Abstract
BRCA1, the breast and ovarian cancer specific tumor suppressor, is a transcriptional repressor as well as a transcriptional activator dependent on the promoter context. In order to identify the genes activated or repressed by BRCA1, we have analyzed microarray results from cells depleted of BRCA1 and revealed a number of genes regulated by BRCA1 on the level of transcription. Among the genes repressed by BRCA1 we have identified Amphiregulin (AREG) and Early Growth Response-1 (EGR1). Results indicate that BRCA1 regulates AREG transcription directly through the binding to the AREG promoter, however we could not detect BRCA1 on the EGR1 promoter, suggesting that EGR1 is indirectly regulated by BRCA1. In an attempt to indentify the mechanism of the AREG transcriptional repression by BRCA1, we have mapped two independent BRCA1-response elements on the AREG promoter located at the positions −202/−182 and +19/+122. BRCA1 depletion leads to induction of AREG protein. Taken together, our data builds the connection between BRCA1 loss of function and AREG up-regulation, a change in gene expression often observed in breast cancer.
Keywords: BRCA1, Amphiregulin (AREG), transcription regulation, breast cancer
Introduction
Loss of function of the tumor suppressor BRCA1 (breast cancer associated 1) protein is responsible for high percentage of familial and sporadic breast cancers. BRCA1 inactivating mutations are found in about fifty percent of patients with familial breast cancer (1, 2), however in sporadic breast cancer mutations in BRCA1 are almost never found. Nevertheless, sporadic cases are often characterized by decreased BRCA1 mRNA and protein level (3), moreover, in many clinical samples, about 14% of sporadic breast cancers, revealed hypermethylated BRCA1 promoter, evidence of epigenetic down regulation (4).
BRCA1 protein participates in key cellular processes including transcription, repair of DNA damage, cell cycle checkpoints, and centrosome dynamics (5-7). The question how BRCA1 mediates its tumor suppressor role is unresolved. BRCA1 is primarily a nuclear protein (1), and one of the first identified roles of BRCA1 was transcriptional stimulation (8, 9). BRCA1 co-purifies with an RNA polymerase II (RNAPII) containing complex (9-11). The mechanism by which the BRCA1 protein regulates transcription has been elucidated. BRCA1 stimulates transcription at a variety of promoters by stabilizing the preinitiation complex (12). BRCA1 can also act as a transcriptional repressor when in the presence of active ubiquitination cofactors by conjugating ubiquitin on RNAPII and preventing the initiation of RNA synthesis (13). It is likely that the specificity of whether BRCA1 stimulates or represses the transcription reaction depends on the factors that recruit the BRCA1 protein to the promoter (12, 13).
How might the BRCA1 function as a transcriptional repressor contribute to its function of tumor suppressor? We hypothesize that the recruitment of BRCA1 to a gene promoter regulates transcription of a factor critical for tumorigenesis. In the current study we have performed gene expression microarray analysis with RNAi-depleted BRCA1 samples and identified genes significantly repressed by BRCA1, including AREG (Amphiregulin) and EGR1 (Early-growth response 1). We found that BRCA1 is present on the AREG promoter. Using reporter assays, we determined that the response to BRCA1 repression mapped to two sites at −202/−182 and +19/+122 positions of the AREG promoter. By contrast, our results suggest that BRCA1 indirectly regulates EGR1 transcription, possibly via AREG protein induction. Taken together, our data demonstrate the connection between BRCA1 tumor suppressor and AREG, a ligand of the epidermal growth factor receptor, which is important in breast cancer development and progression.
Materials and Methods
1. Constructs
Plasmids for the expression of specific shRNA were prepared as described (14). The BRCA1-specific shRNA plasmid was the same one used previously (13). Reporter constructs were based on AREG promoter containing vector described in (15) and prepared by subcloning of the promoter fragments −643/+122, −202/+122, −182/+122, −159/+122, −136/+122, −110/+122, −44/+122, −202/+19, −182/+19, −159/+19, −136/+19, −110/+19 into pGL3 basic vector (Promega, Madison WI, USA). The primer sequences used for subcloning are presented in supplementary material (Supplementary Table 1).
