Abstract
The regulation of autophagy in metazoans is only partly understood, and there is a need to identify the proteins that control this process. The diabetes- and obesity-regulated gene (DOR), a recently reported nuclear cofactor of thyroid hormone receptors, is expressed abundantly in metabolically active tissues such as muscle. Here, we show that DOR shuttles between the nucleus and the cytoplasm, depending on cellular stress conditions, and re-localizes to autophagosomes on autophagy activation. We demonstrate that DOR interacts physically with autophagic proteins Golgi-associated ATPase enhancer of 16 kDa (GATE16) and microtubule-associated protein 1A/1B-light chain 3. Gain-of-function and loss-of-function studies indicate that DOR stimulates autophagosome formation and accelerates the degradation of stable proteins. CG11347, the DOR Drosophila homologue, has been predicted to interact with the Drosophila Atg8 homologues, which suggests functional conservation in autophagy. Flies lacking CG11347 show reduced autophagy in the fat body during pupal development. All together, our data indicate that DOR regulates autophagosome formation and protein degradation in mammalian and Drosophila cells.
Keywords: nuclear co-regulator, protein degradation, GATE16, LC3, fat body
Introduction
Macroautophagy (here referred to as autophagy) is an important cellular pathway for the degradation of long-lived proteins and cytoplasmic organelles (Klionsky, 2007; Mizushima et al, 2008). A central process in autophagy is the formation of autophagosomes, during which portions of the cytoplasm are surrounded by a membranous structure and then fused with lysosomes to form autolysosomes, in which degradation occurs (Mizushima & Klionsky, 2007).
Under conditions of extracellular nutrient scarcity, unicellular eukaryotes catabolize cytoplasmic components by autophagy to maintain ATP concentrations (Kourtis & Tavernarakis, 2009), and support survival (Tsukada & Ohsumi, 1993). Multicellular organisms have maintained this pro-survival function of autophagy (Boya et al, 2005) and have acquired further layers of complexity. Thus, in metazoan species, autophagy is repressed by anabolic growth factors (Lum et al, 2005; Melendez & Neufeld, 2008) in parallel to the promotion of cell proliferation and the increase in cellular uptake of nutrients (Kamada et al, 2000). As a result, a diminished availability or activity of growth factors causes a reduced uptake of nutrients and an increased rate of autophagy. Furthermore, stimulation of autophagy is relevant for protein and organelle turnover, helping to eliminate superfluous or damaged organelles and aggregate-prone proteins, both during tissue remodelling and during tissue homeostasis (Kourtis & Tavernarakis, 2009). Autophagy has been visualized directly in several tissues and organ systems in Drosophila (Neufeld, 2008). The larval fat body is a particularly interesting organ, as a high rate of autophagy takes place there during the prepupal period, providing essential nutrients and energy for the developing tissues of the adult fly (Rusten et al, 2004; Scott et al, 2004).
The regulation of autophagy in metazoans is only partly understood, and a relevant task is to identify further proteins that contribute to this control. Here we provide evidence that the novel protein diabetes- and obesity-regulated gene (DOR) has an essential role in autophagy. We have recently reported that DOR is abundantly expressed in insulin-sensitive tissues and highly repressed in the muscle of obese diabetic rats. DOR is a nuclear protein located in promyelocytic leukaemia nuclear bodies under basal conditions (Baumgartner et al, 2007). Here, we report that DOR is a shuttling protein that moves from the nucleus to the cytoplasm under conditions characterized by the activation of autophagy or cellular stress. The activation of autophagy drives DOR into autophagosomes, in which it promotes the formation of these organelles and regulates protein degradation in mammalian cells. CG11347, the DOR Drosophila homologue, is also required for the activation of autophagy in the fat body during prepupal development, thereby showing further that the role of DOR in autophagy has been conserved through evolution.
