sdo1p localization studies, which show that Sdo1p is found in the cytosol, as well as the nucleus (26,30). In addition, we observed that Sdo1p-GFP not only co-localizes with Hoechst stain, a DNA marker, but is also found in other areas of the cell (Supplementary Material, Fig. S5). We conclude that portions of the cellular pools of both Btn1p and Sdo1p interact, and therefore that the protein–protein interaction between CLN3 and SBDS is evolutionarily conserved.
The sdo1-Δ/sdo1-Δ deletion strain
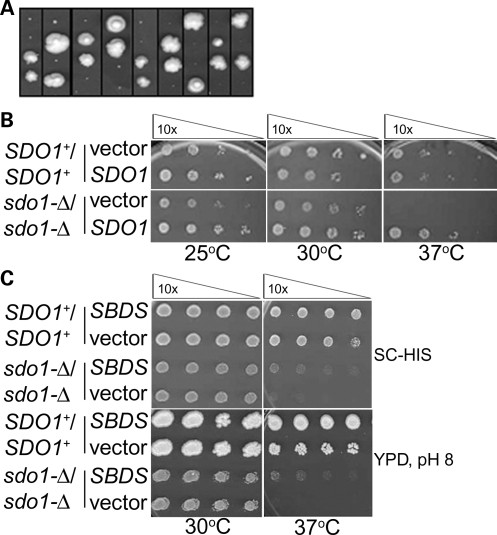
A diploid SDO1 deletion strain (sdo1-Δ/sdo1-Δ) was created in order to study the consequences resulting from the loss of Sdo1p. It was previously shown that a haploid deletion of SDO1 resulted in a viable strain that exhibited a slow-growth phenotype (26). This result was recapitulated here (Fig. 3A). The sdo1-Δ/sdo1-Δ diploid exhibited a slow-growth phenotype similar to that of the haploid deletion, and this phenotype was complemented by plasmid-borne expression of SDO1 (Fig. 3B). Yeast can accumulate mitochondrial DNA mutations (ρ-), which cause a petite phenotype that also manifests as slow growth similar to that seen for the sdo1-Δ/sdo1-Δ strain. However, sdo1-Δ/sdo1-Δ was viable when grown on a non-fermentable carbon source, indicating that the slow-growth phenotype was due specifically to the absence of SDO1, not to mitochondrial dysfunction (Supplementary Material, Fig. S6).
Figure 3.
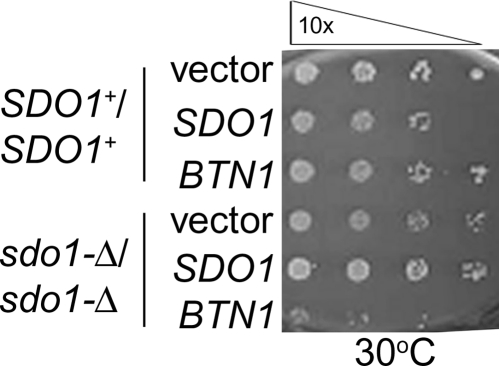
The SDO1 deletion exhibits a slow-growth phenotype that is partially complemented by SBDS. (A) Tetrad analysis of SDO1/sdo1-Δ. The SDO1+/sdo1-Δ diploid strain (Open Biosystems; B4741/B4742) was sporulated and dissected (42). Each column represents the colonies arising from the four tetrad spores. From each tetrad, the two pinpoint colonies were confirmed to be sdo1-Δ, and the two larger colonies were confirmed to be SDO1+ by PCR genotyping. (B) Serial dilutions of SDO1+/SDO1+ (Open Biosystems; B4741/B4742) and sdo1-Δ/sdo1-Δ derived from the B4741/B4742 parental strain with vector or expressing SDO1. Mid-log phase cells grown in selective media were diluted 10-fold with water in each column and plated using a 36-pin replicator. Slow growth and temperature sensitivity in sdo1-Δ/sdo1-Δ are complemented by SDO1. (C) Serial dilutions of parental SDO1+/SDO1+ (Open Biosystems; B4741/B4742) and sdo1-Δ/sdo1-Δ cells of the same background with either vector control or expressing SBDS. Cells were grown in selective media and plated as described earlier on either synthetic complete glucose-histidine (SC-his) or rich media at pH 8. Human SBDS partially complements sdo1-Δ/sdo1-Δ.
We sought to complement the sdo1-Δ/sdo1-Δ slow-growth phenotype with expression of human SBDS to determine whether Sdo1p and SBDS are functional homologs. Accordingly, SBDS expression partially complemented sdo1-Δ/sdo1-Δ, indicating that Sdo1p and SBDS are functional homologs (Fig. 3C).
sdo1-Δ/sdo1-Δ cells have decreased vacuolar pH and V-ATPase activity
The absence of Btn1p (btn1-Δ) in minimal media results in alterations in the coupling of vacuolar H+ transport and V-ATPase activity, in turn modifying the regulation of vacuolar pH at different phases of growth (13). Specifically, btn1-Δ has a decreased vacuolar pH during early growth that continues to elevate above normal at later growth points (29). We therefore tested whether sdo1-Δ/sdo1-Δ have alterations in vacuolar pH and V-ATPase function. Vacuolar pH and V-ATPase activities were measured in rich media (YPD) instead of minimal media (YNB), the condition in which the V-ATPase was characterized in btn1-Δ, due to an inability of sdo1-Δ/sdo1-Δ to grow in minimal media. Even in YPD, vacuolar pH was significantly decreased in sdo1-Δ/sdo1-Δ cells. The vacuolar pH of sdo1-Δ/sdo1-Δ was 5.9, whereas the vacuolar pH of SDO1+/SDO1+ was 6.3 (Table 1).
Table 1.