2. Cell culture
HeLa cell line (ATCC cell line CCL-2, East Greenwich, RI, USA) was maintained in DMEM media supplemented with 10% Bovine Serum, 100 I.U./ml Penicillin, 100 μg/ml Streptomycin. SUM149PT cell line (Asterand, Detroit, MI, USA) was maintained in Ham’s F-12 medium supplemented with 5% Fetal Bovine Serum, 5 μg/ml insulin, 2 μg/ml hydrocortisone and 5 μg/ml gentamicin. All the cell lines were maintained in a humidified incubator at 37° C and 5% CO2.
3. Preparation of cDNA for Microarray and TaqMan Analysis
HeLa cells were cotransfected (Lipofectamine; Invitrogen, Carlsbad, CA, USA) with 5 μg of shRNA expression plasmid (14) and 20 ng of pBabe-puromycin plasmid. BRCA1 shRNA (gccacaggaccccaagaaugag) was targeted to the 3′- untranslated region of the BRCA1 mRNA. The control shRNA was targeted against a mutant GFP construct (gggccauggcacguacggcaag). Puromycin selection (2 μg/ml) was applied 24 h after transfection, and cells were harvested at 72 h post transfection. BRCA1 siRNA used to eliminate the possibility of off-target effect was targeted to the position 2616 on mRNA (GGU UUC AAA GCG CCA GUC AdTdT). The control siRNA was targeted against luciferase (UCG AAG UAU UCC GCG UAC GdTdT). RNA was prepared with either Tri Reagent (Molecular Research, Cincinnati, OH, USA) and further purified over RNeasy columns (Qiagen, Valencia, CA, USA) or by using Dynabeads mRNA direct kit (Invitrogen, Carlsbad, CA, USA). Microarray analysis was performed on separate samples at the Harvard Biopolymers Facility (Boston, MA) and at the Microarray Shared Resource in Comprehensive Cancer Center (Ohio State University, Columbus, OH, USA) on the Affymetrix (Santa Clara, CA, USA) HG-U133_plus_2 chip. The use of different RNA purification strategies and of different microarray facilities, resulted in identification of the most robust changes in gene expression secondary to BRCA1 depletion. For TaqMan assays, cDNA was reverse-transcribed from mRNA using the Superscript II kit (Invitrogen, Carlsbad, CA, USA). Primers for AREG, EGR1, Jun, BRE, XRCC4, DDX58, KRT17, GDF15 and CCL5 were purchased from the Applied Biosystems (Foster City, CA, USA). The real-time PCR reactions were done in nine replicates. GAPDH and TFRC were used as controls. Calculations of ΔΔCt value was done as described previously (16).
4. Chromatin immunoprecipitation (ChIP)
The cells were treated with 1% formaldehyde for 20 minutes at 37° C in the incubator, washed three times with 1X PBS buffer and lysed in a warm buffer (30° C) containing 50 mM Tris HCl pH 8.1, 10 mM EDTA, 1% SDS, protease inhibitor cocktail and 1 mM PMSF. The lysate was incubated on ice for 20 minutes and sonicated to obtain approximately 500 bp DNA fragments. The samples were diluted 10-fold into dilution buffer (part of Chromatin Immunoprecipitation (ChIP) Assay Kit (17-295), cat. # 16-153, Upstate (Millipore), Lake Placid, NY, USA), pre-cleared with ssDNA/protein A beads (part of Chromatin Immunoprecipitation (ChIP) Assay Kit (17-295), cat. # 16-157, Upstate (Millipore), Lake Placid, NY, USA) and incubated with the antibodies at 4° C overnight. The BRCA1-specific antibody and matched pre-immune serum have been described before (17). Sp1 (SC-59, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and Histone H3 antibodies (06-755, Upstate (Millipore), Lake Placid, NY, USA) were used. The beads were added and incubated for one hour, washed one time with the high salt solution (part of Chromatin Immunoprecipitation (ChIP) Assay Kit (17-295), cat. # 16-155, Upstate (Millipore), Lake Placid, NY, USA), low salt solution (part of Chromatin Immunoprecipitation (ChIP) Assay Kit (17-295), cat. # 16-154, Upstate (Millipore), Lake Placid, NY, USA), LiCl solution (part of Chromatin Immunoprecipitation (ChIP) Assay Kit (17-295), cat. # 16-156, Upstate (Millipore), Lake Placid, NY, USA) and three times with 1 X TE buffer pH 8.0. The samples were eluted from the beads in 1% SDS and 0.1 M NaHCO3 solution and treated with 0.2 M NaCl for six hours at 65° C, then with 0.1 mg/ml proteinase K (in the buffer containing 10 mM EDTA and 70 mM Tris-HCl pH=6.5) for one hour at 45° C and extracted using PCR clean-up kit (Qiagen, Valencia, CA, USA). Alternatively, the samples were prepared using ChIP-IT™ Express Enzymatic kit (Active Motif, Rixensart, Belgium). The samples were analyzed by real-time PCR using SYBR Green dye (BioRad, Hercules, CA, USA) and the primers described in supplementary materials Table 2. Real-time PCR reactions were run on the equipment provided by Plant-Microbe Genomics Facility (Ohio State University, Columbus, OH, USA). Results were analyzed using the methods described previously (BioRad, Bulletin 2916).
