Abstract
Autophagy is the cellular homeostatic pathway that delivers large cytosolic materials for degradation in the lysosome. Recent evidence indicates that autophagy mediates selective removal of protein aggregates, organelles and microbes in cells. Yet, the specificity in targeting a particular substrate to the autophagy pathway remains poorly understood. Here, we show that the mitochondrial protein Nix is a selective autophagy receptor by binding to LC3/GABARAP proteins, ubiquitin-like modifiers that are required for the growth of autophagosomal membranes. In cultured cells, Nix recruits GABARAP-L1 to damaged mitochondria through its amino-terminal LC3-interacting region. Furthermore, ablation of the Nix:LC3/GABARAP interaction retards mitochondrial clearance in maturing murine reticulocytes. Thus, Nix functions as an autophagy receptor, which mediates mitochondrial clearance after mitochondrial damage and during erythrocyte differentiation.
Keywords: GABARAP, LC3, mitophagy, Nix, selective autophagy
Introduction
Mounting evidence suggests that selective degradation of bulky cytosolic substrates, such as protein aggregates, damaged organelles and intracellular microbes, is mediated by macroautophagy (autophagy hereafter; Kirkin et al, 2009b). Its main hallmark is the formation of double-membrane vesicles—autophagosomes—that engulf cargo and deliver it to lysosomes for degradation (Xie & Klionsky, 2007; Nakatogawa et al, 2009). Ubiquitination has been proposed as a signal for selective autophagy (Kirkin et al, 2009b). Receptor proteins, p62 and neighbour of BRCA1 gene 1 (NBR1), which interact with both ubiquitin (Ub) and autophagosome-specific Ub-like (UBL) proteins, Atg8-family proteins LC3/GABARAP, have been identified (Komatsu et al, 2007; Pankiv et al, 2007; Kirkin et al, 2009a).
Specific elimination of damaged mitochondria by autophagy (mitophagy) has been demonstrated convincingly (Elmore et al, 2001; Priault et al, 2005; Kim et al, 2007; Narendra et al, 2008). In mammals, mitophagy is important in programmed mitochondrial clearance during differentiation of reticulocytes and T lymphocytes (Schweers et al, 2007; Kundu et al, 2008; Sandoval et al, 2008; Zhang et al, 2009b). The mitochondrion-localized proteins Bcl2/E1B 19 kDa-interacting protein 3-like protein (BNIP3) and Nix (also known as BNIP3L) have been implicated in the removal of mitochondria during an autophagic response (Zhang & Ney, 2009). Thus, hypoxia-induced autophagy depends on both BNIP3 and Nix, and can be mimicked by overexpression of their respective BH3 domains (Bellot et al, 2009). Furthermore, Nix is indispensable for programmed elimination of mitochondria during reticulocyte maturation. Interestingly, in Nix-deficient reticulocytes autophagy per se is functional, but mitochondria fail to enter the autophagosomes (Schweers et al, 2007; Sandoval et al, 2008). Moreover, knockout of autophagy-specific genes (Ulk1, Atg5 and Atg7) does not abolish fully programmed mitochondrial clearance in reticulocytes (Matsui et al, 2006; Kundu et al, 2008; Zhang et al, 2009b), suggesting the existence of autophagy-independent pathways for programmed mitochondrial clearance in this cell lineage.
Selective autophagy implies the targeted formation of autophagosomes around a particular substrate (for example, mitochondrion). At the molecular level, selectivity can be mediated by the binding of autophagy receptors to membrane-anchored, autophagy-specific UBL proteins. This idea has been supported by the identification of several receptors that bind to UBL proteins, coupling their substrate conjugation to a biological response (Kirkin & Dikic, 2007). The common binding site for Atg8-family proteins contains the W/YxxL/I core motif and is referred to as the LC3-interacting region (LIR; Komatsu et al, 2007; Pankiv et al, 2007; Kirkin et al, 2009a). Here, we describe the identification of Nix as a mammalian mitophagy receptor.
