Abstract
Epigenetic silencing of a fraction of ribosomal DNA (rDNA) requires association of the nucleolar chromatin-remodelling complex NoRC to 150–250 nucleotide RNAs (pRNA) that originate from an RNA polymerase I promoter located in the intergenic spacer separating rDNA repeats. Here, we show that NoRC-associated pRNA is transcribed from a sub-fraction of hypomethylated rRNA genes during mid S phase, acting in trans to inherit DNA methylation and transcriptional repression of late-replicating silent rDNA copies. The results reveal variability between individual rDNA clusters with distinct functional consequences.
Keywords: epigenetic silencing, NoRC, non-coding RNA, rDNA, spacer promoter
Introduction
Mammalian ribosomal RNA (rRNA) genes are alternating modules of intergenic spacers (IGSs) and transcribed regions that encode rRNA precursors (pre-rRNA). In rats, mice, Drosophila and Xenopus, the IGS contains one or more RNA polymerase I (Pol I) promoters with high homology to the core region of the main ribosomal DNA (rDNA) promoter (Labhart & Reeder, 1984; de Winter & Moss, 1986; Kuhn & Grummt, 1987; Grimaldi & di Nocera, 1988). Transcripts originating from spacer promoters are co-directional with pre-rRNA synthesis and have been shown to enhance transcription from the main rDNA promoter, possibly by delivering Pol I (Grimaldi & di Nocera, 1988; Putnam & Pikaard, 1992). Furthermore, intergenic spacer rRNA (IGS rRNA) has been shown to have a crucial function in rDNA silencing. In mice, intergenic transcripts originating from a promoter located approximately 2 kb upstream from the pre-rRNA start site are processed into a heterogeneous population of 150–250 nucleotide RNAs, dubbed promoter RNA (pRNA) as their sequence matches the rDNA promoter (Mayer et al, 2006). The pRNA associates with nucleolar remodelling complex (NoRC), a sucrose non-fermenting protein 2 homologue (SNF2h)-containing chromatin-remodelling complex, which induces long-term transcriptional silencing of a fraction of rRNA genes (Strohner et al, 2001; Santoro et al, 2002; Santoro & Grummt, 2005). NoRC-dependent heterochromatin formation and rDNA silencing depends on the association of NoRC with pRNA (Mayer et al, 2006, 2008).
As the steady-state level of IGS rRNA is very low, we considered which mechanisms might account for the under-representation of IGS transcripts. Several models are conceivable, which are not necessarily exclusive: the spacer promoter could be extremely weak, IGS transcripts could be rapidly degraded, the synthesis of IGS rRNA could be restricted to a small fraction of rDNA repeats, or IGS rRNA could be synthesized during a defined time during the cell cycle. Here, we show that spacer transcripts are synthesized from a fraction of unmethylated rRNA genes and mediate CpG methylation and heterochromatin formation at late-replicating, silent rDNA repeats. The results support the idea that mammalian rDNA arrays are functionally distinct and intergenic transcripts from a specific subclass of rDNA repeats regulate the epigenetic state of rRNA genes in trans.