Vacuolar pH, as measured by pH-dependent BCECF fluorescence, is decreased in sdo1-Δ/sdo1-Δ and increased in cells overexpressing BTN1 and in the presence of the ionophore CCCP
| Strain | Strain background | pH | Standard deviation | P-value |
|---|---|---|---|---|
| SDO1+/SDO1+ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | 6.33 | 0.04 | |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | 5.92 | 0.08 | <0.0001 |
| btn1-Δ/btn1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 btn1::KAN/btn1::KAN | 6.25 | 0.06 | <0.004 |
| Vector | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | 6.20 | 0.05 | |
| Overexpressed BTN1 | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | 6.50 | 0.08 | <0.005 |
| CCCP | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | 6.50 | 0.10 | <0.005 |
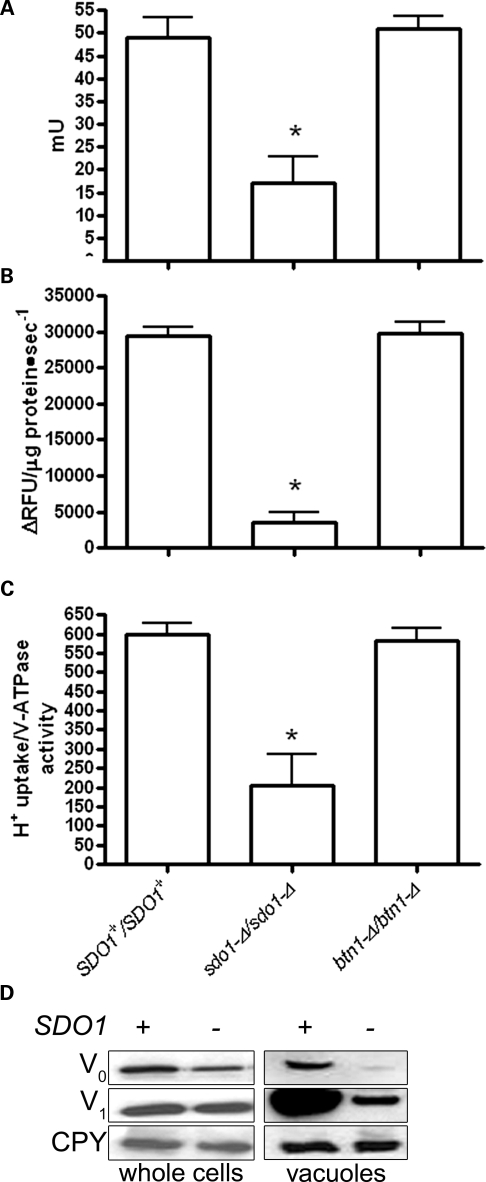
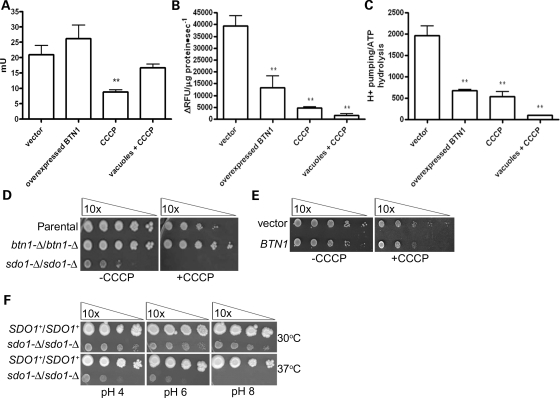
Vacuolar pH is primarily maintained by the V-ATPase, which transports protons via ATP hydrolysis (31–33). We report a significant decrease in V-ATPase-dependent ATP hydrolysis, and an even more drastic decrease in vacuolar H+ transport in sdo1-Δ/sdo1-Δ (Fig. 4A and B). These changes are also manifest as decreased coupling of V-ATPase hydrolysis with H+ transport in sdo1-Δ/sdo1-Δ (Fig. 4C). We had previously described that btn1-Δ results in a down-regulation of V-ATPase-dependent ATP hydrolysis, most likely in order to maintain normal H+ transport across the vacuolar membrane and to compensate for the alteration in pH caused by the lack of Btn1p (13). The V-ATPase is regulated through the assembly and disassembly of its V1 and V0 subunits (33). There was no evidence to suggest that either the levels of V-ATPase subunits or the assembly of this complex was altered in btn1-Δ. Similar to btn1-Δ, the assembly of the subunits appeared to be normal in sdo1-Δ/sdo1-Δ (Supplementary Material, Fig. S7). However, closer examination of the V-ATPase by immunoblotting for Vph1p and Vma1p, components of the V0 and V1 subunits, respectively, revealed a decrease in both proteins at the vacuole and a decrease in Vph1p in whole cell preparations (Fig. 4D).
Figure 4.
Alterations in the V-ATPase in sdo1-Δ/sdo1-Δ. Cells that were grown in YPD media were homogenized and subjected to differential centrifugation to isolate an enriched vacuolar fraction. (A) Concanamycin A-dependent ATPase activity in vacuoles isolated from sdo1-Δ/sdo1-Δ in rich media at mid-log phase is significantly decreased (P < 0.001). Rates are expressed as milliunits per milligram of protein (1 mU corresponds to 1 nmol of substrate transformed per minute). (B) Concanamycin A-dependent proton pumping in vacuoles isolated from sdo1-Δ/sdo1-Δ cells in rich media at mid-log phase is significantly decreased (P < 0.001). Rates are expressed as changes in relative fluorescent units per microgram of protein per second. (C) V-ATPase coupling calculated by the ratio between the mean of the H+ translocation and ATP hydrolysis activities. Coupling is decreased in sdo1-Δ/sdo1-Δ (P < 0.001). (D) sdo1-Δ/sdo1-Δ cells have decreased levels of Vph1p V-ATPase subunit in vacuoles and whole cells. Whole cell and vacuole protein preparations (10 μg each) from SDO1+/SDO1+ (Open Biosystems; B4741/B4742) and sdo1-Δ/sdo1-Δ derived from the B4741/B4742 parental strain were immunoblotted using anti-Vph1p, anti-Vma1p and anti-CPY (bottom), a loading control.
It was unclear whether sdo1-Δ/sdo1-Δ cells are synthesizing less V0, or degrading more V0. Alternatively, Vph1p may have decreased translation since Sdo1p is involved in ribosome maturation, or Vph1p may be differentially translated in response to alterations at the vacuole (34). To test this, we performed comparative real-time reverse transcription (RT) PCR to quantify the levels of VPH1 and VMA1 mRNA. Consistent with the protein levels, the amount of VPH1 mRNA is significantly decreased, whereas VMA1 mRNA is normal in sdo1-Δ/sdo1-Δ (Table 2 and Fig. 4D). Although experimental confirmation is needed, we speculate that VPH1 is transcriptionally repressed in sdo1-Δ/sdo1-Δ to correct for the decreased vacuolar pH. Since the protein levels and mRNA levels of VMA1 are normal in whole cell preparations, the decrease in Vma1p in the sdo1-Δ/sdo1-Δ vacuole-enriched fraction is probably a consequence of the absence of Vph1p in this membrane as the V0 subunit tethers the V1 subunit to the vacuole (Table 2 and Fig. 4D).
Table 2.
VPH1 mRNA levels, as determined by quantitative real-time RT–PCR, are decreased in sdo1-Δ/sdo1-Δ
| Query strain | Strain background | Control strain | Query gene | Fold change | P-value |
|---|---|---|---|---|---|
| btn1-Δ/btn1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 btn1::KAN/btn1::KAN | BTN1+/BTN1+ | VPH1 | NC | |
| btn1-Δ/btn1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 btn1::KAN/btn1::KAN | BTN1+/BTN1+ | VMA1 | 2.85 decrease | <0.0001 |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | SDO1+/SDO1+ | VPH1 | 3.75 decrease | <0.0001 |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | SDO1+/SDO1+ | VMA1 | NC |
Query gene Ct values were normalized to ORC5 (a component of the origin recognition complex). Ct values, fold changes and statistics were calculated using REST© software (49).