Results And Discussion
DOR exits the nucleus by autophagy activation
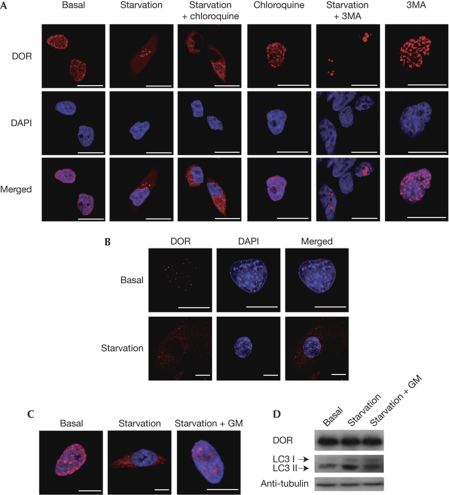
The nuclear cofactor DOR is localized mainly in promyelocytic leukaemia nuclear bodies in 1C9 myoblasts and in DOR-transfected HeLa cells under basal conditions (Fig 1A; Baumgartner et al, 2007). DOR shows a relatively short half-life (4.2±0.3 h; supplementary Fig S1 online) and is degraded through proteasomal activity. Thus, DOR abundance is increased by inhibitors of proteasomes such as MG132 or lactacystin (supplementary Fig S1 online).
Figure 1.
DOR moves out of the nucleus in response to the activation of autophagy. (A) The intracellular distribution of DOR is modified by starvation. HeLa cells were transiently transfected with DOR and incubated for 1 h with DMEM (basal), HBSS (starvation), 50 μM chloroquine or 10 mM 3MA. The intracellular localization of DOR was analysed by immunofluorescence and is shown in red. The nuclei are shown in blue (DAPI staining). Scale bars, 10 μm. (B) Endogenous DOR leaves the nucleus under starvation. C2C12 myoblasts were incubated for 30 min at 37°C with DMEM (basal) or HBSS (starvation). DOR is shown in red and nuclei are in blue (DAPI staining). Scale bars, 5 μm. (C,D) De-activation of autophagy returns DOR to the nucleus. HeLa cells were transiently transfected with DOR and incubated for 1 h with DMEM (basal) or HBSS (starvation). Cells were then incubated with normal DMEM for an additional hour (starvation+GM). The intracellular distribution of DOR was determined by immunofluorescence. DOR is shown in red and the nuclei are in blue. Scale bars, 5 μm. Cell lysates were also obtained and western blot assays were performed with specific antibodies. 3MA, 3-methyladenine; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle medium; DOR, diabetes- and obesity-regulated gene; HBSS, Hank's balanced salt solution; LC3, microtubule-associated protein 1A/1B-light chain 3.
Incubation of DOR-transfected HeLa cells with cellular stressors caused DOR to leave the nucleus (supplementary Fig S2 online). Activation of autophagy by amino-acid starvation (here referred to as starvation) or by rapamycin treatment of DOR-transfected HeLa or C2C12 muscle cells—which show endogenous DOR expression—caused DOR to leave the nucleus and re-localize to cytoplasmic punctate structures (Figs 1A,B and 2A; supplementary Fig S3 online).