Results
Spacer transcripts and pre-rRNA are differently regulated
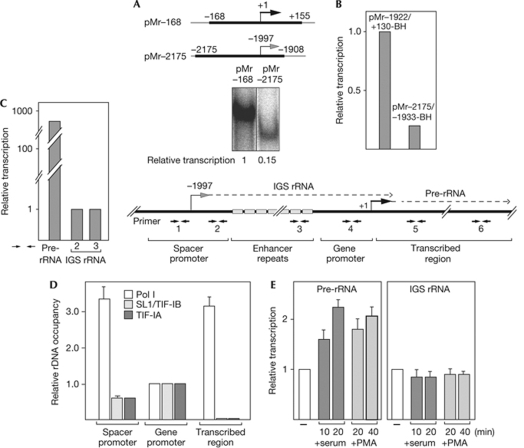
Previous studies have demonstrated that silencing of a fraction of rDNA repeats requires the association of NoRC with RNA from a Pol I promoter that is located in the IGS approximately 2 kb upstream from the pre-rRNA transcription start site (Mayer et al, 2006). To elucidate the mechanisms that regulate transcripts originating from the spacer promoter, we compared the activity of the spacer promoter with the main rRNA gene promoter. In cell-free transcription assays, templates harbouring the spacer promoter (pMr−2175) promoted the synthesis of specific run-off transcripts, although with lower efficiency than templates comprising the pre-rRNA promoter (pMr−168; Fig 1A). A similar difference in transcription activity was observed in transient transfection experiments with artificial minigenes harbouring the main rDNA promoter (pMr−1922/+130-BH) or the spacer promoter (pMr−2175/−1933-BH; Fig 1B). Significantly, the steady-state level of cellular IGS rRNA was almost 1,000-fold lower than pre-rRNA (Fig 1C), suggesting that spacer transcripts are either synthesized from a minor fraction of rRNA genes or are rapidly degraded. To distinguish between both possibilities, we compared the association of the Pol I transcription machinery with the spacer and the main gene promoter by chromatin immunoprecipitation (ChIP) assays. Consistent with the in vitro transcription experiments and the Pol I reporter assays in Fig 1A,B, binding of the promoter selectivity factor SL1 (also known as transcription initiation factor TIF-IB) and the Pol I-associated factor TIF-IA was slightly decreased at the spacer promoter (Fig 1D). By contrast, Pol I occupancy was elevated at the spacer promoter. As Pol I loading does not correlate with the transcriptional output, Pol I is either stalled at the spacer promoter or IGS rRNA is synthesized from a minor fraction of rRNA genes, or is rapidly degraded.
Figure 1.
The steady-state level of IGS rRNA is under-represented in cellular RNA. (A) In vitro transcription. Run-off transcripts from pMr−168 and pMr−2175 linearized with NdeI. The scheme depicts the regions of rDNA contained in pMr−168 and pMr−2175. (B) Transcription of reporter genes pMr−1922/+130-BH and pMr−2172/−1933-BH in NIH3T3 cells. Values were normalized to expression levels of a co-transfected luciferase reporter gene and to GAPDH mRNA. (C) Steady-state levels of IGS rRNA and pre-rRNA. RNA from exponentially growing NIH3T3 cells was subjected to RT–qPCR; values were normalized to GAPDH mRNA. The scheme depicts the structural organization of the 5′-terminal part of murine rDNA, including the transcription start sites of IGS rRNA and pre-rRNA (dotted lines), and the primer pairs used for qPCR amplification (numbered arrows). The light grey boxes are the variable number of enhancer repeats between the spacer promoter and the main gene promoter. (D) Chromatin immunoprecipitation. Polyclonal antibodies against Pol I or the respective transcription factors were used to show the association of Pol I (RPA116), SL1/TIF-IB (TAFI110) and TIF-IA with different regions of rDNA. DNA was amplified with the indicated primer pairs. Data show the ratio of rDNA in the immunoprecipitates and input rDNA (b/i). Error bars indicate ±s.d. values of three experiments. (E) RT–qPCR of IGS rRNA and pre-rRNA after mitogenic stimulation of serum-starved NIH3T3 cells by serum or PMA (0.1 μM) for the indicated times. Values were normalized to GAPDH mRNA. Error bars indicate ±s.d. values of three independent experiments. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IGS, intergenic spacer; mRNA, messenger RNA; Pol I, RNA polymerase I; PMA, phorbol 12-myristate 13-acetate; pre-rRNA, rRNA precursor; rRNA, ribosomal RNA; RT–qPCR, reverse transcriptase quantitative PCR; TAF, TATA box-associated factor; TIF, transcription initiation factor.
