Abstract
RNA-directed DNA methylation (RdDM) in plants requires two RNA polymerase (Pol) II-related RNA polymerases, namely Pol IV and Pol V. A genetic screen designed to reveal factors that are important for RdDM in a developmental context in Arabidopsis identified DEFECTIVE IN MERISTEM SILENCING 4 (DMS4). Unlike other mutants defective in RdDM, dms4 mutants have a pleiotropic developmental phenotype. The DMS4 protein is similar to yeast IWR1 (interacts with RNA polymerase II), a conserved putative transcription factor that interacts with Pol II subunits. The DMS4 complementary DNA partly complements the K1 killer toxin hypersensitivity of a yeast iwr1 mutant, suggesting some functional conservation. In the transgenic system studied, mutations in DMS4 directly or indirectly affect Pol IV-dependent secondary short interfering RNAs, Pol V-mediated RdDM, Pol V-dependent synthesis of intergenic non-coding RNA and expression of many Pol II-driven genes. These data suggest that DMS4 might be a regulatory factor for several RNA polymerases, thus explaining its diverse roles in the plant.
Keywords: IWR1, Pol IV, Pol V, RNA-directed DNA methylation, RNA polymerase
Introduction
RNA-directed DNA methylation (RdDM) is a small RNA-mediated epigenetic modification that is highly developed in plants. In addition to conserved DNA methyltransferases and RNA interference (RNAi) proteins—DICER-like 3 (DCL3), Argonaute 4 and RNA-dependent RNA polymerase (Pol) II—RdDM requires several plant-specific proteins. These include subunits of two RNA Pol II-related RNA polymerases, namely Pol IV and Pol V; the sucrose non-fermenting 2-like chromatin-remodelling protein DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1) and the structural maintenance of chromosomes hinge-domain-containing protein DEFECTIVE IN MERISTEM SILENCING 3 (DMS3; Pikaard et al, 2008; Matzke et al, 2009).
Pol IV and Pol V are distinguished by their unique largest subunits—nuclear RNA polymerase 4, largest subunit (NRPD1) and nuclear RNA polymerase 5, largest subunit (NRPE1), respectively—and act at different steps of the RdDM pathway: Pol IV is needed to produce and amplify the small interfering RNA (siRNA) trigger, whereas Pol V generally acts downstream from this step to facilitate de novo methylation at the siRNA-targeted site. Roles for RdDM in plant development, nucleolar dominance, transposon silencing and stress adaptation have been proposed previously (Pikaard et al, 2008; Matzke et al, 2009). Whether and how the activities of Pol IV and Pol V are integrated with Pol II transcription to carry out these proposed functions is unknown.
To identify new factors that are important for RdDM in a developmental context, we designed a transgene silencing system based on an enhancer that is active in shoot and root meristem regions. In a forward genetic screen using this system, we previously identified five dms mutants: dms1/drd1, dms2/nrpe2a, dms3, dms5/nrpe1 (Kanno et al, 2008) and dms6/dcl3 (Daxinger et al, 2009). These mutants all seem normal when cultivated under standard growth conditions. Here, we report the identification and characterization of a new mutant, dms4, which atypically displays not only defects in RdDM, but also in plant development. We present evidence suggesting that DMS4 is probably a regulatory factor for several RNA polymerases.
Results And Discussion
In the transgene silencing system, the meristem enhancer is targeted for methylation by 24 nucleotide (nt) primary siRNAs that are generated by DCL3 processing of a hairpin RNA transcribed by Pol II (Fig 1). Pol V-mediated methylation of the targeted enhancer region leads to silencing of a downstream gene encoding green fluorescent protein (GFP ). Pol IV-dependent secondary siRNAs, which foster spreading of methylation into the downstream region, are made in this system but are dispensable for GFP silencing (Fig 1; Kanno et al, 2008; Daxinger et al, 2009). We identified the dms4-1 mutant in a population of M2 seedlings, which represent the first generation when a recessive mutation can be homozygous. In addition to recovery of GFP expression in meristem regions (Fig 2A), the dms4-1 mutant flowers late and displays a pleiotropic phenotype (Fig 2B–K).