Sdo1p regulates Btn1p activity
Overexpression of BTN1 in the sdo1-Δ/sdo1-Δ strain caused an exacerbation of the poor growth that is present in sdo1-Δ/sdo1-Δ cells (Fig. 5). Conversely, overexpression of SDO1 in the btn1-Δ/btn1-Δ strain did not noticeably alter btn1-Δ/btn1-Δ growth (Supplementary Material, Fig. S8). Also, an sdo1-Δ/btn1-Δ haploid double-deletion strain appeared to be phenotypically similar to sdo1-Δ, in that the double-deletion mutant exhibited the same slow-growth phenotype seen in sdo1-Δ (data not shown). These observations, along with sdo1-Δ/sdo1-Δ resulting in altered vacuole pH and regulation of V-ATPase suggest that Sdo1p could work upstream of Btn1p, and that Sdo1p could regulate Btn1p function or monitor vacuolar pH.
Figure 5.
BTN1 overexpression is toxic to sdo1-Δ/sdo1-Δ. Serial dilutions of SDO1+/SDO1+ parental strain (Open Biosystems; B4741/B4742) or sdo1-Δ/sdo1-Δ cells of the same background with vector control or overexpressing BTN1. Mid-log-phase cells grown in selective media were diluted 10-fold with water in each column, and plated using a 36-pin replicator.
We postulated that the phenotypes observed upon BTN1 overexpression would mimic those phenotypes that are observed in the sdo1-Δ/sdo1-Δ strain. To this end, the effect of BTN1 overexpression on vacuolar pH, V-ATPase-dependent ATP hydrolysis and H+ transport was analyzed to determine whether the vacuolar phenotypes seen in sdo1-Δ/sdo1-Δ could be due to the misregulation of Btn1p. An increased level of BTN1 expression raised vacuolar pH from 6.2 to 6.5 (Table 1). In addition, H+ transport activity was increased, whereas V-ATPase activity remained the same (Fig. 6A and B). These differences in the V-ATPase resulted in a significant decrease in the coupling between the ATP hydrolysis and H+ transport by this complex in cells overexpressing BTN1 (Fig. 6C). These results suggest that the increased vacuolar pH observed in cells overexpressing BTN1 is a consequence of a decrease in the ability of the V-ATPase complex to pump protons into the vacuole.
Figure 6.
Overexpression of BTN1 mimics sdo1-Δ/sdo1-Δ. Cells (Open Biosystems; B4741/B4742) that were grown in selective media were homogenized and subjected to differential centrifugation to isolate an enriched vacuolar fraction. (A) Concanamycin A-dependent ATPase activity in vacuoles isolated from cells overexpressing BTN1 in minimal media at mid-log phase is significantly decreased (P < 0.001). Rates are expressed as milliunits per milligram of protein (1 mU corresponds to 1 nmol of substrate transformed per minute). (B) Concanamycin A-dependent proton pumping in vacuoles isolated from cells overexpressing BTN1 in minimal media at mid-log phase is significantly decreased (P < 0.001). Rates are expressed as changes in relative fluorescent units per microgram of protein per second. (C) V-ATPase coupling calculated by the ratio between the mean of H+ translocation and ATP hydrolysis activities. Coupling is decreased in cells overexpressing BTN1 (P < 0.001). (D) Serial dilutions of SDO1+/SDO1+, btn1-Δ/btn1-Δ or sdo1-Δ/sdo1-Δ on rich media with or without 5 μm CCCP. sdo1-Δ/sdo1-Δ is sensitive, whereas btn1-Δ/btn1-Δ is resistant to 5 μm CCCP. (E) Serial dilution of wild-type cells (Open Biosystems; B4741/B4742) with vector or overexpressing BTN1 on minimal media with or without 5 μm CCCP. Cells overexpressing BTN1 are sensitive to CCCP. (F) Serial dilutions of SDO1+/SDO1+ (Open Biosystems; B4741/B4742) or sdo1-Δ/sdo1-Δ of the parental control background on rich media (YPD) with pH 4, 6 or 8 at 30 or 37°C. The sdo1-Δ/sdo1-Δ strain displays a pH-dependent, 37°C temperature-sensitive phenotype on rich media.
We have previously suggested that Btn1p is involved in regulating the coupling of ATP hydrolysis and H+ transport by the V-ATPase complex, allowing H+ leakage from the vacuolar lumen to the cytosol in the early phase of growth when high amounts of ATP can be used by the V-ATPase as an energy source to pump protons into the vacuole (13). Thus, high amounts of Btn1p in vacuolar membranes could lead to incremental leakage of H+ from the vacuolar lumen and lower V-ATPase-dependent H+ uptake. The partial loss of the membrane potential and pH gradient across the vacuolar membrane could affect the assembly between the two V-ATPase sectors. Therefore, decreased assembly of the V-ATPase in cells overexpressing BTN1 could be secondary to H+ leakage instead of being a direct action of Btn1p over the V-ATPase complex. We therefore measured vacuolar pH and V-ATPase activities for cells expressing normal levels of BTN1 in presence of the proton gradient uncoupler carbonyl cyanide m-chlorophenil hydrazone (CCCP). The vacuolar pH measured under these conditions increased to a similar degree to the pH measured in cells overexpressing BTN1 (Table 1). In addition, CCCP treatment led to a dramatic decrease of the vacuolar H+ uptake, and ATP hydrolysis was only 50% of the activity measured in BTN1+ without CCCP treatment (Fig. 6A and B). The coupling of the V-ATPase calculated from both activities was similar to the coupling observed in cells overexpressing BTN1 (Fig. 6C). Isolated vacuoles from cells with normal BTN1 levels were also incubated with CCCP. Whereas the H+ transport was as low as the CCCP treated cells, ATP hydrolysis was normal, suggesting that the observed decay of this activity in CCCP-treated cells could be a long-term effect of the treatment with the uncoupler. Interestingly, sdo1-Δ/sdo1-Δ was sensitive to CCCP, whereas btn1-Δ/btn1-Δ had increased growth over the parental control strain (Fig. 6D). Like sdo1-Δ/sdo1-Δ, BTN1 overexpression caused decreased growth in the presence of CCCP (Fig. 6E).
In summary, overexpression of BTN1 causes similar vacuolar and pH-dependent effects as deletion of SDO1. Overexpression of BTN1 caused decreased growth in sdo1-Δ/sdo1-Δ presumably due to the absence of Btn1p regulation, whereas a lack of Btn1p regulation mirrors the uncoupling action of CCCP. Finally, sdo1-Δ/sdo1-Δ did not grow at pH 8, where there is a clear need to regulate vacuolar pH through an active V-ATPase complex (Fig. 6F). These results suggest that Btn1p and Sdo1p function in the same pathway.