Previous studies have revealed that growth factors stimulate Pol I transcription by extracellular-signal-regulated kinase-dependent and ribosomal-S6-kinase-dependent phosphorylation of TIF-IA and the upstream binding factor (UBF; Zhao et al, 2003; Stefanovsky et al, 2006). To examine whether the synthesis of IGS rRNA is responsive to the growth-dependent TIF-IA, we monitored transcripts from the spacer and the main gene promoter after overexpression of TIF-IA. Transcription from both promoters was stimulated by ectopic TIF-IA (supplementary Fig S1 online), supporting the idea that the basal Pol I transcription machinery directs the synthesis of IGS rRNA and pre-rRNA. Nevertheless, the level of cellular IGS transcripts seems to be regulated by different mechanisms than pre-rRNA. As shown in Fig 1E, mitogenic stimulation of serum-starved NIH3T3 cells by serum or phorbol 12-myristate 13-acetate increased pre-rRNA synthesis; however, it did not affect the level of IGS rRNA. This indicates that the spacer promoter and the main rDNA promoter are differentially sensitive to mitogenic signals.
IGS rRNA levels transiently increase in mid S phase
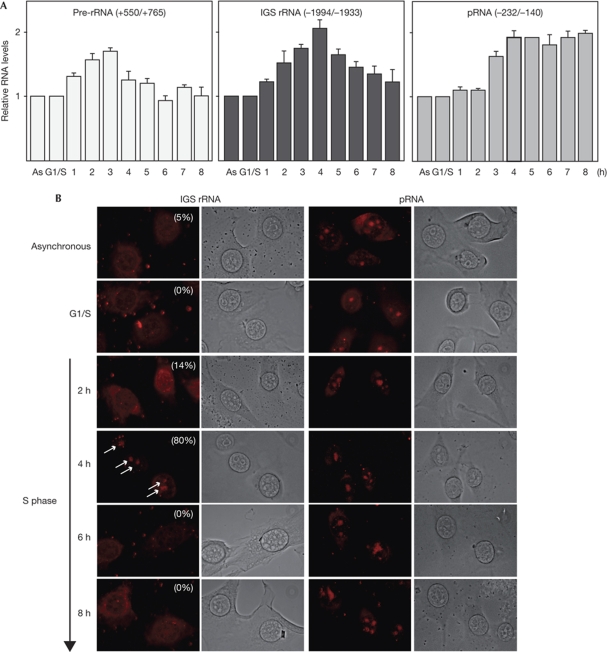
The synthesis of pre-rRNA fluctuates during cell-cycle progression, steadily increasing from G1 to G2 phase and being shut off during mitosis (Klein & Grummt, 1999). By contrast, the steady-state level of IGS rRNA was almost identical in G0, G1/S and G2/M cells, demonstrating that the synthesis and turnover of spacer transcripts does not change during the cell cycle (supplementary Fig S2 online). To uncover a potential link between IGS rRNA synthesis and rDNA replication, we synchronized NIH3T3 cells by treatment with aphidicolin, and monitored pre-rRNA and IGS rRNA levels after release into the S phase. The aim of this experiment was to link transcription of IGS rRNA to replication timing. Previous studies have established that hypomethylated, active rRNA genes replicate early (2–3 h after release), whereas CpG-methylated, silent rRNA genes replicate later, at about 5–6 h after entry into S phase (Li et al, 2005; supplementary Fig S3 online). Consistent with active rDNA copies replicating early, the level of pre-rRNA doubled at 2–3 h after release from the aphidicolin block (Fig 2A, left panel). Notably, the level of unprocessed spacer transcripts (IGS rRNA, middle panel) and processed spacer transcripts (pRNA, right panel) increased at 4 h, a first indication that IGS rRNA is synthesized from other rDNA repeats than pre-rRNA. Consistent with previous results showing that IGS rRNA needs to be processed into pRNA that is stabilized by binding to NoRC (Mayer et al, 2006), IGS rRNA decreased in the second half of S phase, whereas pRNA remained unchanged. Different levels of IGS rRNA and pRNA were also observed in fluorescence in situ hybridization (FISH) experiments (Fig 2B). Although there is an obvious difference in the amount of IGS rRNA determined by reverse transcriptase PCR (RT–PCR) and by in situ hybridization, IGS rRNA is clearly enriched in nucleoli 4 h after entry into S phase, indicating that elevated levels of IGS rRNA precede replication of silent rDNA copies. The pRNA, however, which is an integral component of NoRC and is required for silencing a fraction of rDNA repeats, was permanently present in nucleoli.