Results
Nix interacts with Atg8/LC3/GABARAP proteins
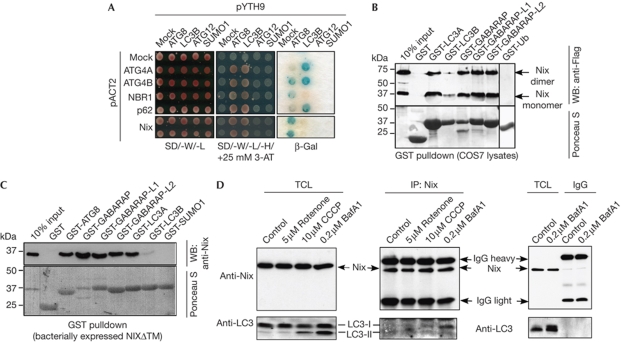
To search for proteins that interact with autophagy-specific UBL proteins, we used Saccharomyces cerevisiae (Sc)Atg8 as bait in yeast two-hybrid screens. One Atg8-interacting clone encoded the full-length Nix protein previously linked to autophagy (Zhang & Ney, 2009). This interaction was confirmed by transformation of the isolated Nix-encoding vector into yeast strains expressing ScAtg8 (Fig 1A). Whereas ATG4A, ATG4B and p62 interacted preferentially with LC3B in this assay, Nix, similarly to NBR1 (Kirkin et al, 2009a), interacted strongly with Atg8 but not LC3B (Fig 1A). The Nix:Atg8 interaction was specific as Nix failed to interact with Atg12 and small ubiquitin-like modifier 1 (SUMO1) in this assay (Fig 1A).
Figure 1.
Nix interacts with Atg8/LC3/GABARAP proteins. (A) Yeast clones harbouring empty bait vector (pYTH9, mock) or those encoding ScAtg8, LC3B, Atg12 and SUMO1 were transformed with empty prey vector (pACT2, mock) or those encoding fragments of ATG4A, ATG4B, NBR1, p62 and full-length Nix. Interaction was assessed by yeast growth on the SD-W/-L/-H medium with a β-gal assay. (B) GST pulldown assays using cell extracts of Flag-Nix-expressing COS7 cells and immobilized GST or the indicated GST fusions. Coprecipitated Nix was detected with Flag antibodies. (C) Purified GST-tagged NixΔTM was cleaved from the GST moiety by thrombin, purified and used for precipitation by GST fusion proteins. Precipitated proteins were analysed by western blotting (WB) with Nix antibodies. Ponceau S staining was used to visualize GST-fusion proteins. (D) HeLa cells were treated as indicated and endogenous proteins were immunoprecipitated (IP) from cell extracts using Nix antibody and analysed by using Nix and LC3 antibodies. β-gal, β-galactosidase; BafA1, bafilomycin-A1; CCCP, carbonyl cyanide m-chlorophenyl hidrazone; GST, glutathione-S-transferase; IgG, immunoglobulin G; NixΔTM, recombinant Nix lacking a TM; SUMO1, small ubiquitin-like modifier 1; TCL, total cell lysate; TM, transmembrane domain.
The in vitro interaction of Nix with the mammalian Atg8 protein family was investigated further in biochemical assays. Flag-Nix appeared on SDS-polyacrylamide gel electrophoresis gels as two distinct protein species of approximately 40 and 70 kDa (monomeric and dimeric forms; Fig 1B; Imazu et al, 1999). Both Nix forms interacted with Atg8-family proteins, but not with Ub in glutathione-S-transferase (GST) pulldown assays (Fig 1B). The Nix transmembrane (TM) domain is responsible for dimerization (Sulistijo & MacKenzie, 2006; Bocharov et al, 2007). Recombinant Nix lacking a TM (NixΔTM) was used to show direct interaction of Nix with human Atg8 homologues (Fig 1C). In both assays, Nix interacted strongly with all tested LC3/GABARAP proteins except for LC3B, which showed much weaker binding.