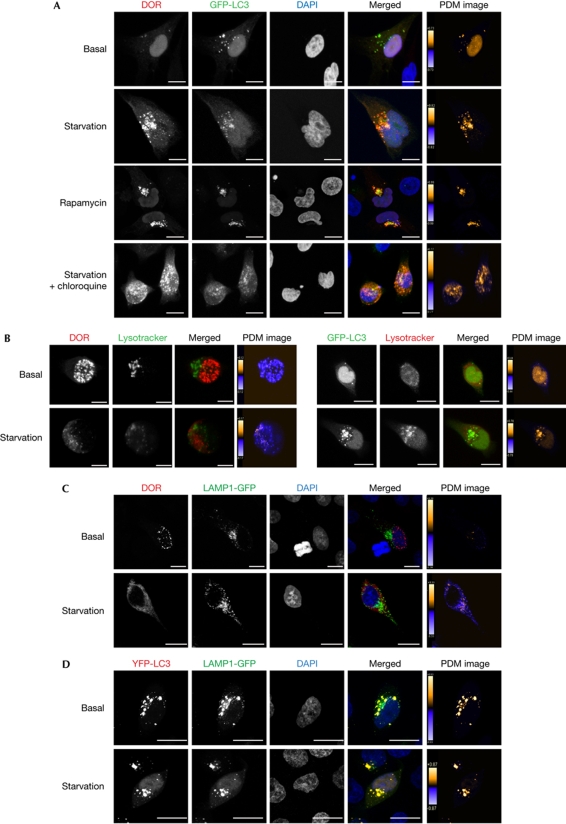
Figure 2.
DOR localizes to autophagosomes when autophagy is activated. (A) Confocal images of HeLa cells transiently transfected with DOR and GFP-LC3, and incubated for 1h with DMEM, HBSS (starvation), 2 μM rapamycin or HBSS containing 50 μM chloroquine. Nuclei were labelled with DAPI. Scale bars, 10 μm. (B,C) Confocal images of HeLa cells transiently transfected with DOR or GFP-LC3 and incubated with 50 nM Lysotracker in DMEM or HBSS. Scale bars, 10 μm. (D) Confocal images of HeLa cells transiently transfected with DOR and LAMP1-GFP, or with YFP-LC3 and LAMP1-GFP. Scale bars, 10 μm. Contrast-corrected merge RGB pictures and the Z-projection of Product of the differences from the mean (PDM) images are shown in all panels. Colour scales of PDM images have different maximal values, so the different conditions are not comparable from these projections. Product of the differences from the mean (PDM) values closer to 1 show reliable colocalized pixels. DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle medium; DOR, diabetes- and obesity-regulated gene; GFP, green fluorescent protein; HBSS, Hank's balanced salt solution; LAMP1, lysosomal-associated membrane protein 1; LC3, microtubule-associated protein 1A/1B-light chain 3.
We next analysed DOR localization during autophagy activation (Fig 1A; supplementary Fig S3 online). The abundance of punctate DOR structures in the cytosol on activation of autophagy was increased further when starved HeLa cells were additionally treated with chloroquine, which blocks the fusion of autophagosomes with lysosomes (Boya et al, 2005; Paludan et al, 2005). This finding suggests that these structures are related to lysosomes, possibly autophagosomes. Adding chloroquine to cells did not reveal any cytosolic DOR. Similarly, inhibition of basal autophagy with the inhibitor 3-methyladenine did not alter the nuclear localization of DOR; however, 3-methyladenine prevented DOR from leaving the nucleus in starved HeLa cells. The movement of DOR in response to starvation was reversible, as re-addition of amino acids for 1 h returned the protein back to the nucleus (Fig 1C; supplementary Fig S3 online), whereas total DOR abundance was not modified (Fig 1D). Starvation increased the abundance of microtubule-associated protein 1A/1B-light chain 3 (LC3) I and II isoforms, which was prevented by the reversal of starvation (Fig 1D).
DOR localizes to early autophagosomes
We next analysed whether DOR colocalized with autophagosomes. HeLa cells were transfected with green fluorescent protein (GFP)-LC3 and DOR, and immunofluorescence microscopy was performed with cells subjected to a range of conditions. Under basal conditions, DOR and GFP-LC3 were detected mainly in the nucleus, whereas during starvation, DOR and GFP-LC3 were in cytosolic punctate structures (Fig 2A; supplementary Fig S4 online). Image analysis indicated a substantial colocalization of both proteins in starvation—with or without chloroquine—and in the presence of rapamycin (Fig 2A; supplementary Fig S4 online).