Figure 2.
The level of IGS rRNA transiently increases during mid S phase. (A) Levels of pre-rRNA, IGS rRNA and pRNA during S-phase progression. NIH3T3 cells arrested at G1/S were released into S phase and transcription was measured in 1 h intervals by RT–qPCR. Values were normalized to 28S rRNA. Error bars indicate ±s.d. values of three independent experiments. (B) RNA FISH showing IGS rRNA and pRNA in asynchronous cells or in NIH3T3 cells that were synchronized by aphidicolin treatment and released into S phase. IGS rRNA was visualized by hybridization with a probe containing rDNA sequences from −554 to −447, pRNA with a probe comprising rDNA sequences from −232 to −140. Signals were visualized with extravidin–Cy3 conjugate. A total of 100 cells were counted; the percentage of IGS rRNA-positive cells is indicated in the images. Arrows mark nascent IGS rRNA in nucleoli. As, asynchronous cells; FISH, fluorescence in situ hybridization; IGS, intergenic spacer; Pol I, RNA polymerase I; pre-rRNA, rRNA precursor; pRNA, promoter RNA; rRNA/DNA, ribosomal RNA/DNA; RT–qPCR, reverse transcriptase quantitative PCR.
The decline in IGS rRNA, but not pRNA, in late S phase indicates that cells transcribe more IGS RNA than they accumulate, implying the existence of an active RNA degradation mechanism. In support of this, knockdown of a subunit of the exosome, ExoSC3, led to a 3.8-fold increase in the level of IGS rRNA and a 2.2-fold increase in that of pRNA without affecting the level of pre-rRNA. This indicates that cells carefully regulate the level of spacer transcripts to produce the amount of pRNA required for propagation of the heterochromatic state of silent rRNA genes (supplementary Fig S4 online).
IGS rRNA is transcribed from a subclass of rRNA genes
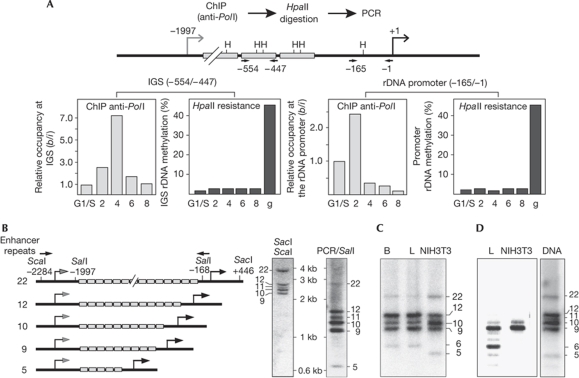
To determine whether IGS rRNA derives from hypomethylated or from CpG-methylated rDNA repeats, we performed ChIP-chop assays; that is, we precipitated chromatin-associated Pol I and monitored CpG methylation of co-precipitated rDNA by methylation-sensitive restriction analysis. Consistent with enhanced pre-rRNA synthesis occurring in early S phase, the association of Pol I with the main rDNA promoter increased 2 h after release from the aphidicolin block (Fig 3A, right panels). Pol I occupancy in the IGS was enhanced after 4 h (Fig 3A, left panels), a further indication that both promoters are differentially regulated and are not active at the same time. To analyse whether IGS transcripts originate from hypomethylated or hypermethylated rDNA repeats, the immunoprecipitated DNA was digested with HpaII, and resistance to HpaII digestion was measured by quantitative PCR (qPCR). As expected, Pol I was associated with hypomethylated rDNA promoters that are sensitive to HpaII digestion. Similarly, Pol I occupied hypomethylated IGS sequences, supporting the view that IGS rRNA is transcribed from a subfraction of unmethylated rRNA genes to propagate the heterochromatic state of silent rDNA repeats through cell division.
Figure 3.