Alterations in ribosome maturation affect the vacuole
Sdo1p participates in the TIF6 ribosome maturation pathway, which requires Sdo1p, Efl1p and Tif6p for proper transport of 60s ribosomal subunits out of the nucleus (26,35–37). We investigated whether the vacuolar defects observed in sdo1-Δ/sdo1-Δ are unique to this deletion, or whether the observed vacuolar changes are a general response to defects in ribosome export. There are several mutations in TIF6 that are reported to suppress the sdo1-Δ/sdo1-Δ growth defects (26). Of the two TIF6 mutants (kindly provided by Allen J. Warren, Cambridge, UK) that were tested, both suppressed the decreased vacuolar pH that was observed in sdo1-Δ/sdo1-Δ (Table 3). Additionally, vacuolar pH, V-ATPase-dependent ATP hydrolysis and H+ pumping, V-ATPase protein levels and pH-dependent growth in efl1-Δ/efl1-Δ were similar to sdo1-Δ/sdo1-Δ (data not shown), indicating that the status of the vacuole depends upon an intact ribosome maturation pathway.
Table 3.
Decreased vacuolar pH, as measured by pH-dependent BCECF fluorescence, of sdo1-Δ/sdo1-Δ is restored by Tif6p-dominant suppressor mutants
| Strain | Strain background | Plasmid | Vacuolar pH | Standard deviation | P-value |
|---|---|---|---|---|---|
| SDO1+/SDO1+ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | Vector | 5.77 | 0.06 | |
| SDO1+/SDO1+ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | pTIF6 | 5.72 | 0.09 | |
| SDO1+/SDO1+ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | pTIF6-V192F | 5.79 | 0.07 | |
| SDO1+/SDO1+ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 | pTIF6-I58T | 5.73 | 0.10 | |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | Vector | 5.52 | 0.08 | <0.002 |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | pTIF6 | 5.52 | 0.09 | <0.002 |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | pTIF6-V192F | 5.82 | 0.10 | |
| sdo1-Δ/sdo1-Δ | MATα/MATa ura3-Δ0/ura3-Δ0 his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2+met15-Δ0/MET15 sdo1::KAN/sdo1::KAN | pTIF6-I58T | 5.71 | 0.06 |
DISCUSSION
Mutations in CLN3 cause JNCL, but the exact function of CLN3 is unknown (1). We demonstrate that SBDS is an interacting partner of CLN3. This interaction is conserved in yeast, as demonstrated by the interaction between Btn1p and Sdo1p, the yeast homologs of CLN3 and SBDS, respectively. This interaction is functionally relevant due to alterations in the regulation of the V-ATPase and vacuolar pH in sdo1-Δ/sdo1-Δ, btn1-Δ, and upon BTN1 overexpression (13). We propose that Sdo1p regulates Btn1p activity, and by extension, that SBDS may regulate CLN3 (Fig. 7). Our results demonstrate that overexpression of BTN1 mimics deletion of SDO1. Consequently, the absence of SDO1 causes altered Btn1p activity, suggesting that Sdo1p regulates Btn1p function, perhaps through the ability of Sdo1p to bind the phosphoinositides PI(4,5)P2 and PI4P (38). It is interesting that the absence and particularly overexpression of Btn1p can alter coupling of the ATP hydrolysis and H+ transport. More remarkable is the fact that either overexpression of Btn1p or loss of Sdo1p in the absence or the presence of the ionophore CCCP results in the same effect, specifically the uncoupling of ATP hydrolysis and H+ transport. Although Sdo1p has been shown to be involved in ribosomal maturation, we argue that an additional role for this protein is to regulate vacuolar function through Btn1p.
Figure 7.
Proposed model of the role of the Sdo1p–Btn1p interaction. A condition such as pH change causes the following events: (1) the interaction of Sdo1p with Btn1p causes the inhibition of Btn1p (2), which in turn influences the coupling of V-ATPase and H+ transport (3), thus altering vacuole pH and possibly other vacuolar functions. Sdo1p may mediate maintenance of vacuolar processes from the ribosome through interacting or signaling to Btn1p (4).
Sdo1p participates in ribosomal maturation, but Btn1p is unlikely to be directly involved in this process. It is pertinent to highlight that it appears that only small amounts of Btn1p and Sdo1p appear to interact. Furthermore, the slow-growth phenotype observed in sdo1-Δ/sdo1-Δ does not occur in btn1-Δ/btn1-Δ. However, a link between CLN3 and RNA metabolism has already been suggested (39). A suppressor screen in Drosophila melanogaster overexpressing CLN3 identified an RNA-binding protein that, when mutated, enhanced the phenotype observed upon CLN3 overexpression alone. Thus, the idea that a component of the ribosome maturation pathway such as Sdo1p/SBDS regulates Btn1p/CLN3 function and that TIF6 suppresses the vacuolar phenotypes observed in the SDO1 deletion strain suggests that perhaps the ribosome monitors vacuolar/lysosome function or homeostasis (Fig. 7).
Such cross-talk has not previously been reported; however, the ability to regulate the high energy demands of the vacuole/lysosome through regulated feedback to the RNA transcription and protein translation machinery would work to benefit the responsiveness of cells. For example, differential translation controlled by ribosome subunit assembly in response to environmental cues has been reported (34). This kind of response would optimize the cell's ability to balance energy demands, in order for the cell to mount the appropriate reaction or compensation to a change or stimulus. For example, human SBDS has recently been implicated as a general stress response protein, as SBDS-depleted cells are sensitive to DNA damage and ER stress, and SBDS interacts with many proteins including parts of the ribosome and DNA repair machinery (40). We propose that through the SBDS/Sdo1p and CLN3/Btn1p interaction, the status of the lysosome/vacuole can be maintained in response to an insult to the cell, such as a change in pH or nutrient levels.
Juvenile Batten disease and Shwachman–Bodian–Diamond syndrome are pathologically very different diseases. Our data support a role for SBDS functioning upstream of CLN3. The loss of CLN3 at the lysosome results in an inability to regulate this organelle, whereas the loss of SBDS results in an inability to regulate CLN3 activity at the lysosome. Thus the effect on the lysosome is more severe in the absence of CLN3 than SBDS. Mutations in CLN3 lead to a devastating progressive neurodegenerative disease, and the true effect of the loss of CLN3 in other tissues may be masked by the severity of neurological decline. The neurological effects are less pronounced in Shwachman–Bodian–Diamond syndrome, and may be a consequence of perturbations in CLN3 regulation, rather than a more marked effect on lysosomal function (41). The loss of SBDS, which is clearly involved in more cellular processes, likely gives rise to a more systemic disease due to alterations in regulating ribosomal maturation in multiple tissues.