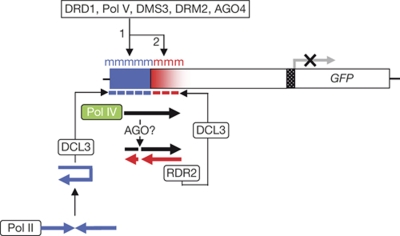
Figure 1.
Transgene system and stepwise RdDM. The target enhancer (blue bar) and downstream region (shaded red bar) undergo stepwise RdDM. In the first step, 24 nt primary siRNAs (blue dashes), which are derived by DCL3 processing of a Pol II-generated hairpin RNA, induce primary RdDM (blue ‘m') at the target enhancer region. This requires the SNF2-like factor DRD1, Pol V, DMS3 (a structural maintenance of chromosomes hinge-domain-containing protein; Kanno et al, 2008), AGO4 (Daxinger et al, 2009) and presumably the de novo methyltransferase DRM2. In the second step, methylated DNA is directly or indirectly transcribed by Pol IV, producing an enhancer-associated ‘nascent' RNA (solid black arrow) that is turned over in a pathway involving RDR2 and DCL3 to generate 24 nt secondary siRNAs (red dashes), which trigger secondary RdDM (red ‘m') in the downstream region. Whether AGO slicing of the ‘nascent' RNA (broken black arrow) is required to produce aberrant RNA substrates for RDR2 is unknown. Primary RdDM is sufficient to silence expression of the downstream GFP reporter gene (Daxinger et al, 2009). AGO, ARGONAUTE; DCL3, DICER-LIKE 3; DMS3, DEFECTIVE IN MERISTEM SILENCING 3; DRD1, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1; DRM2, DOMAINS REARRANGED METHYLTRANSFERASE 2; GFP, green fluorescent protein; RdDM, RNA-directed DNA methylation; RDR2, RNA-DEPENDENT RNA POLYMERASE 2; siRNA, small interfering RNA; SNF2, sucrose non-fermenting 2.
Figure 2.
Effects of dms4 mutations on GFP silencing and plant development. (A) Mutations in the DMS4 gene release GFP silencing in shoot and root apical meristems (SAM and RAM, respectively). High fluorescence is seen in the hypocotyl at this stage of development (Kanno et al, 2008). (B) Phenotypes of adult dms4-1 mutant (right) and wild-type plants (left). Although flower morphology seems normal in the dms4 mutants, (C,D) floral inflorescences of wild-type plants are less closely packed than (E,F) inflorescences of the dms4 mutant. (G) Short internode distances and disturbed phyllotaxy (orange arrows) of the dms4 mutant; normal spiral phyllotaxy is shown inset. (H) dms4-1 seedlings (right) are pale and grow more slowly than wild-type seedlings (left), suggesting a chloroplast defect. (I) Rosette leaves of wild-type plants and (J) the dms4 mutant at the time of bolting. (K) Young plants of the wild-type (left) and dms4-1 and dms4-3 mutants (middle and right, respectively). DMS4, DEFECTIVE IN MERISTEM SILENCING 4; dms4-1, seedling homozygous for the dms4-1 allele; dms4-1 × dms4-3, heteroallelic combination; GFP, green fluorescent protein; T+S, wild-type seedling containing target and silencer loci.
To characterize molecularly the dms4-1 mutant, we examined DNA methylation, accumulation of various RNAs and the expression of several RdDM targets. DNA methylation at the target enhancer and downstream region (Fig 3A) and at 5S ribosomal DNA repeats (supplementary Fig S1 online) is reduced in dms4-1 plants. Deep sequencing of small RNAs revealed a moderate decline in the abundance of 24 nt primary siRNAs in the dms4-1 mutant compared with wild-type plants (Fig 3B). By contrast, Pol IV-dependent 24 nt secondary siRNAs are almost lost completely in the dms4-1 mutant (Fig 3B). The accumulation of several endogenous 24 nt siRNAs in the dms4-1 mutant mirrors largely the pattern observed in nrpe1 mutants (supplementary Fig S2A,B online) and is consistent with a previous suggestion that endogenous 24 nt siRNA populations are mixtures of primary and secondary siRNAs (Daxinger et al, 2009). The level of the Pol V-dependent intergenic non-coding transcript IGN5 (Wierzbicki et al, 2008) is also reduced substantially in the dms4-1 mutant (Fig 3C). Known targets of RdDM, which are de-repressed in drd1 and nrpe1 mutants (Huettel et al, 2006) are similarly affected in the dms4-1 mutant (Fig 4). Thus DMS4 function is not restricted to the transgene enhancer but contributes generally to the RdDM pathway.