IGS rRNA is transcribed from a subclass of rRNA genes. (A) IGS rRNA is transcribed from hypomethylated rRNA genes. Light bars show ChIP data monitoring Pol I occupancy at the main gene promoter and the IGS promoter during S-phase progression. Data show Pol I-associated rDNA compared with input DNA (b/i), normalized to Pol I occupancy in G1/S cells. Dark bars show the percentage of HpaII-resistant immunoprecipitated rDNA. As a control, methylation of genomic DNA (g) from asynchronous cells is shown. Methylation of rDNA is shown as the ratio of HpaII-resistant immunoprecipitated rDNA and undigested total rDNA. The positions of HpaII sites (H) and primers used to amplify the IGS and the rDNA promoter are indicated in the scheme (top). (B) Southern blot analysis. DNA from NIH3T3 cells was digested with SacI and ScaI and hybridized with an IGS-specific probe. As a control, IGS rDNA (−2175/−1) was amplified by PCR and digested with SalI (right lane). The scheme on the left depicts part of the 5′-terminal region of rDNA variants containing a different number of enhancer repeats (light boxes). Arrows represent the PCR primers used to amplify spacer sequences. Nucleotide positions refer to mouse rDNA sequence BK000964. (C) PCR of IGS (from −2175 to −1) using 50–100 ng of genomic DNA from mouse brain (B), mouse liver and NIH3T3 cells. (D) IGS rRNA originates from a subclass of rDNA repeats. RNA from mouse liver and NIH3T3 cells was subjected to RT–PCR using the Expand Long Template PCR Kit (Roche) and primers that amplify rDNA sequences from −1994 to −20. Amplification of the respective region of genomic DNA from NIH3T3 cells is shown on the right. ChIP, chromatin immunoprecipitation; IGS, intergenic spacer; Pol I, RNA polymerase I; rRNA, ribosomal RNA; RT–PCR, reverse transcriptase PCR.
So far, one mouse and one human rDNA repeat has been sequenced (Third Party Annotation accession numbers BK000964 and U13369), suggesting that all rDNA repeats are identical. However, restriction fragment length polymorphism was noted in the 5′-end of mouse rRNA genes, which was attributed to a variable number of enhancer repeats located between the spacer promoter and the promoter-proximal terminator T0 (Arnheim & Kuehn, 1979; Tseng et al, 2008). The existence of rDNA variants with a different number of enhancer repeats enabled us to examine whether IGS transcripts derive from a specific subset of rRNA genes. Southern blot analysis of genomic DNA from NIH3T3 cells revealed distinct lengths of IGSs (Fig 3B). Cloning and sequencing indicated that these rDNA variants have identical spacer and promoter sequences but contain a variable number (9, 10, 11, 12 and 22) of enhancer repeats. A similar restriction fragment length polymorphism in the IGS could be detected after amplification of sequences between the spacer and gene promoter both in NIH3T3 cells and in mouse tissues (Fig 3C).
To examine whether IGS rRNA is transcribed from a specific subclass of rRNA genes, we analysed the length of IGS rRNA by RT–PCR using RNA from NIH3T3 cells and mouse liver (Fig 3D). Amplification of IGS rRNA predominantly yielded DNA fragments containing nine enhancer repeats. An additional band comprising six enhancer elements was observed in mouse liver, supporting the view that IGS rRNA is transcribed from a specific subset of rRNA genes to facilitate heterochromatin formation and silencing of a fraction of rDNA repeats in trans.
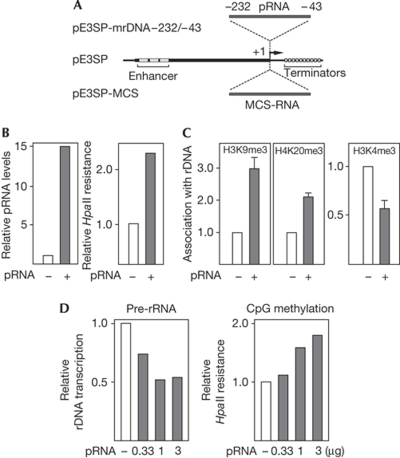
To verify that IGS rRNA functions in trans, we artificially raised the level of spacer transcripts by transfecting NIH3T3 cells with pE3SP-mrDNA−232/−43, a Pol I-driven vector that comprises rDNA sequences from −232 to −43 (Fig 4A). On transfection, a 10–15-fold increase in spacer transcripts was observed (Fig 4B). Consistent with previous results showing that pRNA is required for NoRC-dependent heterochromatin formation and transcriptional silencing, elevated levels of spacer RNA triggered hypermethylation of rRNA genes. In addition, heterochromatic histone marks (H3K9me3 and H4K20me3) increased, whereas rDNA promoter occupancy of the euchromatic histone mark H3K4me3 decreased (Fig 4C). Similarly, elevation of cellular pRNA levels by transfection with in vitro-synthesized pRNA-205/-1 led to hypermethylation of the rDNA promoter and decreased pre-rRNA synthesis, indicating that spacer transcripts mediate heterochromatin formation and silencing of a fraction of rDNA repeats in trans (Fig 4D).