In conclusion, we have validated a novel, evolutionarily conserved interaction, which will help define the function of two disease-associated proteins, CLN3 and SBDS.
MATERIALS AND METHODS
Yeast two-hybrid method
Stratagene's Cytotrap yeast two-hybrid method was used to screen the C-terminus of CLN3. Basepairs 1521–1676 (amino acids Ala388 to STOP) of CLN3 (GenBank accession number NM_001042432) were cloned into the pSos vector and screened against a human fetal brain cDNA library (Stratagene, La Jolla, CA, USA). The pSos-CLN3 was co-transformed with the pMyr cDNA library into the Cdc25H yeast strain, and growth in the presence or absence of galactose at 37°C was evaluated. Suppression of the inherent temperature sensitivity of the Cdc25H strain indicated an interaction between CLN3 and a candidate protein. The library DNA was screened again through the system to eliminate false-positive interactions.
Co-immunoprecipitation
Protein was expressed for 36 h in NIH/3T3 fibroblasts and then extracted by incubating the cell pellet in ice-cold non-denaturing lysis buffer (50 mm Tris–Cl, pH 7.5, 300 mm NaCl, 5 mm, EDTA, 1% Triton X-100). The post-nuclear supernatant was pre-cleared using protein A-conjugated agarose beads. Cell lysates were incubated with anti c-myc antibody (Cell Signaling, Danvers, MA, USA), followed by protein A-conjugated agarose beads. The immunoprecipitate was washed with wash buffer (50 mm Tris–Cl, pH 7.5, 300 mm NaCl, 5 mm EDTA, 0.01% Triton X-100), resuspended in Laemmli buffer and boiled. Immunoprecipitates and lysates were subjected to sodium dodecyl sulfate (SDS)–PAGE, followed by western blotted using either an anti-V5 antibody (Invitrogen, Carlsbad, CA, USA) or anti-myc antibody (Cell Signaling).
Immunofluorescence
NIH/3T3 culture grown on a poly-d-lysine-coated coverglass was transfected with the constructs described. Protein was expressed for 36 h and fixed for 30 min at room temperature (4% PFA/4% sucrose phosphate buffer, pH 7.5). Cells were permeabilized (PBS, 0.1% Triton X-100), and blocked in Blotto (4% non-fat dried milk, PBS, 0.1% Triton X-100). Anti-V5 and anti-myc antibodies were incubated overnight at 4°C. Cells were washed and incubated in the ALEXA-conjugated secondary antibodies (Molecular Probes, Carlsbad, CA, USA). Mounted cells were visualized using confocal microscopy.
Plasmids
CLN3 was cloned into pBudCE4 (Invitrogen), and SBDS was cloned into pBudCE4. For overexpression of BTN1 or SDO1, the ORF was amplified from yeast chromosomal DNA and cloned into the multicopy vector pRS426. The plasmid containing human SBDS was created by cloning SBDS into the multicopy vector pESC-HIS for expression in yeast. The PGAL1:BTN1-V5-6xHIS chimera was made by amplifying the BTN1 ORF from yeast chromosomal DNA and insertion in-frame into the 2 μm pYES2.1 vector (Stratagene). For the construction of the PGAL1:SDO1-MYC chimera, the SDO1 ORF was amplified from yeast chromosomal DNA and inserted into pESCHIS (Stratagene). The BTN1-V5 plasmid was constructed by ligating the V5 tag into a pRS316 plasmid already containing the BTN1 ORF without its stop codon plus 1 kb upstream of the ORF. The BTN1-eSapphire construct that was used in microscopy was provided by D. Wolfe. An eSapphire tag was inserted into a plasmid (pRS316) already containing the BTN1 ORF without its stop codon plus 1 kb upstream. Plasmids containing wild-type and TIF6 mutants were provided by Alan J. Warren (Cambridge, UK). The pTIF6-V192F and pTIF6-I58T plasmids contain mutations in TIF6 and are cloned into the pRS316 vector (26).
Yeast strains
The intrachromasomally GFP-tagged SDO1 was purchased from Open Biosystems (30). All other strains were purchased from Open Biosystems unless stated (Open Biosystems; B4741/B4742: MATα/a his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0). Each haploid yeast strain was of the B4741 background (Open Biosystems; MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). To make the diploid strain that is homozygous for the SDO1, BTN1, or the EFL1 deletion, tetrad spores derived from heterozygous diploid (Open Biosystems; B4741/B4742: MATα/a his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 MET15/met15Δ0 ura3Δ0/ura3Δ0 with sdo1::KAN/SDO1, btn1::KAN/BTN1 or efl1::KAN/EFL1) (42) were separated. Deletions were tested for amino acid auxotrophy, mated and diploids were selected. Deletions were also confirmed by PCR. In all experiments, mutant strains were compared with the wild-type parental strain of the same auxotrophic background.
Media
Yeast media was prepared as described in Adams et al. (42). Yeast strains without plasmids were grown in rich media (YPD) which contains 1% yeast extract, 2% peptone and 2% dextrose, pH 6.4. Strains containing plasmids requiring uracil or histidine auxotrophic marker selection were grown in synthetic complete media lacking either uracil or histidine. Synthetic complete media contained 6.7 mg/ml yeast nitrogen base without amino acids, 5 mg/ml ammonium sulfate, 2% dextrose and all amino acids except asparagine, glutamine, proline, alanine, cysteine and glycine, pH 5.6–5.8.
Growth phenotyping
For tetrad analysis, the SDO1+/sdo1-Δ diploid strain (Open Biosystems) was sporulated in sporulation media (1% potassium acetate, 0.1% yeast extract, 0.05% dextrose and auxotrophic amino acids) with vigorous shaking (42). Tetrad walls were digested using glucuronidase, and spores were separated on YPD plates using a micromanipulator attached to a Singer Instruments MSM system tetrad dissection scope. Each resulting colony was genotyped using gene-specific primers via PCR. For plate phenotypes, strains were grown in YPD or synthetic complete selection media, harvested and washed twice with sterile water. Cells were resuspended to 3 × 108 cells/ml in water and diluted 10-fold for each column. A 36-pin replicator was used to spot cells onto the media followed by incubation at 30°C.
Affinity purification
Yeast harboring the BTN1-V5-6HIS and SDO1-MYC constructs were grown to mid-log phase and induced in galactose. Cells were harvested and resuspended in non-denaturing extraction buffer (50 mm sodium phosphate, pH 7, 300 mm NaCl, 1% Triton X-100 and protease inhibitors). Cells were bead-beaten and centrifuged at 3000g for 5 min at 4°C. Cell lysate was incubated with 50 μl Talon cobalt resin, and Btn1p-V5-6HIS was purified as per the manufacturer's protocol (BD Biosciences). Fractions from the column were TCA-precipitated, solubilized in Lammaeli buffer and subjected to western analysis.