Although almost all DOR colocalizes with GFP-LC3, some GFP-LC3 structures seem to have no DOR, indicating that DOR might associate only with a subset of LC3 structures. Indeed, DOR did not colocalize with lysosomal-associated membrane protein 1 (LAMP1)-GFP or with Lysotracker under any of the studied conditions (Fig 2B,C), which indicates that DOR is a marker of early autophagosomes. By contrast, a substantial colocalization of GFP-LC3 and Lysotracker or yellow fluorescent protein (YFP)-LC3 and LAMP1-GFP was detected (Fig 2B,C).
We also provide support for the physical interaction of DOR with components of the autophagy machinery such as LC3 or Golgi-associated ATPase enhancer of 16 kDa (GATE16) through pulldown, immunoprecipitation and two-hybrid assays (supplementary Fig S5 online).
DOR stimulates autophagy
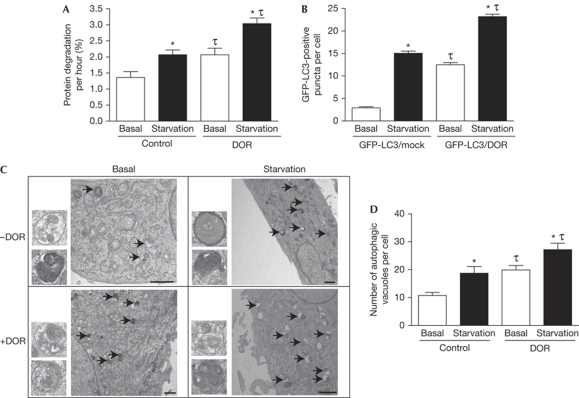
DOR protein is a marker of early autophagosomes when autophagy is activated, yet it does not colocalize with autolysosomes, which suggests that DOR might have a new regulatory role in autophagy. Next, we hypothesized that DOR enhances the degradation of proteins through autophagy. To test this, we measured the degradation of middle–long half-life proteins in control and DOR-transfected HeLa cells (Ogier-Denis et al, 1996). Under basal conditions, DOR gain-of-function cells showed a greater rate of protein degradation (52% increase; Fig 3A). Amino-acid starvation caused a marked increase in protein degradation (52% increase), and DOR gain-of-function together with starvation further activated this process (Fig 3A). To determine whether, under these conditions, DOR increased the number of autophagosomes, cells were transfected either with an irrelevant protein (thyroid hormone receptor TRα1) or with DOR and GFP-LC3. DOR gain-of-function markedly enhanced the number of GFP-LC3-positive puncta per cell both under basal conditions (4.3-fold) and in response to amino-acid starvation (40% increase; Fig 3B). The effect of DOR gain-of-function on autophagosome formation was substantiated further by electron microscopy. Control cells had a relatively low number of autophagosomes, which was visualized by transmission electron microscopy, whereas starvation increased their abundance (Fig 3C,D). DOR gain-of-function caused an increase in the number of autophagosomes under basal conditions and starvation caused a further increase (Fig 3C,D). DOR gain-of-function did not alter the cell viability analysed when assayed under basal conditions or after autophagy activation (data not shown). A NES motif was identified in DOR (residues 32–40), and the mutation (L36A/I37A/I38A/L40A) nullified the capacity of DOR to leave the nucleus in response to stress. This specific DOR mutant blocked the capacity of cells to respond to rapamycin by increasing the number of GFP-LC3-positive puncta (supplementary Fig S6 online).
Figure 3.
