Despite a significant body of literature identifying the main regulators of autophagy in model organisms, there are gaps in our understanding of the precise molecular and biochemical modulators that control this adaptive process in metazoans. Two studies in this issue of EMBO reports provide new insight into the autophagic process. Mauvezin and colleagues (2010) show that a nuclear protein, known as DOR, is a new player in stress-induced autophagy, and the Dikic lab reports that the mitochondrial protein Nix is a selective autophagy receptor that mediates mitochondrial clearance (Novak et al, 2010).
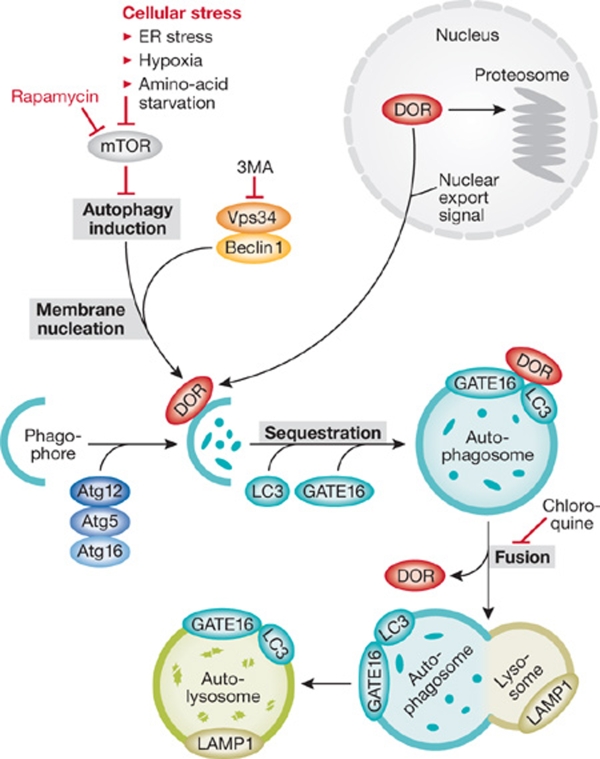
Autophagy is a conserved catabolic cellular process in which macromolecules and organelles are degraded, either as a means to eliminate long-lived, damaged, or faulty cellular components, or in response to stresses such as hypoxia, endoplasmic reticulum stress, or nutrient deprivation. Cells can use the products of autophagic degradation to generate energy or other essential proteins required for cell survival. When cellular conditions are favourable, autophagy is negatively regulated by the nutrient sensor mammalian target of rapamycin (mTOR). In the presence of stress, mTOR is inhibited and autophagy is induced (Fig 1). In this setting, beclin 1 forms a complex with Vps34, which is a class III phosphatidylinositol 3-kinase (PI3K). Through a poorly defined process, this complex initiates the nucleation of the autophagosome membrane. The autophagy-related (Atg) proteins Atg5, Atg12 and Atg16 also have a role in this process, by promoting the formation of the autophagosome precursor, the phagophore. The proteins LC3 and GATE16 localize to the autophagosomal membrane during autophagosome formation and remain bound as it fuses with a lysosome and becomes an autolysosome, the end organelle where autophagic substrates are degraded. Two widely used drugs are known to inhibit autophagy: chloroquine, which blocks the fusion of autophagosomes with lysosomes, and 3-methyladenine (3MA), an inhibitor of class III PI3K (Fig 1).
Figure 1.
Incorporation of DOR into the autophagy pathway. The autophagy pathway is explained in detail in the main text. Upon cellular stress, DOR is exported from the nucleus and possibly aids the beclin 1/Vps34 complex in the nucleation of the autophagosome. LC3 and GATE16 localize to the autophagsome during its formation and form a complex with DOR. DOR is detached from the autophagsome before it becomes an autolysosome, in which the targeted cellular components are degraded.
Although our understanding of autophagy has increased substantially in recent years, there are still many unknowns about the identity of the components of the autophagic pathway, the molecular signals that regulate it, and the mechanisms that target specific cellular components to be degraded. In a breakthrough article in this issue of EMBO reports, the Zorzano group successfully identify a previously unknown component of the autophagic pathway, the nuclear cofactor DOR (diabetes- and obesity-regulated gene), thereby providing additional insight into this evolving field (Mauvezin et al, 2010). In addition, Novak and colleagues identify a mitochondrial receptor involved in the specific targeting of this organelle for mitophagy (Novak et al, 2010).
DOR: a key regulator of autophagy
DOR was identified originally by the Zorzano group as a protein expressed in nuclear bodies of insulin-sensitive tissues (Baumgartner et al, 2007). Now, the authors characterize a novel function of DOR in autophagy regulation. In response to cellular stress or activation of autophagy, DOR exits the nucleus and relocates to punctate cytoplasmic structures. A large series of well-executed experiments further demonstrates that cytoplasmic DOR localizes to early autophagosomes and interacts directly with the autophagosome-membrane-associated proteins LC3 and GATE16, but not autolysosome-associated protein LAMP1 (Fig 1). Why does DOR fail to remain associated with the autophagosome after fusion to the lysosome? The answer is unclear, but the fact that it does not remain associated suggests that DOR could function as a cofactor to target material to the autophagosome in a retrieval–recycling manner, similar to Atg and Rab proteins (Congcong & Klionsky, 2007; Schwartz et al, 2008). In the absence of cellular stress, neither chloroquine nor 3MA perturbed the nuclear localization of DOR, indicating that a specific activation of autophagy by cellular stress is crucial to release DOR from the nucleus.