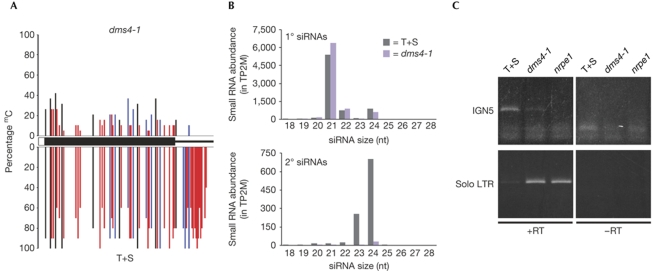
Figure 3.
DNA methylation and RNA accumulation in the dms4 mutant. (A) DNA cytosine methylation (CG, black; CNG, blue; CNN, red; where N is A, T or C) is induced at the targeted enhancer (thick black bar) in wild-type plants (T+S) by primary siRNAs produced from the hairpin RNA trigger. Methylation triggered by Pol IV-dependent secondary siRNAs (Daxinger et al, 2009) is present at the downstream region (black line in white bar). In the dms4-1 mutant, methylation is reduced at the targeted enhancer and is nearly eliminated in the downstream region. (B) Histograms showing the abundance of siRNAs as measured in two sequencing by synthesis libraries, one each from wild-type (T+S) and dms4-1 mutant. The y-axis indicates the sum of the normalized abundance of each matching small RNA in TP2M. Top: a histogram of siRNA abundances matching the target enhancer region (primary siRNAs). Primary siRNAs (21, 22 and 24 nt) result from the redundant action of several DCL enzymes acting on the hairpin RNA trigger. RdDM is triggered by DCL3-generated 24 nt siRNAs (Daxinger et al, 2009). Bottom: a histogram of siRNA abundances matching in the 150 bp downstream from the target enhancer (secondary siRNAs). Pol IV-dependent 24 nt secondary siRNAs result from turnover of an enhancer-associated nascent RNA (Fig 1). The 24 nt primary siRNAs are reduced in abundance from 894 TP2M in wild type to 606 TP2M in the dms4-1 mutant, a decline of 32.2%; the 24 nt secondary siRNAs go from 702 TP2M in wild type to 31 TP2M in the dms4-1 mutant, a decline of 95.6%. (C) A Pol V-dependent transcript, IGN5 (Wierzbicki et al, 2008), is present in wild-type T+S plants, reduced in the dms4-1 mutant and eliminated in an nrpe1 mutant. By contrast, a Pol II-dependent solo LTR transcript (Wierzbicki et al, 2008) is reactivated in the two mutants owing to release of silencing. The nrpe1-12 allele was used in (C). DCL, DICER-LIKE; LTR, long terminal repeat; nrpe1, mutant defective in the largest subunits of Pol V; RT; reverse transcriptase; siRNA, small interfering RNA; T, wild-type plants containing the target locus; TP2M, transcripts per two million; T+S, wild-type plants containing target and silencer complexes.