Immunopurification
Cell extract as prepared earlier was incubated with 1:400 anti-c-MYC antibody (Cell Signaling), followed by incubation with protein A-agarose beads. Lamaelli buffer was incubated with the beads at 65°C for 5 min to solubilize the proteins for subsequent western analysis.
Fractionation and endogenous co-immunopurification
Mid-log-phase cells were made into protoplasts, lysed using a Dounce homogenizer in lysis buffer (0.2 m sorbitol, 50 mm Tris–Cl, pH 7.6, 1 mm EDTA and protease inhibitors), and unlysed cells were separated by centrifugation at 500g for 5 min. The supernatant was centrifuged at 13 000g for 15 min (plasma membrane, mitochondria and vacuoles). The supernatant was spun at 100 000g to yield the endosomes and Golgi, and cytosol and vesicles. Fifty microgram was analyzed by SDS–PAGE and western blotting; 500 μg protein with 1% Triton X-100 was incubated with 1:400 anti-GFP antibody (Molecular Probes), and again incubated with Protein G-Agarose. Protein for subsequent western analysis was eluted using Lamaelli buffer at 65°C.
Yeast cell imaging
Cells were visualized under the 100X objective using an epifluorescent microscope (Olympus BX61, Melville, NY, USA), a CoolSNAP HQ CCD camera (Photometrics, Tucson, AR, USA) and IPLab 4.0 acquisition software (BD Biosciences, Rockville, MD, USA). Image deconvolution was performed using the Autodeblur software package (Media Cybernetics, Bethesda, MD, USA), and overlays of fluorescent images were performed using ImageJ (NIH) and Photoshop software.
Fractionation
Mid-log-phase cells were made into spheroplasts, resuspended in 20 mm Tris–MES, pH 6, 0.6 m sorbitol and protease inhibitors, homogenized and centrifuged to remove intact cells. The supernatant was centrifuged at 12 000g in 10 ml 20 mm Tris–MES, pH 6, 0.6 m sorbitol. The resulting pellet (A) contained a crude mitochondrial fraction. Pellet A was resuspended in 20 mm Tris–MES, pH 6, 0.6 m sorbitol ultracentrifuged at 100 000g for 1 h. The top layer is the mitochondrial associated membranes, whereas the middle layer was enriched for mitochondria. The supernatant from the 12 000g spin was centrifuged at 20 000g to yield the microsomal fraction in the pellet. It was centrifuged at 32 500g for 45 min and the resulting pellet contained rough ER, and again the supernatant was centrifuged at 100 000g for 1 h. The resulting pellet contained smooth ER. Vacuoles were isolated using the methods of Ohsumi and Anraku (43). Each enriched fraction was analyzed by SDS–PAGE and immunoblotting.
Vacuolar pH
Vacuolar pH measurements were performed as described previously (44,45). Yeast cells were grown to mid-log phase in YPD and harvested by centrifugation. Cells harboring plasmids were grown in selective media to early-log phase, harvested and grown to mid-log phase in YPD. Two aliquots from each strain of 4 × 107 cells were incubated and resuspended in 200 μl YPD. One aliquot was incubated with 50 μm BCECF-AM for 20 min at 30°C, whereas the other was used as a background control. Cells were collected again by centrifugation, washed three times and resuspended in 1 ml water. Cell suspensions of 100 μl were used for fluorescent measurements. Fluorescence absorbances were measured using a Spectra Max M5 multimode microplate reader (Molecular Devices) at 440 and 490 nm excitation wavelengths, and background fluorescence was measured using the BCECF-free cultures. The values were normalized to cell density to calculate pH. For each experiment and yeast strain, a calibration curve of fluorescence intensity versus pH was plotted by incubating yeast cells in 50 mm MES, 50 mm HEPES, 50 mm KCl, 50 mm NaCl, 0.2 m ammonium acetate, 10 mm NaN3, 10 mm 2-deoxyglucose, 50 μm carbonyl cyanide m-chlorophenylhydrazone, and titrated to pH 4–8 using NaOH. To estimate vacuolar pH, absorbance values for each strain were compared with the corresponding calibration curve.
V-ATPase measurements
ATP hydrolysis was performed as described previously (46). Vacuolar membranes (10 μg, above) were suspended in 250 μl of assay mixture which contained 100 mm Tris–MES, 80 mm KCl, 6 mm MgCl2, 5 mm sodium azide (inhibits mitochondrial ATPase), 0.2 m ammonium molybdate (inhibits acid phosphatases) and 100 μm sodium orthovanadate (inhibits plasma membrane ATPase). The reaction was started with the addition of 2 mm Na-ATP and incubated for 20 min at 30°C. The reaction was stopped by the addition of 500 μl of 2% sulfuric acid, 0.5% ammonium molybdate and 0.5% SDS solution and 50 μl of 10% ascorbic acid. After 5 min incubation at room temperature, the absorbance was measured at 750 nm. The activity measured in the presence of 1 μm concanamycin A (a specific inhibitor of the vacuolar ATPase) was subtracted from the activity measured in the absence of concanamycin A to calculate the V-ATPase activity. The proton transport activity into the lumen of isolated vacuoles was measured by monitoring the formation of a pH gradient across the vacuolar membrane by fluorescence quenching of 9-amino-6-chloro-2-methoxyacridine (ACMA) in the presence or absence of 1 μm concanamycin A (47). Vacuoles (25 μg, above) were added to a cuvette containing 2 ml of reaction buffer [10 mm BisTrisPropene (BTP)-MES, pH 7, 25 mm KCl, 2 mm MgSO4, 10% glycerol and 2 μm ACMA]. The reaction was started by the addition of 1 mm ATP in BTP, pH 7.5, and fluorescence emission was recorded at 480 nm after excitation at 412 nm.
Quantitative real-time reverse-transcription PCR
RNA was extracted by bead-beating protoplasts in the presence of Trizol reagent (Invitrogen). After DNAse I treatment, cDNA was made using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Reactions without reverse transcriptase were included. Primers were designed using Beacon Designer 7 (Premier Biosoft, Palo Alto, CA, USA) (rtORC5f: GTA TGG CTG GAA CCT GTT GAG TTG; rtORC5r: GGA TCG TAA TCT GTG GTG GGA ATG; rtVMA1f: GTC AGC ACA GAG CCC ACA AAA G; rtVMA1r: AGA CAA ACG GCG GAC ACT ACG; rtVPH1f: ACA TCC CAC AGG TGA CGC ATA AC; rtVPH1r: GGA GCA GCG AGG CGA TAC C). ORC5 was selected for normalization using GeNORM software (48). Of several housekeeping genes that were tested, ORC5 mRNA did not change between the different mutant strains under different experimental conditions. Reactions were performed using an Mx3005p thermocycler with MxPro QPCR software (Stratagene), and Power SYBR Green PCR Master Mix (Applied Biosystems) using the following cycling conditions: step 1: 95°C for 10 min; step 2: 95°C for 30 s and 60°C for 1 min for 40 cycles; step 3: 95°C for 1 min, 55°C for 30 s and 95°C for 30 s. Specificity of the amplified product was determined by melt-curve analysis. Relative gene expression was calculated using the REST-XL program and expressed as fold change versus control or wild-type (49).