To confirm the in vivo interaction between Nix and LC3/GABARAP, we performed co-immunoprecipitation experiments with endogenous proteins. We found that Nix co-precipitated weakly with LC3 under normal growth conditions (Fig 1D), but interacted more strongly with LC3 under conditions that induce mitochondrial stress (rotenone or carbonyl cyanide m-chlorophenyl hidrazone (CCCP) treatment) or block autophagy (bafilomycin-A1 treatment; Fig 1D).
Nix contains LIRs
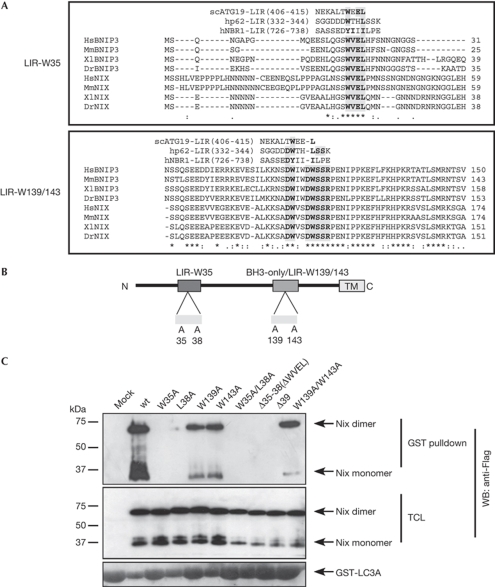
The known autophagy receptors bind to Atg8 family members through a linear sequence motif known as the LIR. Its core consensus sequence is W/YxxL/I (Kirkin et al, 2009b). By using multiple species alignment of Nix and its relative BNIP3, we predicted several putative LIR sequences in Nix (Fig 2A). We found one WxxL motif near the amino-terminal of Nix (amino acids 35–38 in murine Nix) that is well conserved between Nix and BNIP3 (Fig 2A,B). This motif closely resembles the LIR found in ScAtg19 protein (Noda et al, 2008). The other potential LIR is located adjacent to an atypical BH3-like domain (Fig 2A,B). It has low homology to the W/YxxL/I core LIR consensus, but instead contains a double DW motif (amino acids 138–143) that is similar to p62 (DDWTHL) and NBR1 (EDYIIIL; Pankiv et al, 2007; Kirkin et al, 2009a). To distinguish between the two putative LIR sequences, we refer to the N-terminal motif as LIR-W35 and to the BH3 domain-specific one as LIR-W139/143 (Fig 2B).
Figure 2.
Identification of Nix-LIRs. (A) Amino-acid sequences of Nix and related BNIP3 protein from the indicated species were aligned using the ClustalW2 program (EMBL-EBI, Cambridge, UK). The published LIR sequences were aligned manually alongside Nix/BNIP3 for comparison. The shaded regions indicate highly conserved residues in the two putative LIRs. Consensus symbols * (identical residues), : (conserved substitution) and . (semi-conserved substitution) are valid for the Nix/BNIP3 alignment only. (B) A schematic of Nix showing its domain organization and the point mutations used in this study to identify LIRs. (C) Lysates of COS7 cells transfected with Flag-wt-Nix or the indicated mutants were incubated with immobilized GST-LC3A. The coprecipitated proteins were detected by western blotting (WB) with Flag antibodies. BNIP3, Bcl2/E1B 19kDa-interacting protein 3-like protein; Dr, Danio reio; GST, glutathione-S-transferase; Hs, Homo sapiens; LIR, LC3-interacting region; Mm, Mus musculus; wt, wild type; Xl, Xenopus laevis.