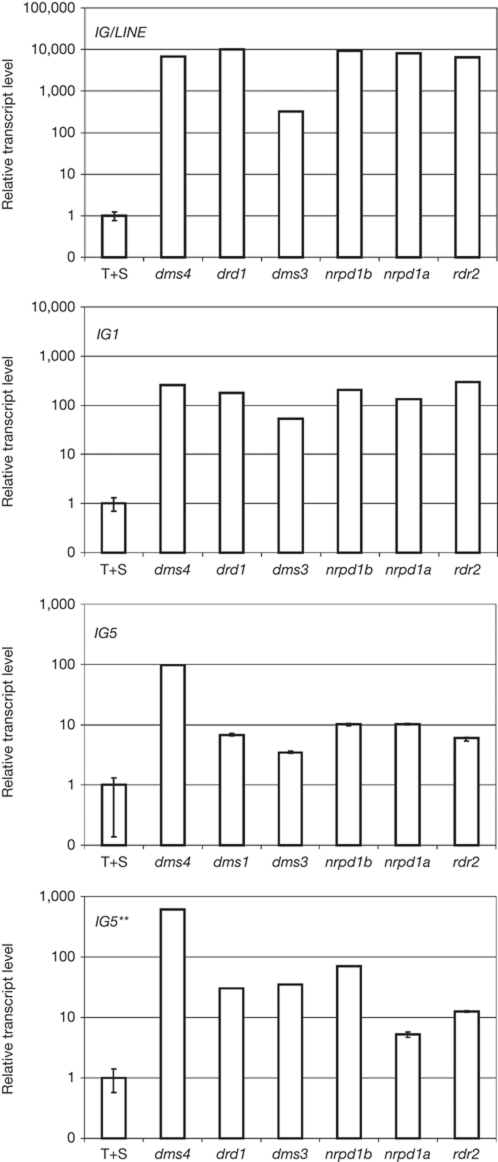
Figure 4.
Transcription of RdDM targets in the dms4-1 mutant. Intergenic transcripts (IG/LINE, IG1, IG5 and IG5**) identified in the drd1 mutant (Huettel et al, 2006) were analysed by quantitative RT–PCR in dms4-1 plants and other mutants defective in RdDM (drd1, dms3, nrpe1, nrpd1 and rdr2; Kanno et al, 2008). A similar level of target de-repression was observed for all mutants. Transcription data are shown in a log scale. UPL7 (At3g53090) is the internal reference gene. Expression levels are relative to wild-type plants (T+S). Error bars, s.d.±mean. Error bars are not visible in some cases because of the log scale. RdDM, RNA-directed DNA methylation; RT–PCR, reverse transcriptase PCR; T+S, wild-type plants containing target and silencer constructs; UPL7, ubiquitin protein ligase 7.
By using genetic mapping with co-dominant markers (Konieczny & Ausubel, 1993) and co-expression data (http://www.bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi), we identified At2g30280 as a potential DMS4 candidate gene. Sequencing of At2g30280 in the dms4-1 mutant revealed a G-to-A conversion at the splice-site acceptor of the sixth intron (supplementary Fig S3A,B online). A second allele, dms4-2, has a premature stop codon at amino-acid position 147 (supplementary Fig S3B online). We obtained a third allele, dms4-3 (SAIL_1144_B05; supplementary Fig S3A online), from a T-DNA insertion collection. The homozygous dms4-2 and dms4-3 mutants and the heteroallelic dms4-1/dms4-3 mutant resemble phenotypically the homozygous dms4-1 mutant (Fig 2K; data not shown) and release GFP silencing (Fig 2A). The aberrant morphology and release of GFP silencing observed with three independent alleles of dms4 show that these phenotypes are due to mutations in the same gene. This was confirmed by a complementation test in which the wild-type DMS4 complementary DNA (cDNA) largely restored a normal phenotype and GFP silencing (data not shown).
At2g30280 is a previously uncharacterized polypeptide, 346 amino acids in length, which lacks conserved domains and is annotated as ‘similar to zinc-finger-protein-related' (http://www.arabidopsis.org). Encoded by a single-copy gene in Arabidopsis, in which it is predicted to be a ubiquitously expressed nuclear protein (http://www.arabidopsis.org), At2g30280 has homologues in grape, rice and moss (supplementary Fig S4 online). Amino-acid sequence alignments revealed that DMS4 is similar to budding yeast (Saccharomyces cerevisiae) IWR1/YDL115C and related, uncharacterized proteins in fission yeast, Drosophila melanogaster, Caenorhabditis elegans and humans (supplementary Fig S4 online).
IWR1 is a putative transcription factor that interacts physically with a number of subunits of yeast Pol II (Krogan et al, 2006; Collins et al, 2007). The IWR1 homologue in Drosophila co-localizes with Pol II on polytene chromosomes (Krogan et al, 2006). An epistasis analysis in fission yeast revealed positive interactions between IWR1 and several RNAi components (Roguev et al, 2009), consistent with a requirement for Pol II in RNAi-mediated heterochromatin formation (Verdel et al, 2009). Apart from their presumed roles as Pol II transcription factors (Pieró-Chova & Estruch, 2009), the specific functions of IWR1-like proteins are unknown. In yeast, IWR1 might be important for modulating transcription in response to stress, as indicated by the hypersensitivity of the iwr1 mutant to K1 killer toxin (Pagé et al, 2003) and other stress conditions (Aouida et al, 2004). Transforming a yeast iwr1 mutant with DMS4 cDNA partly complements the K1 hypersensitivity (Table 1), suggesting some functional conservation.