Figure 4.
pRNA functions in trans. (A) Scheme of pE3SP-mrDNA−232/−43 used to express ectopic pRNA. The vector pE3SP contains 5′-terminal rDNA sequences (−639/−1) including three enhancer elements (grey boxes) fused to 3′-terminal Pol I transcription terminators (Brenz Verca et al, 2007). The rDNA sequences or unrelated sequences from the MCS of pBluescript SK+ were inserted downstream from the Pol I transcription start site (+1) to yield pE3SP-mrDNA−232/−43 or pE3SP-MCS. (B) Ectopic pRNA induces hypermethylation of rDNA in trans. NIH3T3 cells were transfected with pE3SP-MCS (white bars) or pE3SP-mrDNA−232/−43 (grey bars). The graph on the left illustrates the level of ectopic pRNA relative to cellular pRNA. The graph on the right shows the ratio of HpaII-resistant rDNA to total rDNA in cells overexpressing pRNA or MCS RNA. (C) Ectopic pRNA triggers heterochromatin formation in trans. ChIP assays monitoring the indicated histone modifications (H3K9me3, H4K20me3 and H3K4me3) at the rDNA promoter after transfection of NIH3T3 cells with pE3SP-MCS (white bars) or pE3SP-mrDNA−232/−43 (grey bars). (D) Synthetic pRNA induces hypermethylation and rDNA silencing. NIH3T3 cells were transfected with the indicated amounts of in vitro transcribed MCS RNA (white bars) or pRNA-205/-1 (grey bars). Left panel: pre-rRNA synthesis was monitored by RT–qPCR and normalized to GAPDH mRNA. Right panel: data represent the ratio of HpaII-resistant rDNA to total rDNA in pRNA-transfected compared with MCS RNA-transfected cells. ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MCS, multiple cloning site; Pol I, RNA polymerase 1; pre-rRNA, rRNA precursor; pRNA, promoter RNA; rDNA, ribosomal DNA; RT–qPCR, reverse transcriptase quantitative PCR.
Discussion
Recently, Tseng et al (2008) identified several mouse rDNA variants that are genetically distinct and show a tissue-specific expression pattern, challenging the idea that rDNA is an array of identical, co-regulated repetitive units. Our study supports the idea of the existence of rDNA variants with distinct functions. We show that the spacer promoter of a specific set of hypomethylated rRNA genes directs the synthesis of IGS rRNA in early S phase. These spacer transcripts are processed during mid to late S phase to yield pRNA that is indispensable for NoRC-dependent rDNA silencing (Mayer et al, 2006). Although the spacer promoter and the main gene promoter have some sequence homology and Pol I is enriched at the spacer promoter, IGS transcripts are rare and cannot be detected on northern blots, indicating that IGS rRNA is rapidly degraded unless stabilized by binding to NoRC. In turn, NoRC requires the association with pRNA for binding to chromatin and heterochromatin formation.
Notably, timing of IGS rRNA transcription and processing into pRNA correlates with NoRC binding to silent rRNA genes during mid to late S phase; that is, the time when NoRC associates with newly replicated silent genes (Li et al, 2005). Thus, the cell carefully tunes the timing of IGS rRNA transcription/processing to inherit rDNA silencing during cell division, suggesting that replication timing represents a ‘window of opportunity' to propagate the epigenetic state and transcriptional activity of individual rDNA clusters, allowing activators or repressors to bind selectively to newly replicated chromatin.