DOR stimulates autophagy. (A) DOR gain-of-function accelerates the degradation of proteins of middle–long half-life. Untreated HeLa cells and those transiently transfected with DOR were incubated in DMEM or HBSS. *Significant effects of starvation, P<0.01; τsignificant effects caused by DOR overexpression, P<0.01. (B) DOR overexpression enhances autophagosomal formation. LC3-GFP-positive vacuoles were counted in 100 transfected HeLa cells (with GFP-LC3 and TRα1, or GFP-LC3 and DOR). *Significant effects of starvation, P<0.001; τsignificant effects of DOR overexpression, P<0.001. (C,D) HeLa cells transiently transfected with empty pCDNA3 or DOR were incubated in DMEM (basal) or in HBSS (starvation). Cells were processed by transmission electron microscopy. Arrows indicate autophagosomes. Insets are autophagosomes (diameter, 407±63 nm). Scale bars, 1 μm. Autophagosomes were counted in 50 randomly chosen transfected cells. *Significant effects of starvation, P<0.05; τsignificant effects of DOR overexpression, P<0.05. DMEM; Dulbecco's modified Eagle medium; DOR, diabetes- and obesity-regulated gene; GFP, green fluorescent protein; HBSS; Hank's balanced salt solution; LC3, microtubule-associated protein 1A/1B-light chain 3.
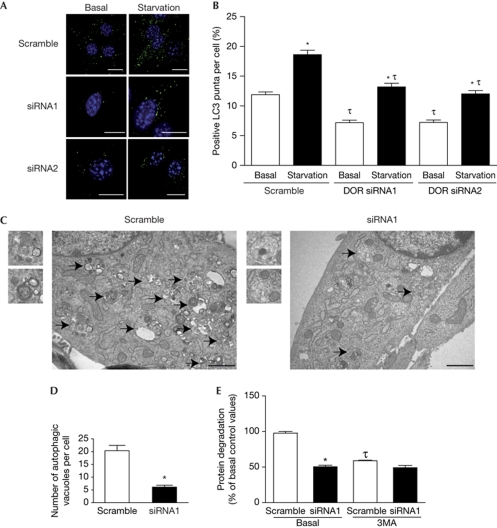
Next, we analysed the impact of DOR loss-of-function on autophagosome abundance. C2C12 muscle cells were infected with lentiviruses encoding scramble or DOR short interfering RNAs (siRNAs). Lentiviral-mediated expression of siRNA1 or siRNA2 caused an 80% and 70% reduction in DOR protein expression, respectively (data not shown). LC3 was detected by immunofluorescence assays in scramble or siRNA C2C12 myoblasts. Control cells showed an increased number of LC3-positive puncta in response to amino-acid starvation (Fig 4A,B). DOR knockdown with siRNA1 or siRNA2 caused a marked decrease in LC3-positive puncta under basal conditions (45% reduction) and during starvation (Fig 4A,B). siRNA1-induced DOR repression also caused a decrease in the number of autophagosomes (75% reduction; Fig 4C,D) and a substantial inhibition of protein degradation (Fig 4E). Together, our data indicate that DOR positively regulates autophagosome formation and protein degradation in cells.
Figure 4.
DOR loss-of-function reduces autophagy. (A,B) C2C12 myoblasts were previously infected with lentiviruses encoding scrambled RNA, DOR siRNA1 or DOR siRNA2. Endogenous LC3 was detected by immunofluorescence. LC3-positive puncta were counted in 100 cells per group under basal conditions or starved for 20 min. Scale bars, 10 μm. *Significant effects of starvation, P<0.001; τsignificant effects caused by DOR knockdown, P<0.001. (C,D) Scramble or DOR siRNA1 C2C12 cells were processed by transmission electron microscopy. Arrows indicate autophagosomes. Insets are autophagosomes. Scale bars, 1 μm. Autophagosomes were counted in 50 randomly chosen cells. *Significant effects of DOR repression, P<0.001. (E) Scramble or DOR siRNA1 C2C12 cells were incubated in DMEM with and without 10 mM 3MA. *Significant effects of DOR knockdown, P<0.01; τsignificant effects caused by 3MA, P<0.01. 3MA, 3-methyladenine; DMEM, Dulbecco's modified Eagle medium; DOR, diabetes- and obesity-regulated gene; LC3, microtubule-associated protein 1A/1B-light chain 3.