5. Reporter gene assay
HeLa cells on 6-well plate were cotransfected (Lipofectamine; Invitrogen, Carlsbad, CA, USA) with 1.6 μg of shRNA expression plasmid targeting either BRCA1 or, as a control, mutant GFP (14), 1.6 μg promoter-reporter construct and 66 ng of pBabe-puromycin plasmid. Puromycin selection (2 μg/ml) was applied 24 h after transfection, and 72 h after transfection cells were lysed using lysis buffer from luciferase assay system kit (Promega, Madison WI, USA). The fluorescent units were normalized to the sample protein content measured using BCA protein assay kit (Pierce, Rockford, IL, USA). The transfection efficiency was checked by contransfection of the GFP expression plasmid.
Results
Inhibition of BRCA1 reveals a number of repressed and activated genes in cancer-associated pathways
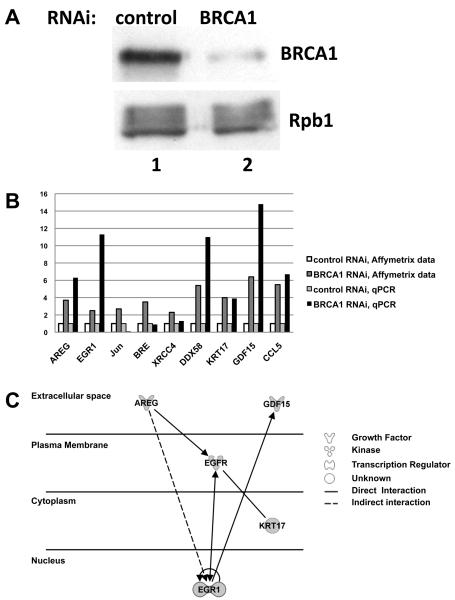
In an effort to determine which genes might be contributing to cancer development and progression when BRCA1 function is lost, we performed an Affymetrix array analysis by comparing the gene expression pattern of HeLa cells in which the BRCA1 protein was depleted using RNAi (Figure 1A).
Figure 1. Affymetrix data identify a number of genes repressed by BRCA1, among them AREG and EGR1 and the pathway analysis reveals EGR1 as a downstream target for AREG.
A. shRNA-mediated BRCA1 depletion in HeLa cells. HeLa cells were transfected with a plasmid expressing shRNA specific for GFP (control, lane 1) or for BRCA1 (lane 2). Cells were harvested 72 h after transfection, and Western blots were probed with antibodies against BRCA1 (top panel) and RNA polymerase II large subunit (Rpb1, loading control, bottom panel).
B. Validation of the data obtained from the Affymetrix microarray analysis by real-time PCR. The controls in Affymetrix microarray data and in the real time PCR were normalized to 1 and shown in white and light grey. The Affymetrix microarray data and the real-time PCR data for the samples where BRCA1 was depleted shown in dark gray and black, respectively. X axis shows the genes used in the study, Y axis shows the fold change relative to the control.
C. Pathway analysis of the genes identified by Affymetrix microarray and validated by quantitative real-time PCR. A biological circuit with AREG taken from Ingenuity Pathway Analysis software (IPA) of microarray results is shown. AREG, GDF15, EGR1, and KRT17 all had significant changes in expression following depletion of BRCA1, and EGFR did not significantly change expression however was included in the regulatory module due to the pathway analysis. The position of each gene on the figure corresponds to its cellular localization. The dashed line illustrates the indirect connections between the molecules, the solid lines indicate direct connection between molecules. The line without arrows shows interaction, the lines with arrows indicate activation.