The finding that transcripts from the spacer promoter have an indispensable function in epigenetic silencing of rDNA is in apparent disagreement with previous studies showing that, in Xenopus and mice, spacer promoters enhance pre-rRNA synthesis in cis, suggesting that spacer promoters form a functional unit in conjunction with adjacent enhancers to activate transcription (de Winter & Moss, 1986; Tower et al, 1989; Paalman et al, 1995; Caudy & Pikaard, 2002). This implies that the spacer promoter has a dual function in rDNA transcription, regulating the readout of rDNA both in cis and in trans. It is probable that loop formation between the spacer promoter and the gene promoter activates Pol I transcription at specific rRNA genes (R.S., unpublished data), whereas the production of IGS rRNA from other repeats is required for NoRC-dependent rDNA silencing. These results reveal a further level of complexity in the regulation of rDNA transcription, suggesting that different gene clusters are independently regulated and have distinct functions.
Methods
Plasmids. The plasmid pMr−168 contains rDNA sequences from −168 to +155 with respect to the transcription start site. The plasmid pMr−2175 contains spacer sequences from −2175 to −1905. The rDNA reporter plasmids are fusions of 5′-terminal rDNA fragments containing the pre-rRNA gene promoter (pMr−1922/+130-BH) or the spacer promoter (pMr−2175/−1933-BH) with a 3′-terminal fragment including two terminator elements. Reporter transcripts were monitored by RT–qPCR using primers that amplify pUC sequences present between the promoter and the terminators. pE3SP-mrDNA−232/−43, rDNA sequences from −232 to −43 were inserted into pE3SP (Brenz Verca et al, 2007). To synthesize pRNA in vitro, pT7mr−205/−1 was used, a pTOPO® vector containing mouse rDNA sequences from −205 to −1 fused to the T7 bacteriophage promoter.
Transfection and RNA analysis. NIH3T3 cells were transfected with plasmid DNA by using the calcium phosphate co-precipitation method or with synthetic pRNA (1 μg) by using the Mirus TransIT TKO reagent (Mobitec, Göttingen, Germany). Cellular IGS rRNA and pre-rRNA were measured by RT–PCR with random hexameric primers or a primer encompassing rDNA sequences from −20 to −1. For RNA-FISH, NIH3T3 cells were fixed for 15 min with 4% formaldehyde and permeabilized with 0.5% Triton X-100 for 5 min. Cells were incubated at 37°C overnight with a biotin-labelled RNA probe (5 ng/μl) in 50% formamide, 2 × saline sodium citrate, transfer RNA (1 mg/ml), 1% bovine serum albumin and 10% dextran sulphate. Signals were visualized with extravidin–Cy3 conjugate (Sigma, Deisenhofen, Germany). The IGS rRNA was visualized by hybridization with a biotin-labelled RNA probe encompassing sequences from −554 to −447; pRNA was detected using a probe comprising sequences from −232 to −140 with respect to the transcription start site.
In vitro transcription assays. The assays (25 μl) contained 50 ng of template DNA (pMr−168 or pMr−2175 linearized with NdeI), 30 μg of nuclear extract proteins from exponentially growing FM3A cells, 12 mM 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (HEPES)–KOH (pH 8.0), 0.1 mM EDTA, 5 mM MgCl2, 80 mM KCl, 10 mM creatine phosphate, 12% (v/v) glycerol, 0.66 mM each of ATP, GTP and CTP, 0.01 mM UTP and 0.5 mCi [α-32P]UTP (5,000 Ci/mmol). After incubation for 60 min at 30°C, RNA was extracted and analysed on 4.5% polyacrylamide gels. To synthesize pRNA, a DNA fragment containing the T7 promoter fused to mouse rDNA sequences from −205 to −1 was used as a template in T7 RNA polymerase-driven transcription assays. Control RNA (MCS-RNA) was synthesized to generate T7 RNA polymerase from pBluescript II SK+/EcoRI.