The fly DOR homologue is required for autophagy
The Drosophila genome contains a homologue of DOR (CG11347, hereafter referred to as dDOR) and two homologues of Atg8 (Atg8a and Atg8b; Rusten et al, 2004). dDOR is predicted to interact with Atg8a and Atg8b by high-throughput two-hybrid screening (Giot et al, 2003), thereby suggesting that the interaction between DOR and Atg8 is conserved from humans to flies.
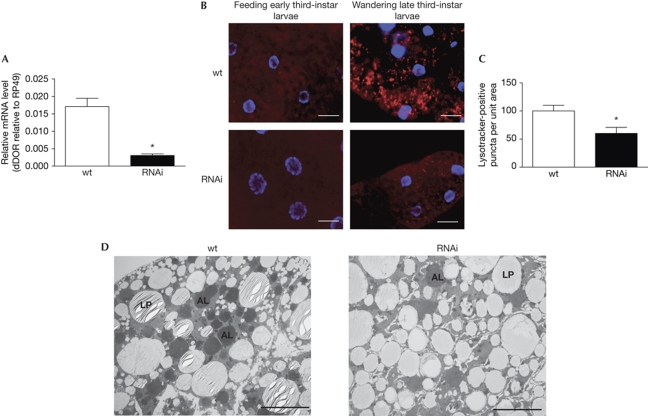
To test whether dDOR controls autophagy during development, we used three fly strains from the Vienna Drosophila RNAi Center (VDRC): two strains harbouring inducible RNAi expression constructs against the third exon of dDOR (T1 and T4) and one against the fourth exon (T2). Results shown here are from the T4 line, but similar results were obtained with the other strains. By crossing the T4 RNAi line to the ubiquitous tubulin-GAL4 driver—to express the RNAi construct in the entire animal—and extracting RNA from the resulting larvae, we confirmed that the RNAi constructs reduced dDOR expression levels by 95% compared with that of control flies (TubG4 × w1118; Fig 5A). Larvae in which dDOR was knocked down pupated normally and without delay, but died later as pupae (data not shown).
Figure 5.
CG11347 is required for autophagy activation in the fat body of Drosophila. (A) Decrease of mRNA levels in TubG4, UAS-CG11347-RNAi animals compared with TubG4 controls. All measurements were normalized to rp49. *Significant change, P<0.05. (B) Lysotracker staining of fat bodies. No Lysotracker staining is observed in feeding early third-instar larvae, whereas Lysotracker-positive granules (red) accumulate in fat body cells of wandering late third-instar controls (upper panels, TubG4/+). dDOR knockdown animals (TubG4, UAS-CG11347-RNAi T4, lower panels) show decreased staining of Lysotracker-positive granules in wandering late third-instar larvae. Nuclei are shown in blue. Scale bars, 50 μm. (C) The number of Lysotracker-positive puncta per unit area are shown for each genotype (control and RNAi), normalized to the value for control cells processed in parallel. *Significant change, P<0.05. (D) Transmission electron microscopy images of fat body cells from wandering late third-instar larvae. Developmental autophagy results in the accumulation of large autolysosomes (AL) in control (TubG4/+) larvae, whereas only a few autolysosomes are seen in CG11347 RNAi fat body cells of the same age. Control and RNAi fat bodies present lipid droplets (LP) with a ‘striped' appearance, which is an electron microscopy artefact. Scale bars, 5 nm. dDor, CG11347, a homologue of DOR; DOR, diabetes- and obesity-regulated gene; mRNA, messenger RNA; wt, wild type.