Our study revealed the large number of genes, which were either up- or down-regulated following depletion of BRCA1, and we chose 2.3-fold change as a reasonable cut off for further investigation. The list of these genes is shown in Table 3 of supplementary materials. Ingenuity Pathway Analysis software (IPA) was used to analyze the lists of genes and reveal their contribution to disease, to identify genetic networks and pathways that include the affected genes. Analysis of the gene list (253 genes), using IPA software, revealed that 85 genes are expressed in mammary gland and 153 genes are expressed in breast cancer cell lines (Table 4 of supplementary materials). Moreover, IPA revealed the involved biological functions and diseases. The distribution of the genes according to their function and their role in the pathological processes is presented in the Table 1. The pathological processes that involve cancer (cell cycle, cell growth, and cell movement, and gene expression) are all consistent with known functions of BRCA1 in controlling cell cycle progression after DNA damage (18), DNA repair (19), and control of centrosomes (18, 20).
Table 1.
The distribution of the genes up or down regulated more than 2.3 fold time in response to BRCA1 silencing according the top networks, diseases and disorders to which they might be involved and their function in the cell
| The distribution of the genes up or down regulated more than 2.3 fold time in response to BRCA1 silencing according to the networks to which they might be involved | ||
|---|---|---|
| Top networks | Score | Molecules |
| Immune Response, Viral Function, Nucleic Acid Metabolism | 46 | 26 |
| Cardiac Hypertrophy, Cardiovascular Disease, Developmental Disorder |
39 | 23 |
| Cancer, Cell Cycle, Gastrointestinal Disease | 34 | 21 |
| Cancer, Cellular Growth and Proliferation, Hematological Disease | 32 | 20 |
| Cardiovascular System Development and Function, Cell Morphology, Cellular Development |
30 | 19 |
| Cancer, Cellular Movement, Neurological Disease | 21 | 15 |
| Amino Acid Metabolism, Cancer, Small Molecule Biochemistry | 21 | 15 |
| Cardiovascular System Development and Function, Molecular Transport, Small Molecule Biochemistry |
20 | 14 |
| Cell Signaling, Nucleic Acid Metabolism, Small Molecule Biochemistry |
18 | 13 |
| Organ Morphology, Reproductive System Development and Function, Amino Acid Metabolism |
18 | 13 |
| Cancer, Cell Death, Gene Expression | 18 | 13 |
| Drug Metabolism, Molecular Transport, Nucleic Acid Metabolism | 16 | 12 |
| The distribution of the genes up or down regulated more than 2.3 fold time in response to BRCA1 silencing according to the diseases and disorders to which they might be related | ||
|---|---|---|
| Diseases and Disorders | p-value | molecules |
| Cancer | 1.45E-08 -2.79E-03 | 100 |
| Viral function | 2.66E-06 - 1.57E-03 | 9 |
| Organism injuries and abnormalities | 2.84E-06 - 2.47E-03 | 29 |
| Dermatological diseases and conditions | 6.25E-06 - 3.91E-04 | 22 |
| Genetic disorder | 6.25E-06 - 2.70E-03 | 12 |
| The distribution of the genes up or down regulated more than 2.3 fold time in response to BRCA1 silencing according to their function in the cell | ||
|---|---|---|
| Molecular and cellular functions | p-value | molecules |
| Cell death | 2.27E-08 – 2.79E-03 | 67 |
| Cellular growth and proliferation | 2.86E-07 – 2.79E-03 | 69 |
| Cellular development | 2.66E-06 – 2.72E-03 | 25 |
| Gene expression | 9.07E-06 – 1.73E-03 | 34 |
| Cell cycle | 1.50E-05 – 2.79E-03 | 34 |
The distribution of the genes according to the diseases and disorders to which they might be related is also shown in Table 1. 100 of these genes are involved in cancer development and progression. The functional analysis of the genes reveals that they are involved in cell growth and proliferation (69 molecules), cell death (67 molecules) as well as in cellular development, gene expression and cell cycle regulation (Table 1). These results in Table 1 illustrate how BRCA1 loss can contribute to cancer development and progression. Since the changes in gene expression due to BRCA1 depletion are consistent with known processes, functions, and diseases associated with BRCA1, investigation of specific genes from this dataset is supported.