In contrast to untreated cells, HeLa cells transfected transiently with DOR showed greater rates of protein degradation and had higher numbers of LC3-GFP-positive puncta per cell, as well as an increased accumulation of autophagosomes, both under basal and starvation conditions. The role of DOR in mediating autophagy was confirmed through the generation of a number of DOR mutants devoid of their nuclear export signal, which were confined to the nucleus and defective in their ability to stimulate autophagosome formation in response to starvation. When DOR expression was suppressed by siRNA in C2C12 myoblasts, cells had fewer LC3-GFP-positive puncta, fewer autophagosomes, and a substantial inhibition of protein degradation compared with control cells. Together, this provides convincing evidence for the positive role of DOR in stress-induced autophagy.
Interestingly, the class III PI3K (Vps34) inhibitor 3MA prevented starvation, rapamycin-mediated autophagy and DOR export into the cytoplasm. This result seems to contradict previous evidence that Vps34 is essential for the stimulating effects of mTOR on insulin (Byfield et al, 2005) and the established role of Vps34 in the early step of autophagosome nucleation (Levine et al, 2008), although another study suggests that Vps34 can promote autophagy without affecting Tor signalling (Juhasz et al, 2008). One could speculate that DOR might act as a bridge to integrate signals between mTOR and Vps34 during autophagy; whether this is true, however, needs further study.
In addition, a well-characterized model was used to establish a role for DOR in vivo. During insect morphogenesis and development, autophagy is activated to remove superfluous cells in the fat body, salivary gland and midgut of the larvae. Having identified a Drosophila homologue of DOR (dDOR), siRNA was used to suppress the expression of dDOR in larval fat bodies. Although the larvae were largely normal, the pupae underwent late-stage cell death. Control larvae in the wandering stage—during which autophagy is highly induced—had a greater number of Lysotracker-positive puncta in the fat body, as well as more autophagosomes and autolysosomes than the larvae deficient in dDOR. It should be noted that the DOR characterized in this article is not deep orange, the Drosophila Vps18 homologue that has also been implicated in autophagy, and which is unluckily also known as Dor. Collectively, these experiments demonstrate that DOR has a role in regulating autophagy, which is conserved in humans, mice and Drosophila.
Opening new questions
Autophagy can promote both cell death and cell survival; it is therefore surprising that despite the crucial role of DOR in controlling autophagy, this issue was not addressed fully by Mauvezin and colleagues. Pupae lacking dDOR undergo cell death at a late stage, which suggests that in this system autophagy might promote pupae survival. Also, as mentioned above, the association of DOR only with autophagosomes raises interesting questions about its potential shuttling properties. One possibility could be that DOR functions as a cofactor to target autophagic-targeted material to the nucleating autophagosomes. This is consistent with the role of Vps34 in this early step of autophagosome formation and the observation that DOR nuclear exit in response to amino-acid starvation is abolished by 3MA.
How specific cellular constituents are targeted for autophagic degradation remains unclear. Atg32 in yeast and Uth1p in mammalian cells are thought to tag damaged mitochondria to be degraded by autophagy (Okamoto et al, 2009 and references within). In this issue of EMBO reports, the Dikic group shows that Nix—a Bcl2-related family member—is a selective autophagy receptor for mitochondrial clearance that is implicated in mitophagy after organelle damage and during reticulocyte maturation—a process dependent on the degradation of cellular mitochondria (Novak et al, 2010). Whether DOR interacts with Nix or plays a similar role in targeting cellular organelles to autophagosomes would be an interesting topic for future research.
In spite of the remaining questions surrounding the molecular details of DOR and Nix in autophagy, the current studies have contributed an immense body of work to the field by identifying and characterizing the function of two novel regulators of autophagy. This is an important step towards uncovering the missing links in the activating signals and dynamics of the cellular interactions during this process.
References
- Baumgartner BG et al. (2007) PLoS ONE 2: e1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield MP et al. (2005) J Biol Chem 280: 33076–33082 [DOI] [PubMed] [Google Scholar]
- Congcong H, Klionsky DJ (2007) Autophagy 3: 271–274 [DOI] [PubMed] [Google Scholar]
- Juhasz G (2008) J Cell Bio 4: 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B et al. (2008) Autophagy 4: 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C et al. (2010) EMBO Rep 11: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I et al. (2010) EMBO Rep 11: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K et al. (2009) Dev Cell 17: 87–97 [DOI] [PubMed] [Google Scholar]
- Schwartz SL et al. (2008) J Cell Sci 120: 3905–3910 [DOI] [PubMed] [Google Scholar]