To determine whether Nix binding to Atg8-family proteins depends on one of the putative LIR motifs, we performed a series of GST pulldown experiments with Nix mutants (Fig 2C). Mutation of the conserved W35 residue in the N-terminal LIR to an alanine ablated most of Nix binding to LC3A (Fig 2C). Furthermore, the L38A mutation also abolished binding (Fig 2C). Trp:Ala replacements in the W139/W143 motif impaired the interaction only slightly (Fig 2C). In addition, removal of WVEL either alone (Δ35–38 mutant) or in the context of the larger N-terminal deletion (Δ39 mutant) led to ablation of Nix:LC3A binding (Fig 2C). These results suggest that LIR-W35 is the main Atg8-family protein-interaction site in Nix, whereas LIR-W139/143 has a minor role.
Characterization of Nix-LIR binding to LC3 proteins
Next, we characterized the interaction of the putative Nix-LIRs with Atg8-family proteins by isothermal titration calorimetry (ITC) and nuclear magnetic resonance (NMR). We used short synthetic peptides, corresponding to LIR-W35 (Nix-W35) and LIR-W139/143 (Nix-W139/143; supplementary Fig S1A online). A peptide corresponding to p62 LIR (p62-LIR) was used as reference (Ichimura et al, 2008; Noda et al, 2008). LC3B showed the lowest affinity in the binding assays (Fig 1), so we chose this protein to establish the basal level of Atg8-family interactions with Nix-LIRs. For comparison, we assessed the interaction of Nix-LIRs with LC3A, as a representative of the Atg8 protein family with stronger Nix binding (Fig 1). First, the ITC experiments showed that the Nix-W35 peptide interacted with both LC3B and LC3A, albeit with differing affinities: KD of 91 and 28 μM, respectively (Table 1). Second, an ITC comparison of the interaction between LC3 proteins and Nix-W35 versus Nix-W139/143 peptides showed that LC3 proteins interacted with Nix-W35 much more strongly than with Nix-W139/143: KD values of 91 μM (LC3B) and 28 μM (LC3A) versus 670 μM (LC3B) and 130 μM (LC3A), respectively. Experiments with the reference peptide (p62-LIR) showed that the Nix-LIRs interact much more weakly with Atg8-family proteins than p62 does (KD of 1.5 μM for the p62-LIR:LC3B interaction; Table 1). NMR titration experiments with 15N-labelled proteins confirmed the ITC results (supplementary Fig S1B,C online).
Table 1.
KD values acquired by ITC analyses of the interaction of Nix-LIR peptides with LC3 proteins
| LC3B | LC3A | |
|---|---|---|
| p62-LIR | 1.5 μM | NA |
| Nix-W35 | 91 μM | 28 μM |
| Nix-W139/W143 | 670 μM | 130 μM |
| ITC, isothermal titration calorimetry; LIR, LC3-interacting region; NA, not available. | ||
Nix recruits GABARAP-L1 to depolarized mitochondria
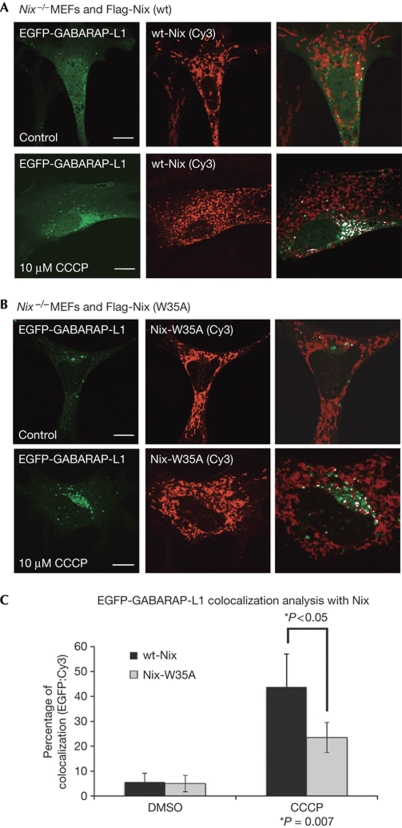
Previous studies showed that Nix localizes to the outer mitochondrial membrane where it affects mitochondrial integrity (Zhang & Ney, 2009). To see if Nix recruits LC3/GABARAP to stressed mitochondria, we performed a series of colocalization studies of cells cultured under normal growing conditions or treated with mitochondrial poison, CCCP, to induce mitochondrial stress and autophagy (Chen et al, 2007). For these studies we used GABARAP-L1 because of its strong interaction with Nix in our previous assays (Fig 1B).