Table 1.
Expression of the Arabidopsis thaliana DMS4 gene partly complements K1 toxin hypersensitivity of a yeast Δiwr1-null mutant
| Saccharomyces cerevisiae strain | Relevant genotype | K1 toxin sensitivity (%) | |
|---|---|---|---|
| Raffinose | Galactose | ||
| BY4742 (wild type) | IWR1 | 100 | 100 |
| YDL115C | Δiwr1 | 132 | 132 |
| YDL115C | Δiwr1 + DMS4 | 132 | 118 |
| YDL115C | Δiwr1 + DMS3 | 132 | 132 |
| DMS4 and DMS3 represent yeast 2μ episomal vectors expressing the A. thaliana genes DMS4 and/or DMS3 under control of the galactose-inducible GAL1 promoter. | |||
| K1 toxin sensitivity was determined in an agar diffusion assay on MBA plates (at pH 4.7) and is expressed as a percentage of growth inhibition against 104 units of K1 toxin (see Methods). The indicated strain was embedded into MBA containing either raffinose or galactose as its carbon source to ensure DMS4/DMS3 repression or induction, respectively. The resulting zone of growth inhibition was measured after incubating the plates for three days at 20°C (100% sensitivity corresponds to a growth inhibition zone of 20 mm). The mean values of at least six independent experiments are shown; s.d. value of partial complementation of K1 hypersensitivity in the Δiwr1 mutant after DMS4 expression was <4%. | |||
| DMS, DEFECTIVE IN MERISTEM SILENCING; MBA, methylene blue agar. | |||
The dms4 mutant is the first mutant we have identified in a forward screen that has defects in both RdDM and plant development. Other mutants identified in the same screen, including drd1, nrpd2a, nrpe1, dms3 and dcl3, are deficient in core factors of the RdDM machinery, but seem normal when cultivated under standard conditions (Kanno et al, 2008; Daxinger et al, 2009). Therefore, the basic RdDM pathway does not seem to be essential for normal plant development. Moreover, in contrast to the RdDM factors mentioned above, DMS4 has a putative homologue in S. cerevisiae, which lacks RNAi and DNA methylation. These observations raise questions about the function(s) of DMS4 in plants and whether it has a direct or indirect role in RdDM.
One possibility is that DMS4 is needed for Pol II transcription of genes encoding components of the RdDM machinery and/or the precursor of the siRNA trigger. Under this hypothesis, DMS4 would not participate directly in RdDM but would have an indirect effect through its conserved function as a Pol II transcription factor. Although we cannot totally exclude this possibility, it seems unlikely because the expression of most known RdDM components does not change significantly in the dms4-1 mutant (Table 2; supplementary Table 1 online), and for the two that do (DRD1, NRPE1), expression is still at least 50% of the wild-type level (supplementary Fig S5A online). Although the levels of hairpin-derived 24 nt primary siRNAs decrease in the dms4-1 mutant, they still accumulate to relatively high levels that are likely to be sufficient to induce wild-type levels of methylation (Daxinger et al, 2009). Moreover, 21 and 22 nt primary siRNAs do not decrease in the dms4-1 mutant (Fig 3B) even though they are derived from the same hairpin RNA precursor. So it is unlikely that the partial reduction of 24 nt siRNAs is due to reduced Pol II transcription of the precursor RNA. Additional studies are required to determine whether small to moderate reductions of 24 nt siRNAs and various components of the RdDM machinery affect the efficiency of RdDM.
Table 2.