During insect metamorphosis, larval tissues such as the salivary gland, fat body and midgut undergo precisely timed periods of autophagy (Butterworth et al, 1988; Lee et al, 2002; Rusten et al, 2004). As dDOR expression by quantitative PCR is much higher in the fat body than in the salivary gland (data not shown), we chose the fat body as a model system to study autophagy and used Lysotracker as a reporter. At the end of larval development, third- instar larvae cease eating, leave the food and start wandering to find a dry place for pupation. During this wandering stage, autophagy is induced to high levels in the larval fat body and it has been proposed that it participates in the removal of this tissue by programmed cell death events that occur during metamorphosis (Penaloza et al, 2006; Neufeld & Baehrecke, 2008). In agreement with previous observations (Rusten et al, 2004), we detected numerous Lysotracker-positive puncta in the fat body of control larvae at the wandering stage, just before the onset of pupation (Fig 5B). By comparison, few puncta were observed in control larvae in early third instar, when they were still feeding, and there was no activation of autophagy. Compared with control animals, wandering third-instar dDOR knockdown animals showed a 40% decrease in Lysotracker-positive dots in the fat body (Fig 5B,C). Moreover, electron microscopy studies showed an abundance of autophagosomes and autolysosomes in the fat bodies of wandering third-instar control animals, whereas a strong reduction was detected in the dDOR knockdown group (Fig 5D). Together, these data indicate that dDOR is required in Drosophila for developmentally regulated autophagy at the onset of metamorphosis.
Concluding remarks
We propose that DOR is a new regulator of autophagy in mammalian and Drosophila cells. The observations that support this idea are as follows: (i) DOR interacts with LC3 and GATE16, two homologous proteins that participate as lipidated forms in the formation of autophagosomes in mammalian cells; (ii) DOR does not colocalize with LAMP1 protein or with Lysotracker, thereby indicating that it is a marker of early autophagosomes; (iii) DOR does not localize with autophagosomes when HeLa cells are treated with chloroquine, thereby indicating that its role is restricted to induced autophagy; (iv) DOR gain-of-function enhances the abundance of autophagosomes and accelerates protein degradation in HeLa cells; (v) DOR loss-of-function reduces autophagosome formation and protein degradation in muscle cells; and (vi) DOR loss-of-function represses autophagy of the fat body during Drosophila pupation.
Our data also indicate that DOR protein localization is sensitive to cellular stress. In particular, activation of autophagy by either amino-acid starvation or rapamycin treatment—the inhibition of the mammalian target of rapamycin (mTOR)—drives DOR into autophagosomes in a reversible manner. On this basis, we propose that the shuttling of DOR between the nucleus and cytosol is mTOR activity-dependent. Interestingly, the effect of amino-acid starvation on the efflux of DOR from the nucleus was abolished in the presence of 3-methyladenine, an inhibitor of class III phosphatidylinositol 3-kinase (Vps34), which indicates the existence of a functional crosstalk between mTOR and Vps34 in the regulation of the subcellular localization of DOR. In mammals, Vps34 participates in a complex that is essential for an early nucleation step in autophagosome formation (Pattingre et al, 2008). The interaction between Vps34 and mTOR is complex and poorly understood, and although Vps34 is required for insulin-stimulated and amino-acid-stimulated activation of mTOR (Byfield et al, 2005; Nobukuni et al, 2005), it also stimulates autophagy (Petiot et al, 2000).
The DOR protein has a short half-life and undergoes degradation through the proteasome. A functional interaction between autophagy and proteasomal activities has recently been reported (Korolchuk et al, 2009). On the basis of this observation, we propose that DOR is one of the proteins that control autophagy and is regulated by proteasomal activity.
Together with our previous findings, these results show that DOR is a bifunctional protein that operates both in the nucleus and in the cytosol. In the former, DOR acts as a nuclear co-factor, and binds to and co-activates the thyroid hormone receptor TRα1 (Baumgartner et al, 2007). In the latter, DOR participates in autophagosome formation and, consequently, causes enhanced autophagy and accelerated protein degradation. We therefore propose that DOR is a component of the regulatory machinery of autophagy in metazoans.
Methods
Cell lines and transfection. HeLa and C2C12 cells were maintained in Dulbecco's modified Eagle medium (Gibco, Paisley, UK). For transient transfection assays, cells were typically transfected with 6 μg DNA using calcium and phosphate solutions. See supplementary information online.