First, we examined the effect of CCCP on enhanced green fluorescent protein (EGFP)–GABARAP-L1 colocalization with mitochondria in reconstituted Nix−/− primary mouse embryonic fibroblasts (MEFs). Nix−/− MEFs were reconstituted with either wild-type (wt) Nix (Flag-wt-Nix) or the W35A mutant of Nix (Flag-Nix-W35A). Importantly, we observed that both versions of Nix colocalized with mitochondria, indicating that the W35A mutation of Nix does not disrupt its recruitment to mitochondria (Fig 3A,B).
Figure 3.
Nix recruits EGFP-GABARAP-L1 to stressed mitochondria in reconstituted Nix−/− MEFs. Nix−/− MEFs cotransfected with EGFP-GABARAP-L1 and wt-Nix (A), or Nix-W35A (B), were incubated with and without mitochondrial poison CCCP for 3 h. Cells were then fixed and analysed for colocalization between EGFP-GABARAP-L1 and Nix (stained with Flag antibodies). The final panel in each row shows colocalization between Flag-Nix-Cy3 and GABARAP-L1 using the ‘colocalization' highlighter plug-in for ImageJ software (NIH, Bethesda, MA, USA). (C) Quantification was achieved by counting the number of complete colocalizations, EGFP/Cy3 per cell for approximately 30 cells per condition. Results are expressed as a percentage of colocalization (EGFP:Cy3) in control and CCCP-treated cells. Error bars in (C) show the standard deviation (s.d.) of three independent experiments. Statistical analysis was performed using the Mann–Whitney t-test. CCCP, carbonyl cyanide m-chlorophenyl hidrazone; DMSO, dimethyl sulfoxide; EGFP, enhanced green fluorescent protein; MEF, mouse embryonic fibroblast; wt, wild type.
In Nix−/− MEFs cotransfected with EGFP-GABARAP-L1 and Flag-wt-Nix, GABARAP-L1 colocalized infrequently with Nix in mitochondria (Fig 3A); however, on treatment with CCCP there was an increase in puncta with colocalization of GABARAP-L1 and Nix, indicating recruitment of GABARAP-L1 to Nix in mitochondria (Fig 3A,C). Following treatment with CCCP, the ability of Nix-W35A to recruit EGFP-GABARAP-L1 to stressed mitochondria was impaired but not lost completely (Fig 3B,C). Quantification of GABARAP-L1 and Nix (wt or W35A) colocalization showed that there is approximately a 50% reduction in GABARAP-L1 recruitment to Nix-W35A mitochondria, as compared with wt Nix (Fig 3C). We obtained similar results with transfected HeLa cells (supplementary Fig S2A–D online). We also assessed the recruitment of EGFP-GABARAP-L1 to mitochondria in Nix+/+ and Nix−/− MEFs, and observed its impairment in the absence of Nix (supplementary Fig S3 online). These results suggest that recruitment of EGFP-GABARAP-L1 to mitochondria, under stress conditions, is partly dependent on the Nix-W35 LIR motif.