Differential gene expression in the dms4-1 mutant
| DiffRank | ID | Q | A | M |
|---|---|---|---|---|
| 1 | AT5G60250 | 7 × 10–8 | 7.15 | 6.37±0.09 |
| 2 | AT2G41260 | 7 × 10–8 | 7.66 | 8.97±0.15 |
| 3 | AT1G61280 | 7 × 10–8 | 9.54 | 5.32±0.04 |
| 4 | AT5G05340 | 1 × 10–7 | 6.75 | 8.93±0.17 |
| 5 | AT3G57890 | 1 × 10–7 | 16.68 | −4.38±0.04 |
| 6 | AT5G16960 | 2 × 10–7 | 8.83 | −4.78±0.07 |
| 7 | AT1G61800 | 2 × 10–7 | 10.69 | 5.28±0.09 |
| 8 | AT5G42900 | 2 × 10–7 | 12.69 | −4.13±0.07 |
| 9 | AT2G15880 | 2 × 10–7 | 9.46 | −4.39±0.01 |
| 10 | AT4G25810 | 2 × 10–7 | 7.72 | 6.40±0.14 |
| … | ||||
| 4,267 | AT2G40030 (NRPE1) | 0.6% | 8.58 | −0.95±0.16 |
| 6,200 | AT2G16390 (DRD1) | 3% | 8.85 | −0.40±0.06 |
| 7,561 | AT3G43920 (DCL3) | 9% | 6.47 | −0.49±0.12 |
| 17,498 | AT5G14620 (DRM2) | 100% | 8.79 | −0.08±0.11 |
| 18,450 | AT2G27040 (AGO4) | 100% | 17.74 | 0.03±0.05 |
| 19,018 | AT3G49250 (DMS3) | 100% | 9.45 | 0.03±0.10 |
| The table shows an excerpt of the statistics computed to test for differential expression in the dms4-1 mutant compared with the wild type. A comprehensive list is available in the supplementary Table S1 online. Genes (ID) are sorted by the significance of their differential expression (diffRank). Allowing a false discovery rate q<5% identified 6,793 candidate genes (Benjamini–Yekutieli corrected). The table shows the average expression level (A), differential expression (M) and its s.e. on an asymptotic log2 scale. The top 10 ranked genes (supplementary Table S1 online) are shown for comparison with six genes of the RdDM machinery (bold print). Of these, only two (AT2G40030 and AT2G16390) change significantly (q<5%) in the dms4-1 mutant. Their expression levels, however, are only moderately reduced in the mutant (supplementary Fig S5A online). | ||||
| AGO4, ARGONAUTE 4; DCL3, DICER-LIKE 3; DMS3, DEFECTIVE IN MERISTEM SILENCING 3; DRD1, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1; DRM2, DOMAINS REARRANGED METHYLTRANSFERASE 2; RdDM, RNA-directed DNA methylation. | ||||
An alternative hypothesis is that DMS4 has a dual role, carrying out its conserved function as a Pol II transcription factor for genes involved in development and, in addition, participating directly in RdDM by modulating the activity of Pol IV and/or Pol V. A role for DMS4 in Pol II transcription is suggested by a transcriptome analysis that revealed thousands of genes that are differentially regulated in the dms4-1 mutant compared with a drd1 mutant, which is defective only in RdDM but not development (supplementary Table S2 online). These include genes encoding chloroplast proteins (supplementary Table S3; supplementary Fig S5B online) and transcriptional regulators (supplementary Fig S5C online), which might explain the pleiotropic phenotype of dms4 mutants. In addition to loss of Pol V-mediated DNA methylation from RdDM targets, further evidence for a direct influence of DMS4 on Pol IV and Pol V transcriptional activities is the substantial reduction of Pol IV-dependent secondary siRNAs (Fig 3B) and the Pol V-dependent IGN5 transcript (Fig 3D) in the dms4-1 mutant.