Statistical analysis
Statistical significance for V-ATPase activities (n = 3–6) was tested using one-way ANOVA via GraphPad Prism software. Differences in vacuolar pH (n = 3–6) were calculated using Student's t-tests using Microsoft Excel. Statistical analysis of Quantitative RT–PCR results were performed using Pair Wise Fixed Reallocation Randomization Test© via REST© software (49). Reactions from three biological replicates were each performed using three technical replicates.
SUPPLEMENTARY MATERIAL
FUNDING
This work is funded in part by the National Institutes of Health (grant number R01 NS36610), the Batten Disease Support and Research Association, Beat Batten Foundation and the Shwachman Diamond Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank D. Wolfe for providing the Btn1p-eSapphire construct, S. Leistman and J. Weimer for technical assistance in the yeast two-hybrid experiments, C.H. Chan for technical assistance with RT–PCR and A. Getty for critically reading the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.B.D. Consortium. Isolation of a novel gene underlying Batten disease, CLN3. The International Batten Disease Consortium. Cell. 1995;82:949–957. doi: 10.1016/0092-8674(95)90274-0. [DOI] [PubMed] [Google Scholar]
- 2.Jarvela I., Sainio M., Rantamaki T., Olkkonen V.M., Carpen O., Peltonen L., Jalanko A. Biosynthesis and intracellular targeting of the CLN3 protein defective in Batten disease. Hum. Mol. Genet. 1998;7:85–90. doi: 10.1093/hmg/7.1.85. [DOI] [PubMed] [Google Scholar]
- 3.Kyttala A., Ihrke G., Vesa J., Schell M.J., Luzio J.P. Two motifs target Batten disease protein CLN3 to lysosomes in transfected nonneuronal and neuronal cells. Mol. Biol. Cell. 2004;15:1313–1323. doi: 10.1091/mbc.E03-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storch S., Pohl S., Braulke T. A dileucine motif and a cluster of acidic amino acids in the second cytoplasmic domain of the batten disease-related CLN3 protein are required for efficient lysosomal targeting. J. Biol. Chem. 2004;279:53625–53634. doi: 10.1074/jbc.M410930200. [DOI] [PubMed] [Google Scholar]
- 5.Pearce D.A., Sherman F. BTN1, a yeast gene corresponding to the human gene responsible for Batten's disease, is not essential for viability, mitochondrial function, or degradation of mitochondrial ATP synthase. Yeast. 1997;13:691–697. doi: 10.1002/(SICI)1097-0061(19970630)13:8<691::AID-YEA123>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y., Ramirez-Montealegre D., Pearce D.A. A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc. Natl Acad. Sci. USA. 2003;100:15458–15462. doi: 10.1073/pnas.2136651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce D.A., Sherman F. A yeast model for the study of Batten disease. Proc. Natl Acad. Sci. USA. 1998;95:6915–6918. doi: 10.1073/pnas.95.12.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gachet Y., Codlin S., Hyams J.S., Mole S.E. btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell Sci. 2005;118:5525–5536. doi: 10.1242/jcs.02656. [DOI] [PubMed] [Google Scholar]
- 9.Croopnick J.B., Choi H.C., Mueller D.M. The subcellular location of the yeast Saccharomyces cerevisiae homologue of the protein defective in the juvenile form of Batten disease. Biochem. Biophys. Res. Commun. 1998;250:335–341. doi: 10.1006/bbrc.1998.9272. [DOI] [PubMed] [Google Scholar]
- 10.Codlin S., Haines R.L., Burden J.J., Mole S.E. Btn1 affects cytokinesis and cell-wall deposition by independent mechanisms, one of which is linked to dysregulation of vacuole pH. J. Cell Sci. 2008;121:2860–2870. doi: 10.1242/jcs.030122. [DOI] [PubMed] [Google Scholar]
- 11.Codlin S., Haines R.L., Mole S.E. btn1 affects endocytosis, polarization of sterol-rich membrane domains and polarized growth in Schizosaccharomyces pombe. Traffic. 2008;9:936–950. doi: 10.1111/j.1600-0854.2008.00735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osorio N.S., Carvalho A., Almeida A.J., Padilla-Lopez S., Leao C., Laranjinha J., Ludovico P., Pearce D.A., Rodrigues F. Nitric oxide signaling is disrupted in the yeast model for Batten disease. Mol. Biol. Cell. 2007;18:2755–2767. doi: 10.1091/mbc.E06-11-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padilla-Lopez S., Pearce D.A. Saccharomyces cerevisiae lacking Btn1p modulate vacuolar ATPase activity to regulate pH imbalance in the vacuole. J. Biol. Chem. 2006;281:10273–10280. doi: 10.1074/jbc.M510625200. [DOI] [PubMed] [Google Scholar]
- 14.Pearce D.A., Nosel S.A., Sherman F. Studies of pH regulation by Btn1p, the yeast homolog of human Cln3p. Mol. Genet. Metab. 1999;66:320–323. doi: 10.1006/mgme.1999.2819. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y., Chattopadhyay S., Locke S., Pearce D.A. Interaction among Btn1p, Btn2p, and Ist2p reveals potential interplay among the vacuole, amino acid levels, and ion homeostasis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:281–288. doi: 10.1128/EC.4.2.281-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitiello S.P., Wolfe D.M., Pearce D.A. Absence of Btn1p in the yeast model for juvenile Batten disease may cause arginine to become toxic to yeast cells. Hum. Mol. Genet. 2007;16:1007–1016. doi: 10.1093/hmg/ddm046. [DOI] [PubMed] [Google Scholar]
- 17.Ezaki J., Takeda-Ezaki M., Koike M., Ohsawa Y., Taka H., Mineki R., Murayama K., Uchiyama Y., Ueno T., Kominami E. Characterization of Cln3p, the gene product responsible for juvenile neuronal ceroid lipofuscinosis, as a lysosomal integral membrane glycoprotein. J. Neurochem. 2003;87:1296–1308. doi: 10.1046/j.1471-4159.2003.02132.x. [DOI] [PubMed] [Google Scholar]
- 18.Mao Q., Foster B.J., Xia H., Davidson B.L. Membrane topology of CLN3, the protein underlying Batten disease. FEBS Lett. 2003;541:40–46. doi: 10.1016/s0014-5793(03)00284-9. [DOI] [PubMed] [Google Scholar]