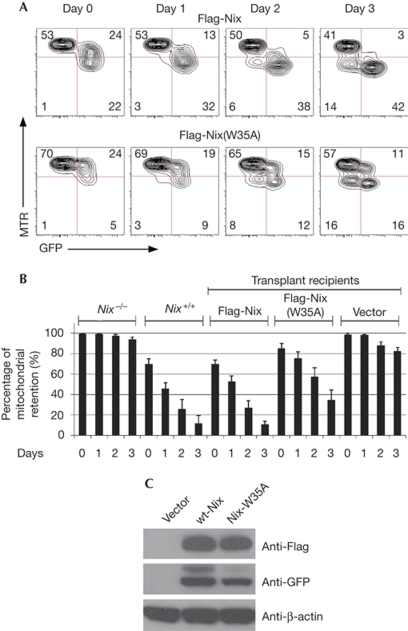
Mitochondrial clearance is partly mediated by LIR-35
Nix deficiency in mice leads to pronounced impairment in mitochondrial clearance during reticulocyte maturation (Schweers et al, 2007; Sandoval et al, 2008). We asked whether mitochondrial clearance in these cells depends on Nix LIR sequences, and hence on the binding of Nix to LC3/GABARAP proteins. To investigate this we used Nix−/− mice (Schweers et al, 2007). We expressed wt-Nix or LIR mutants of Nix in Nix−/− reticulocytes by using retrovirus-mediated gene transfer into Nix−/− bone marrow, followed by transplantation into lethally irradiated recipients. In unstressed mice, reconstitution with either wt-Nix or its LIR mutants (W35A, W139/143A and W35/139/143A) rescued mitochondrial clearance (data not shown). We speculated that mitochondrial clearance was partly defective, but that erythrocytes in vivo could clear their mitochondria, given enough time. To investigate this possibility, we examined mitochondrial clearance in reticulocytes cultured ex vivo. We induced reticulocytosis in transplant recipients with phenylhydrazine (Vannucchi et al, 2001), harvested the reticulocyte-enriched blood and monitored the mitochondrial clearance over three days. Under these conditions, wt-Nix rescued mitochondrial clearance to the same levels seen in Nix+/+ mice. By contrast, LIR-W35A-mutant Nix showed impaired ability to rescue mitochondrial clearance (Fig 4A,B). Western blot analysis of Nix protein and the W35A mutant confirmed similar expression levels of reconstituted Nix protein in transduced erythroid cells (Fig 4C). These results are consistent with a role for LC3/GABARAP binding to Nix in programmed mitochondrial clearance in reticulocytes.
Figure 4.
Mitochondrial clearance is impaired in Nix−/− reticulocytes transduced with Nix-W35A. (A) Flow cytometry of reticulocyte-enriched blood, cultured in vitro for 3 days, stained with MTR. The blood was taken from mice transplanted with Nix−/− bone marrow and reconstituted with wt-Nix and Nix-W35A viruses. Reticulocytes were induced by phenylhydrazine treatment. The viruses also express GFP. (B) Mitochondrial clearance in the GFP-positive fraction of the reticulocyte-enriched blood, as described in panel (A). Mice were reconstituted with wt-Nix, LIR-W35A or empty vector viruses. Data from Nix−/− and Nix+/+ mice are shown for comparison. (C) Anti-Flag, anti-GFP and anti-β-actin western blots of fetal liver cells transduced with Flag-Nix and Flag-W35A-Nix viruses, normalized to total protein. GFP, green fluorescent protein; LIR, LC3-interacting region; MTR, MitoTracker Red; wt, wild type.
Discussion
Here we identify Nix as an interaction partner for autophagy-specific UBL proteins, LC3/GABARAP, implicated in mitophagy. Nix binds to LC3/GABARAP through two potential LIRs that share selected features with the published LIR sequences in other proteins. Of the two LIRs characterized in this study, LIR-W35 is particularly important as its mutation or deletion abolishes Nix:LC3/GABARAP binding in vitro and in vivo. Importantly, while this paper was in preparation, Schwarten et al reported the identification of Nix as a GABARAP-interacting protein. A Nix-derived peptide identical to LIR-W35 was implicated in the GABARAP–Nix interaction and a mutation of W35 to an alanine abolished this binding (Schwarten et al, 2009).
Nix binds relatively weakly to LC3B in comparison with LC3A and GABARAP proteins (Fig 1; Table 1; supplementary Fig S1 online; Schwarten et al, 2009). This suggests that LC3A and GABARAP proteins, rather than LC3B, have a leading role in Nix-dependent mitophagy. Further, Nix recruits GABARAP-L1 to stressed mitochondria in a LIR-W35-dependent manner, suggesting involvement of previously uncharacterized Atg8-family proteins in the clearance of damaged mitochondria. In line with this, ATG4D, an isoform of the LC3/GABARAP-specific cystein protease, which is essential for LC3/GABARAP function, was shown recently to have increased specificity to GABARAP-L1 and to localize to mitochondria (Betin & Lane, 2009).