Inhibition of BRCA1 leads to increased AREG mRNA expression
Loss of BRCA1 protein, as may happen in a breast cancer tumor cell, would be expected to result in a de-repression of genes that the tumor suppressor normally inhibits. Genes that were significantly up-regulated in our dataset and identified using IPA to impact cancer development and progression, were selected from the microarray data and validated further by real-time PCR (Figure 1B). AREG, EGR1, DDX58, KRT17, GDF15 and CCL5 were confirmed by real-time PCR (Figure 1B). The genes BRE, Jun, and XRCC4 were not confirmed by real-time PCR and therefore excluded from the study. AREG, EGR1, DDX58, KRT17, GDF15 and CCL5 were all expressed in breast cancer cell lines according to IPA (Supplementary Table 4).
Among the genes up-regulated in HeLa cells with depleted BRCA1, we have identified AREG and EGR1 to be of particular interest since both genes have been reported to have high levels of expression in breast cancer (21, 22), moreover both genes were upregulated in Affymetrix microarray experiment and validated by quantitative real-time PCR. Affymetrix data revealed 3.7-fold increase in AREG mRNA, and quantitative RT-PCR revealed a 6.3-fold increase in AREG expression after depletion of BRCA1 by RNAi (Figure 1B). Therefore, we conclude that the BRCA1 silencing leads to the increased AREG mRNA expression in HeLa cells. For EGR1 we have observed 2.5-fold increase in expression from Affymetrix data and 11.3 fold increase in quantitative real-time PCR (Figure 1B). In order to eliminate the possibility of off-target effects, we depleted BRCA1 in HeLa cells using an siRNA targeting a different BRCA1 sequence than was specified by the shRNA used in the preceding experiments. The quantitative real-time PCR data revealed a 2.3-fold increase in AREG expression and 2.9-fold increase in EGR1 expression (Supplementary Figure 1); therefore we have concluded that AREG and EGR1 mRNA levels increase in response to BRCA1 depletion. Moreover, IPA analysis revealed that EGR1 is a downstream target for AREG (Figure 1C and Supplementary Figure 2), and indicates how genes identified by microarray and confirmed by quantitative real-time PCR can all be linked: AREG overexpression indirectly leads to EGR1 up-regulation which in turn stimulates Growth Differentiation Factor (GDF15). AREG has been reported to bind to the Epidermal Growth Factor Receptor (EGFR) and to activate it (23). Pathway analysis reveals that EGFR interacts with KRT17 and stimulates EGR1 expression, which in turn stimulates GDF15 (Figure 1C). The stimulation of this module could be initiated by increases in AREG concentration. Our subsequent experiments are focused on AREG and EGR1 since the increases in mRNA levels for these genes ranked among the highest in the Affymetrix data, and both of these genes are linked to breast cancer.
BRCA1 binds to the AREG promoter
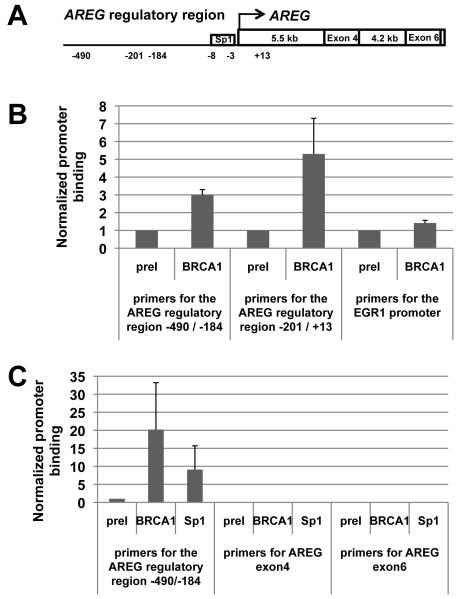
To test whether BRCA1 regulation of AREG is direct, we assayed by the chromatin immunoprecipitation assay (ChIP) for the presence of BRCA1 protein on the AREG promoter. Using either antibodies specific to BRCA1 or a matched pre-immune serum, the quantitative real-time PCR of the immunoprecipitated DNA used primers covering two regions, −490/−184 and −201/+13, of the AREG promoter. We observed a three and five-fold increase relative to the matched pre-immune serum for the promoter regions at −490 to −184 and −201 to +13, respectively relative to the mRNA start site (Figures 2A and 2B). As a positive