- 19.Boocock G.R., Morrison J.A., Popovic M., Richards N., Ellis L., Durie P.R., Rommens J.M. Mutations in SBDS are associated with Shwachman–Diamond syndrome. Nat. Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 20.Shwachman H., Diamond L.K., Oski F.A., Khaw K.T. The syndrome of pancreatic insufficiency and bone marrow dysfunction. J. Pediatr. 1964;65:645–663. doi: 10.1016/s0022-3476(64)80150-5. [DOI] [PubMed] [Google Scholar]
- 21.Bodian M., Sheldon W., Lightwood R. Congenital hypoplasia of the exocrine pancreas. Acta Paediatr. 1964;53:282–293. doi: 10.1111/j.1651-2227.1964.tb07237.x. [DOI] [PubMed] [Google Scholar]
- 22.Donadieu J., Michel G., Merlin E., Bordigoni P., Monteux B., Beaupain B., Leverger G., Laporte J.P., Hermine O., Buzyn A., et al. Hematopoietic stem cell transplantation for Shwachman–Diamond syndrome: experience of the French neutropenia registry. Bone Marrow Transplant. 2005;36:787–792. doi: 10.1038/sj.bmt.1705141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goobie S., Popovic M., Morrison J., Ellis L., Ginzberg H., Boocock G.R., Ehtesham N., Betard C., Brewer C.G., Roslin N.M., et al. Shwachman–Diamond syndrome with exocrine pancreatic dysfunction and bone marrow failure maps to the centromeric region of chromosome 7. Am. J. Hum. Genet. 2001;68:1048–1054. doi: 10.1086/319505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesling C., Oliveira C.C., Castilho B.A., Zanchin N.I. The Shwachman–Bodian–Diamond syndrome associated protein interacts with HsNip7 and its down-regulation affects gene expression at the transcriptional and translational levels. Exp. Cell Res. 2007;313:4180–4195. doi: 10.1016/j.yexcr.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S., Shi M., Hui C.C., Rommens J.M. Loss of the mouse ortholog of the Shwachman–Diamond syndrome gene (Sbds) results in early embryonic lethality. Mol. Cell Biol. 2006;26:6656–6663. doi: 10.1128/MCB.00091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menne T.F., Goyenechea B., Sanchez-Puig N., Wong C.C., Tonkin L.M., Ancliff P.J., Brost R.L., Costanzo M., Boone C., Warren A.J. The Shwachman–Bodian–Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 27.Zanchin N.I., Roberts P., DeSilva A., Sherman F., Goldfarb D.S. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips S.N., Muzaffar N., Codlin S., Korey C.A., Taschner P.E., de Voer G., Mole S.E., Pearce D.A. Characterizing pathogenic processes in Batten disease: use of small eukaryotic model systems. Biochim. Biophys. Acta. 2006;1762:906–919. doi: 10.1016/j.bbadis.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Pearce D.A., Ferea T., Nosel S.A., Das B., Sherman F. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 1999;22:55–58. doi: 10.1038/8861. [DOI] [PubMed] [Google Scholar]
- 30.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 31.Forgac M. Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 1999;274:12951–12954. doi: 10.1074/jbc.274.19.12951. [DOI] [PubMed] [Google Scholar]
- 32.Graham L.A., Powell B., Stevens T.H. Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J. Exp. Biol. 2000;203:61–70. doi: 10.1242/jeb.203.1.61. [DOI] [PubMed] [Google Scholar]
- 33.Kane P.M., Parra K.J. Assembly and regulation of the yeast vacuolar H(+)-ATPase. J. Exp. Biol. 2000;203:81–87. doi: 10.1242/jeb.203.1.81. [DOI] [PubMed] [Google Scholar]
- 34.Komili S., Farny N.G., Roth F.P., Silver P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu U., Si K., Warner J.R., Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becam A.M., Nasr F., Racki W.J., Zagulski M., Herbert C.J. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;266:454–462. doi: 10.1007/s004380100548. [DOI] [PubMed] [Google Scholar]
- 37.Senger B., Lafontaine D.L., Graindorge J.S., Gadal O., Camasses A., Sanni A., Garnier J.M., Breitenbach M., Hurt E., Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 39.Tuxworth R.I., Vivancos V., O'Hare M.B., Tear G. Interactions between the juvenile Batten disease gene, CLN3, and the Notch and JNK signalling pathways. Hum. Mol. Genet. 2009;18:667–678. doi: 10.1093/hmg/ddn396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ball H.L., Zhang B., Riches J.J., Gandhi R., Li J., Rommens J.M., Myers J.S. Shwachman–Bodian–Diamond syndrome is a multi-functional protein implicated in cellular stress responses. Hum. Mol. Genet. 2009;18:3684–3695. doi: 10.1093/hmg/ddp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toiviainen-Salo S., Makitie O., Mannerkoski M., Hamalainen J., Valanne L., Autti T. Shwachman–Diamond syndrome is associated with structural brain alterations on MRI. Am. J. Med. Genet. A. 2008;146A:1558–1564. doi: 10.1002/ajmg.a.32354. [DOI] [PubMed] [Google Scholar]
- 42.Adams A., Gottschling D.E., Kaiser C.A., Stearns T. Methods in Yeast Genetics. Plainview: Cold Spring Harbor Laboratory Press; 1997. pp. 177. [Google Scholar]
- 43.Ohsumi Y., Anraku Y. Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 1981;256:2079–2082. [PubMed] [Google Scholar]
- 44.Ali R., Brett C.L., Mukherjee S., Rao R. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J. Biol. Chem. 2004;279:4498–4506. doi: 10.1074/jbc.M307446200. [DOI] [PubMed] [Google Scholar]
- 45.Plant P.J., Manolson M.F., Grinstein S., Demaurex N. Alternative mechanisms of vacuolar acidification in H(+)-ATPase-deficient yeast. J. Biol. Chem. 1999;274:37270–37279. doi: 10.1074/jbc.274.52.37270. [DOI] [PubMed] [Google Scholar]
- 46.Serrano R. Characterization of the plasma membrane ATPase of Saccharomyces cerevisiae. Mol. Cell Biochem. 1978;22:51–63. doi: 10.1007/BF00241470. [DOI] [PubMed] [Google Scholar]
- 47.Camarasa C., Prieto S., Ros R., Salmon J.M., Barre P. Evidence for a selective and electroneutral K+/H(+)-exchange in Saccharomyces cerevisiae using plasma membrane vesicles. Yeast. 1996;12:1301–1313. doi: 10.1002/(SICI)1097-0061(199610)12:13%3C1301::AID-YEA18%3E3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 48.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034.1-0034.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.