How might DMS4 influence the activities of several RNA polymerases? Yeast IWR1 co-purifies with 11 out of 12 subunits of Pol II, the exception being subunit 12 (http://www.uniprot.org). In addition to unique largest subunits and a shared second largest subunit, Pol IV and Pol V have specific subunits 3, 4, 5 and 7 (Ream et al, 2009; Huang et al, 2009; Lahmy et al, 2009; He et al, 2009a). By contrast, most of the smaller non-catalytic subunits are common to Pol II, Pol IV and Pol V (Ream et al, 2009). DMS4 interactions with common subunits of different polymerases might explain the effect of dms4 mutations on both plant phenotype and RdDM. However, we did not identify RNA polymerase subunits in a yeast two-hybrid screen using DMS4 as bait. Furthermore, direct yeast two-hybrid assays did not detect interactions between DMS4 and the carboxy-terminal domain of the largest subunit of Pol II or between DMS4 and subunits 3, 10, 11 or 12, which are common to Pol II and Pol V (data not shown). Two previous studies failed to co-purify DMS4 in immunoprecipitations using antibodies against NRPE1 (Ream et al, 2009; Huang et al, 2009), suggesting that any associations between DMS4 and Pol V are weak or transient. By contrast, a second transcription factor in the Pol V pathway, KOW transcription facror 1/suppressor of Ty insertions 5-like (Bies-Etheve et al, 2009), co-immunoprecipitates with NRPE1 (Huang et al, 2009) and was also identified in a forward genetic screen (He et al, 2009b).
The DMS4 protein displays a low affinity in a yeast two-hybrid assay for the ‘defective chloroplast and leaves' (DeCL) motif in the C-terminal domains of NRPD1 and NRPE1 (supplementary Fig S6A,B online). Although this affinity potentially enhances interactions of DMS4 with Pol IV and Pol V, it is not yet known whether such associations occur in vivo. Nevertheless, the importance of the C-terminal domains of NRPD1 and NRPE1 in RdDM is supported by loss-of-function mutations that produce truncated proteins or motifs lacking only the region containing the WG/GW and/or DeCL motifs (supplementary Fig S6C online). The possibility that DMS4 might interact with several, functionally diversified proteins is suggested by a previous study demonstrating the strong interaction of yeast IWR1 with replication factors of a plant RNA virus (Li et al, 2008).
The involvement of DMS4 in RdDM reveals a role in epigenetic silencing for a member of the IWR1 family of putative transcription factors that are conserved from yeasts to humans. In addition to its involvement in RdDM, which might involve an association with Pol IV and Pol V, DMS4 also contributes directly or indirectly to Pol II transcription of many nuclear genes that are important for chloroplast function and plant physiology. Investigating the basis of the pleiotropic morphological phenotype of dms4 mutants and how it is linked to the defect in RdDM is likely to provide further insight into how Pol IV and Pol V activities are integrated with other transcriptional pathways.
Methods
Mapping the dms4-1 mutation. Lehle Seeds (Round Rock, TX, USA) performed ethyl methanesulphonate mutagenesis of seeds homozygous for the target and silencer complexes. We screened for mutants by selecting M2 seedlings showing GFP activity in root meristems and double resistance to kanamycin (encoded at the target locus) and hygromycin (encoded at the silencer locus; Kanno et al, 2008). Details of genetic mapping, cloning and sequencing of the dms4 gene and complementation analysis are provided in supplementary information online.
Molecular characterization of the dms4-1 mutant. Analysis of small RNAs by northern blot hybridization, nascent RNA by reverse transcriptase PCR (RT–PCR), and DNA methylation by bisulphite sequencing were performed as described previously (Kanno et al, 2005; Huettel et al, 2006). Primers used for bisulphite sequencing are shown in supplementary Table S4 online.
Pol V transcripts. Total RNA was isolated from 15–17-day-old seedlings using TRIzol reagent (Invitrogen, Lofer, Austria). After RNase-free DNase I treatment, Pol V transcripts were amplified from 250 ng of RNA template using the One-Step RT–PCR kit (Qiagen, Vienna, Austria) according to manufacturer's protocols. Using 60°C as an annealing temperature, PCR was carried out with 33 cycles for solo long terminal repeat and 38 cycles for IGN5, respectively. The primer sequences are shown in supplementary Table S4 online.
Quantitative real-time PCR analysis. The RNA extraction (RNeasy mini kit, Qiagen) and cDNA synthesis (RevertAid H Minus First strand cDNA synthesis kit, MBI Fermentas, St Leon-Rot, Germany) were performed as described previously (Kanno et al, 2005; Huettel et al, 2006). The cDNA was diluted to 75 μl with diethylpyrocarbonate-treated double distilled water, and 2 μl was used in a 20 μl PCR reaction. The mixture was set up with 10 μl of QuantiFast SYBR Green PCR (Qiagen), 2 μl cDNA and 2 μl of each primer (1 μM final concentration). The PCR was performed after a pre-incubation as suggested by the supplier (95°C for 5 min) by 40 two-step cycles of denaturation at 95°C for 10 s, and annealing/extension at 60°C for 30 s. The comparative threshold cycle (Ct) method was used to determine relative RNA levels (User Bulletin no. 2, Applied Biosystems, Vienna, Austria). Primer sequences are shown in supplementary Table S4 online.