control, ChIP assay using antibody specific for histone H3 had an 8.8 fold increase when compared to the negative control pre-immune serum for the region −490 / −184 and 37.3 fold increase when compared to the negative control pre-immune serum for the region −201 / +13 (data not shown). We conclude that BRCA1 is present on the AREG promoter and directly represses AREG transcription. Since the histone H3 is certainly abundant in multiple sites on the AREG promoter, it is not surprising that the ChIP assay revealed a higher-fold increase relative to BRCA1. As an additional positive control we assayed by ChIP using antibodies specific for Sp1 transcription factor since its binding site has been located at the positions −3/−8 of the AREG promoter (15) (Figure 2A). As negative controls, we assayed by ChIP for AREG exon 4 (about 5.5 kb away from transcription start site) and exon 6 (about 9.8 kb away from the transcription start site) regions using antibodies specific for BRCA1 and Sp1. As illustrated on Figure 2C, Sp1 and BRCA1 samples had an increase in relative promoter binding in comparison to the preimmune matched serum on the AREG regulatory region −490/−184. By contrast, neither Sp1 nor BRCA1 bound to the AREG exon 4 and AREG exon 6 regions. Taken together, these results indicate that BRCA1 does indeed bind directly to the proximal promoter region of the AREG gene.
Figure 2. BRCA1 is present on the AREG promoter but not on the EGR1 promoter.
A. Map of human AREG gene showing regulatory elements and exons which were used in ChIP assay study.
B. Relative AREG promoter-BRCA1 and EGR1 promoter-BRCA1 association in HeLa cells. The data derived from the quantitative real-time PCR are shown. Chromatin from HeLa cells was immunoprecipitated with pre-immune serum (negative control) or with BRCA1-specific antiserum. For the AREG promoter the primers for the regions −490/−184 and −201/+13 were used, for the EGR1 promoter the primers for the region −108/−21 were used. Error bars show the S.E.M. from three experiments for the AREG promoter and five experiments for the EGR1 promoter.
C. Positive and negative controls for ChIP assay on AREG promoter. Relative AREG promoter-BRCA1 and AREG promoter-Sp1 association in HeLa cells is illustrated. The data derived from the quantitative real-time PCR are shown. Chromatin from HeLa cells was immunoprecipitated with pre-immune serum (negative control), with BRCA1-specific antiserum or with Sp1-specific antiserum. For the AREG promoter the primers for the region −490/−184 were used, for the negative controls the primers for the regions of AREG exon4 and exon6 were used and each experiment was done three times. For the negative control experiments PCR products were obtained only for the input samples (data not shown). Error bars show the S.E.M. from three experiments.
In contrast to the results with the AREG promoter, ChIP experiments aiming to investigate whether BRCA1 binds the EGR1 promoter revealed the absence of BRCA1 on the EGR1 promoter (Figure 2B), suggesting that the effect of BRCA1 on EGR1 transcript up-regulation is indirect. This result is consistent with analysis using the IPA software, which showed that EGR1 stimulation is a downstream target of increasing AREG protein concentration (Figure 1C and Supplementary Figure 2). AREG is an extracellular ligand that interacts with the EGFR receptor present on the surface of the HeLa cell (24), leading to activation of a number of downstream genes including EGR1, which was significantly increased by BRCA1 depletion.