Cell synchronization. The NIH3T3 cells were arrested at the G1/S boundary by treatment with aphidicolin (2 μg/ml for 20 h), released and labelled in 1 h intervals for 30 min with 30 μM BrdU. Newly replicated DNA was immunoprecipitated with BrdU antibody in the presence of BrdU-labelled Escherichia coli DNA, and measured by semi-quantitative PCR (Li et al, 2005).
ChIP and DNA methylation assays. Sheared cross-linked chromatin (40 μg) was incubated with antibodies at 4°C overnight in 16.7 mM Tris–HCl (pH 8.0), 167 mM NaCl, 0.01% SDS and 1.1% Triton X-100. Immunoprecipitates were captured on protein G/A–Sepharose saturated with salmon sperm DNA (0.25 mg/ml) for 2 h. Protein–DNA complexes were washed twice in a low salt buffer (150 mM NaCl, 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2 and 1% Triton), twice in the same buffer but containing 500 mM NaCl, once in a buffer containing 250 mM LiCl, 10 mM Tris–HCl (pH 8.0), 5 mM EDTA, 0.5% Na-deoxycholate and 0.5% Triton, and twice in Tris–EDTA (TE) buffer. After reversal of the cross-links (6 h at 65°C) and digestion with proteinase K, DNA was extracted and amplified by qPCR. DNA in the immunoprecipitates and input chromatin was normalized to control reactions from mock-transfected cells. To monitor CpG methylation, 1 μg genomic DNA was digested with 5 U HpaII or MspI before PCR amplification with promoter-specific primers (Mayer et al, 2006). Primer sequences are listed in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Marlene Tesch for technical assistance. This study was supported by the Deutsche Forschungsgemeinschaft (Transregio 5, Priority Programme ‘Epigenetics'), the European Union Network ‘Epigenome' and the Fonds der Chemischen Industrie. J.S. was supported by a short-term fellowship from the Federation of European Biochemical Societies.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnheim N, Kuehn M (1979) The genetic behaviour of a cloned mouse ribosomal DNA segment mimics mouse ribosomal gene evolution. J Mol Biol 134: 743–763 [DOI] [PubMed] [Google Scholar]
- Brenz Verca MS, Weber P, Mayer C, Graf C, Refojo D, Kühn R, Grummt I, Lutz B (2007) Development of a species-specific RNA polymerase I-based shRNA expression vector. Nucleic Acids Res 35: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Pikaard CS (2002) Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J Biol Chem 277: 31577–31584 [DOI] [PubMed] [Google Scholar]
- de Winter RF, Moss T (1986) Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell 44: 313–318 [DOI] [PubMed] [Google Scholar]
- Grimaldi G, di Nocera PP (1988) Multiple repeated units in Drosophila melanogaster ribosomal DNA spacer stimulate rRNA precursor transcription. Proc Natl Acad Sci USA 85: 5502–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Grummt I (1999) Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci USA 96: 6095–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Grummt I (1987) A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J 6: 3487–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P, Reeder RH (1984) Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell 37: 285–289 [DOI] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I (2005) The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 12: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I (2008) The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 9: 774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paalman MH, Henderson SL, Sollner-Webb B (1995) Stimulation of the mouse rRNA gene promoter by a distal spacer promoter. Mol Cell Biol 15: 4648–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Pikaard CS (1992) Cooperative binding of the Xenopus RNA polymerase I transcription factor xUBF to repetitive ribosomal gene enhancers. Mol Cell Biol 12: 4970–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2005) Epigenetic mechanisms of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol 25: 2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T (2006) Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell 21: 629–639 [DOI] [PubMed] [Google Scholar]
- Strohner R, Németh A, Jansa P, Hoffmann-Rohrer U, Santoro R, Längst G, Grummt I (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Chou W, Wang J, Zhang X, Zhang S, Schultz RM (2008) Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS ONE 3: e1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Henderson SL, Dougherty KM, Wejksnora PJ, Sollner-Webb B (1989) An RNA polymerase I promoter located in the CHO and mouse ribosomal DNA spacers: functional analysis and factor and sequence requirements. Mol Cell Biol 9: 1513–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan X, Frödin M, Grummt I (2003) ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol Cell 11: 405–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information