The sequence conservation of Nix LIRs across the Chordata phylum suggests that the interaction between Nix and the autophagic machinery has been essential in the course of evolution. Recently, a novel mitochondrial protein, Atg32, was characterized as a selective autophagy receptor for autophagic degradation of stressed mitochondria in yeast (Kanki et al, 2009; Okamoto et al, 2009). As no homologue of Atg32 in higher organisms could be identified, we propose that Nix, and possibly BNIP3, might fulfill its function in higher organisms. Atg32 shares several features with Nix: (i) a carboxy-terminal TM domain that is essential for the targeting of the protein to the outer mitochondrial membrane; (ii) an LIR responsible for binding to Atg8; and (iii) Atg32 is induced by stress cues and recruits Atg8 to stressed mitochondria. Interestingly, the LIR of Atg32 is also only partly required for mitophagy as Atg32 interacts with the Cvt pathway adaptor Atg11 independently of its LIR domain. This suggests that several signals might be essential for efficient mitophagy in yeast.
Programmed mitochondrial clearance in reticulocytes is Nix-dependent (Schweers et al, 2007; Sandoval et al, 2008) and Nix binding to LC3/GABARAP through LIR-W35 might contribute to this process during reticulocyte maturation (Fig 4). The effect of the LIR mutation on Nix-mediated mitochondrial clearance in vivo is partial, indicating that other properties of Nix are also important in this process. Similarly, defects in general autophagy caused by gene targeting of Ulk1, Atg7 and Atg5 also had a partial effect on mitochondrial clearance in reticulocytes (Matsui et al, 2006; Kundu et al, 2008; Zhang et al, 2009b). As, however, ATG5/ATG7-independent autophagy has now been described, more studies are required to understand the role of autophagy in reticulocyte maturation (Nishida et al, 2009).
We envisage that Nix-dependent recruitment of LC3/GABARAP proteins associated with autophagosomes to mitochondria might be one factor in mediating membrane tethering and/or hemifusion of mitochondria with autophagosomes. However, other signals—for example, ubiquitination of mitochondrial proteins or interactions with lysosomal components—might mediate the full incorporation of mitochondria into autophagosomes (supplementary Fig S4 online). This could be the reason why mitochondria align to autophagosomes, but are not incorporated in Nix−/− reticulocytes. Several E3 ligases associated with mitochondria, such as mitochondrial E3 ubiquitin ligase, membrane-associated ring finger 5 or LISTERIN, might participate in ubiquitination of mitochondrial membrane proteins and contribute to mitochondrial clearance through mitophagy (Karbowski et al, 2007; Li et al, 2008; Chu et al, 2009). Furthermore, a recent work showed that translocation of E3 ligase Parkin and its interacting partner PTEN-induced putative kinase 1 to damaged mitochondria is required for efficient mitophagy (Narendra et al, 2008; Dagda et al, 2009).
Nix might mediate mitochondrial clearance in an LC3/GABARAP-independent or autophagy-independent manner. Our model (supplementary Fig S4 online) could explain why a deficiency in autophagy rather than a deficiency in Nix generally has a more subtle effect on mitochondrial clearance: lack of Nix would block both autophagy-dependent and autophagy-independent mechanisms, whereas failure of Nix to interact with autophagosome-localized LC3/GABARAP proteins would only inhibit autophagy-dependent mitochondrial clearance.
Methods
Detailed experimental procedures are described in the supplementary information online.
Protein purifications and biochemical assays. Yeast transformation assay and interaction between recombinant, purified proteins and assays of cellular proteins were performed as described in the supplementary information online.