Immunofluorescence assays. After specific treatments, cells were fixed in 4% paraformaldehyde and washed twice with PBS. Immunocytochemistry assays were performed using DOR (1/150; Baumgartner et al, 2007) and LC3 (clone 5F10; Nanotools) antibodies. Hoechst (1/2,000; Molecular Probes, Eugene, OR, USA) was used to label DNA. See supplementary information online.
Middle–long half-life protein degradation assay. Degradation of stable proteins was measured by following a standard method (Ogier-Denis et al, 1996). See supplementary information online.
Drosophila strains and transgenic lines. w1118, P{GD5124}v41186, P{GD5124}v41183/TM6, v105330 and tubulin-Gal4 were obtained from the Bloomington Stock Center (http://fly.bio.indiana.edu) and from the VDRC. See supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Data
Acknowledgments
We thank the Advanced Digital Microscopy Facility (Institute for Research in Biomedicine, Barcelona), the Unit of Electron Microscopy (Serveis Cientificotècnics, Universitat de Barcelona), I. Castrillón, J.C. Monasterio and M. Alloza for technological assistance. We thank T. Yates for editorial support, E. Knecht for advice on measurements of protein degradation, and S. Lavandero, Z. Elazar and J. Lippincott-Schwartz for providing the plasmids. C.M. is an FPI fellow and V.A.F. an FPU fellow from the Ministerio de Educación y Ciencia (MEC), Spain. This study was supported by research grants from the MEC (SAF2008-03803), the Generalitat de Catalunya (2005SGR00947), and CIBERDEM (Instituto de Salud Carlos III). A.Z. is the recipient of a Science Intensification Award from the University of Barcelona. A.A.T. is supported by a Helmholtz Young Investigator grant. During the revision of this manuscript, Novak et al (Mol Cell Biol, 2009) reported a role of tp53inp2, a human DOR isoform, in autophagy.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baumgartner BG et al. (2007) Identification of a novel modulator of thyroid hormone receptor-mediated action. PLoS ONE 2: e1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P et al. (2005) Inhibition of macroautophagy triggers apoptosis 1. Mol Cell Biol 25: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth FM, Emerson L, Rasch EM (1988) Maturation and degeneration of the fat body in the Drosophila larva and pupa as revealed by morphometric analysis. Tissue Cell 20: 255–268 [DOI] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM (2005) hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 280: 33076–33082 [DOI] [PubMed] [Google Scholar]
- Giot L et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC (2009) Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell 33: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N (2009) Autophagy and cell death in model organisms. Cell Death Differ 16: 21–30 [DOI] [PubMed] [Google Scholar]
- Lee CY, Simon CR, Woodard CT, Baehrecke EH (2002) Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev Biol 252: 138–148 [DOI] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB (2005) Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 6: 439–448 [DOI] [PubMed] [Google Scholar]
- Melendez A, Neufeld TP (2008) The cell biology of autophagy in metazoans: a developing story. Development 135: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ (2007) Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr 27: 19–40 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP (2008) Genetic manipulation and monitoring of autophagy in Drosophila. Methods Enzymol 451: 653–667 [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Baehrecke EH (2008) Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy 4: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T et al. (2005) Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA 102: 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E, Houri JJ, Bauvy C, Codogno P (1996) Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem 271: 28593–28600 [DOI] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C (2005) Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307: 593–596 [DOI] [PubMed] [Google Scholar]
- Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P (2008) Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 90: 313–323 [DOI] [PubMed] [Google Scholar]
- Penaloza C, Lin L, Lockshin RA, Zakeri Z (2006) Cell death in development: shaping the embryo. Histochem Cell Biol 126: 149–158 [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P (2000) Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 275: 992–998 [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H (2004) Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell 7: 179–192 [DOI] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP (2004) Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7: 167–178 [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data