Complementation of K1 killer toxin hypersensitivity. Details of yeast strains and procedures can be found in supplementary information online.
Microarray data analysis. Total RNA was extracted from approximately 12 seedlings each of the drd1-1, drd1-6 and respective wild type (10 days after germination) and dms4-1 mutants (21 days after germination and respective wild type) using an RNeasy mini kit (Qiagen). Owing to delayed growth of the dms4 mutant, the drd1 and dms4 mutant seedlings were all approximately of the same size. The RNA was always isolated around 15:00 hours. Transcriptomes were analysed using 1 μg of total RNA as starting material. Targets were prepared with the one-cycle cDNA synthesis kit followed by biotin labelling with the 3′ IVT labelling kit (GeneChip One-cycle target labelling and control reagents, Affymetrix, High Wycombe, UK) and hybridized to Arabidopsis thaliana hybridization array 1-type GeneChips for 16 h as recommended by the supplier (Gene expression analysis manual, Affymetrix; http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Transcriptome data (CEL files) were submitted to a public repository database (ArrayExpress accession number E-MEXP-2251; user name: Reviewer_E-MEXP-2251; password: 1248175579260). Further information on low-level analysis and transforms, and analysis of differential expression is available in supplementary information online.
Deep sequencing of small RNAs. Details of construction and analysis of small RNA libraries are provided in the supplementary information online. Libraries of small RNAs were sequenced using Illumina's (San Diego, CA, USA) sequecing by synthesis technology, generating 6,670,921 total reads from wild type (with trigger and silencer transgenes) and 5,809,235 from the dms4-1 mutant. These data will be available at http://mpss.udel.edu/at_sbs and through GenBank (Gene expression omnibus accession number: GSE18302).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank J-K Zhu, P Cramer and F Estruch for helpful discussions, and J van der Winden for technical assistance. This study was supported by grants from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF; P20707-B03 and P19929-B26). D.P.K. acknowledges support from the Vienna Science and Technology Fund, Baxter A.G., Austrian Research Centres Seibersdorf, and the Austrian Centre of Biopharmaceutical Technology. Z.J.L. and M.J.S. acknowledge support from the FWF (P19929-B26) and the Deutsche Forschungsgemeinschaft (SCHM 541/11-3), respectively. S.A.S., S.G.R.G. and B.C.M. are supported by the United States National Science Foundation Plant Genome Research Program (award number #0701745, to B.C.M.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aouida M, Pagé N, Leduc A, Peter M, Ramotar D (2004) A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res 64: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T (2009) RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep 10: 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJM, Matzke M (2009) A stepwise pathway for biogenesis of 24 nt secondary siRNAs and spreading of DNA methylation. EMBO J 28: 48–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK (2009a) An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK (2009b) NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 23: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jones AME, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC (2009) An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol 16: 91–93 [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M (2006) Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J 25: 2828–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJM (2008) A structural maintenance of chromosome hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 40: 670–675 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel F (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Krogan NJ et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T (2009) Pol V (Pol IVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci USA 106: 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barajas D, Panavas T, Herbst DA, Nagy P (2008) Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J Virol 82: 6911–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM (2009) RNA-mediated chromatin-based silencing in plants. Targets of RNA-directed DNA methylation. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Pagé N et al. (2003) A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163: 875–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieró-Chova L, Estruch F (2009) The yeast RNA Pol II-associated factor Iwr1p is involved in the basal and regulated transcription of specific genes. J Biol Chem 284: 28958–28967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Haag R, Ream T, Wierzbicki AT (2008) Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13: 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream TS et al. (2009) Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A et al. (2009) Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322: 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L (2009) Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol 53: 245–257 [DOI] [PubMed] [Google Scholar]
- Wierzbicki A, Haag J, Pikaard CS (2008) Noncoding transcription by RNA polymerase IVb/V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.