ITC and NMR spectroscopy. ITC experiments were performed at 25°C using a VP-ITC calorimeter (MicroCal Inc., Northampton, MA, USA) and analysed with the ITC-Origin software (MicroCal Inc.) based on the assumption of one-site binding reactions.
Laser-scanning microscopy. Images of fixed and stained cells expressing fusions of fluorescent proteins cells were acquired by the LSM 510 META laser-scanning microscope (Carl Zeiss MicroImaging, Jena, Germany).
Retrovirus-mediated gene transfer of Nix into Nix−/− mice and analysis of mitochondrial clearance. Retrovirus-mediated gene transfer into Nix−/− mice was performed as described in the supplementary information online. Reticulocytosis was induced by phenylhydrazine treatment and reticulocyte-enriched blood was cultured and stained with MitoTracker Red, as previously described (Zhang et al, 2009a).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary information
Acknowledgments
I.D., A.S.R. and A.O. acknowledge the support from the Deutsche Forschungsgemeinschaft and the Cluster of Excellence ‘Macromolecular Complexes' of the Goethe University Frankfurt (EXC115). A.S.R. and A.O. were supported by a DFG grant (RE1575-1/1). I.D. and J.T. were supported by the Terry Fox Foundation. P.A.N. was funded by a National Institutes of Health grant (DK074519), National Institutes of Health Cancer Center support grant (P30 CA21765) and the American, Lebanese and Syrian Associated Charities. I.N. is an EMBO long-term postdoctoral fellow.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM (2009) Hypoxia-induced autophagy is mediated through HIF-induction of BNIP3 and BNIP3L via their BH3-domains. Mol Cell Biol 29: 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin VM, Lane JD (2009) Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 122 (Part 14): 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV et al. (2007) Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger. J Biol Chem 282: 16256–16266 [DOI] [PubMed] [Google Scholar]
- Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB (2007) Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci 120 (Part 23): 4155–4166 [DOI] [PubMed] [Google Scholar]
- Chu J et al. (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci USA 106: 2097–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ III, Kulich SM, Tandon A, Park D, Chu CT (2009) Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 284: 13843–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ (2001) The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J 15: 2286–2287 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 283: 22847–22857 [DOI] [PubMed] [Google Scholar]
- Imazu T, Shimizu S, Tagami S, Matsushima M, Nakamura Y, Miki T, Okuyama A, Tsujimoto Y (1999) Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene 18: 4523–4529 [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol 178: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ (2007) Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Dikic I (2007) Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol 19: 199–205 [DOI] [PubMed] [Google Scholar]
- Kirkin V et al. (2009a) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I (2009b) A role for ubiquitin in selective autophagy. Mol Cell 34: 259–269 [DOI] [PubMed] [Google Scholar]
- Komatsu M et al. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB (2008) Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3: e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N (2006) Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Commun 339: 485–489 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461: 654–658 [DOI] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC (2005) Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ 12: 1613–1621 [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J (2008) Essential role for Nix in autophagic maturation of erythroid cells. Nature 454: 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, Hersch N, Hoffmann B, Merkel R, Willbold D (2009) Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy 5: 690–698 [DOI] [PubMed] [Google Scholar]
- Schweers RL et al. (2007) NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA 104: 19500–19505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulistijo ES, MacKenzie KR (2006) Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix–helix interactions. J Mol Biol 364: 974–990 [DOI] [PubMed] [Google Scholar]
- Vannucchi AM et al. (2001) Accentuated response to phenylhydrazine and erythropoietin in mice genetically impaired for their GATA-1 expression (GATA-1(low) mice). Blood 97: 3040–3050 [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ney PA (2009) Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 16: 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kundu M, Ney PA (2009a) Mitophagy in mammalian cells: the reticulocyte model. Methods Enzymol 452: 227–245 [DOI] [PubMed] [Google Scholar]
- Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, Ney PA (2009b) Mitochondrial clearance is regulated by Atg7-dependent and independent mechanisms during reticulocyte maturation. Blood 114: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information