BRCA1 repression of AREG transcription maps to two elements of the AREG promoter
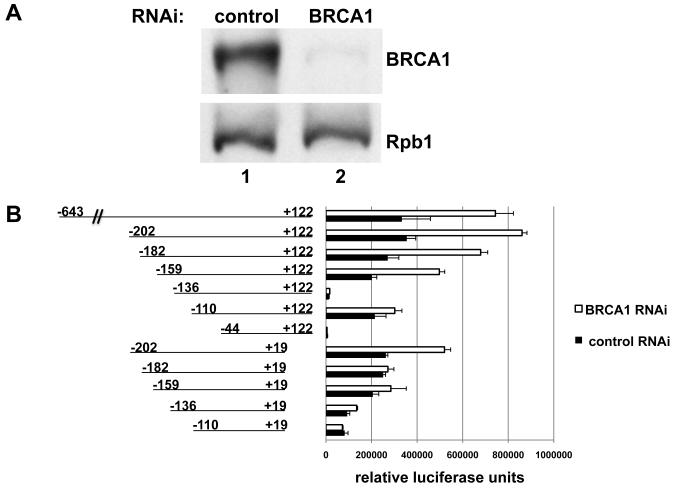
We mapped the sequences of the AREG promoter DNA that are BRCA1-responsive, using a gene expression reporter assay. The reporter construct contained the AREG regulatory sequence −643/+122 relative to the mRNA 5′ end previously described by Plowman et. al., 1990 (15). The reporter gene, luciferase, was placed downstream of the AREG promoter. HeLa cells were transfected with the reporter plasmid, the plasmid that expresses the shRNA that specifies BRCA1 mRNA, and the plasmid carrying puromycin resistance. Cells that received these plasmids were then selected by growth in the presence of puromycin and BRCA1 protein was effectively depleted (Figure 3A and Supplementary Figure 3).
Figure 3. BRCA1 response elements map to positions −202/−182 and +19/+122 of the AREG promoter.
A. shRNA-mediated BRCA1 depletion in HeLa cells. HeLa cells were transfected with a plasmid expressing shRNA specific for GFP (control, lane 1) or for BRCA1 (lane 2), reporter plasmid and puromycin selection plasmid. 24 hours after transfection, puromycin was added to the media to select cells that received plasmid. Cell lysates were harvested 72 hours after transfection, and Western blots were probed with antibodies against BRCA1 (top panel) and RNA polymerase II large subunit (Rpb1, loading control, bottom panel). The western blot for samples transfected with the promoter-reporter construct containing the AREG regulatory sequence −202/+122 is shown. The examples of the western blots showing the samples transfected with other promoter-reporter constructs are shown is supplementary figure 3.
B. Reporter gene assay in HeLa cells. The left panel shows the promoter-reporter constructs used in this study, the right panel illustrates the results derived from the reporter gene assay. The control samples (control RNAi) are colored in black, the test samples (BRCA1 RNAi) are colored in white. Error bars represent S.E.M. of three repeats.
Depletion of BRCA1 led to 2.5-fold de-repression of the reporter containing the AREG regulatory sequence −643/+122 (Figure 3B). The deletion of the region −643/−203 from the promoter-reporter construct revealed that this sequence was not important for the BRCA1 repression and therefore mapped the BRCA1 responsive region to −202/+122. These results for the BRCA1 repression of the reporter containing −202/+122 of the AREG promoter have been validated using an siRNA specific for a different BRCA1 sequence, ruling out off-target effects as causing this repression (Supplementary Figure 4). The comparison of the promoter-reporter constructs containing −202/+122 and −202/+19 fragments of the AREG promoter with the further progressive deletion constructs corresponding to −182/+122 and −182/+19 fragments revealed that BRCA1 depletion can lead to de-repression of the reporters containing −202/+122, −202/+19 and −182/+122 fragments of the promoter but not −182/+19 leading to the conclusion that BRCA1 binds to two regions on the AREG promoter located at −202/−182 and +19/+122 positions. Moreover, these data illustrate that −202/−182 and +19/+122 BRCA1 responsive sites are independent.
BRCA1 regulates transcription on the variety of promoters, however on its own it does not bind DNA with sequence specificity (25, 26). Therefore, we hypothesize that the BRCA1 protein binds indirectly to −202/−182 and +19/+122 DNA elements via other factors specifically bound at these sites, and the identification of these other factors is the subject of continuing investigation.
BRCA1 represses AREG protein levels
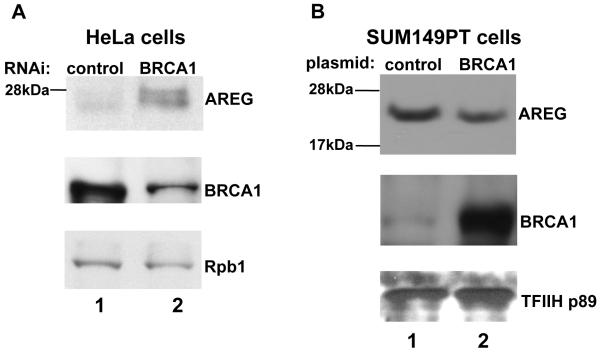
The previous experiments have shown that BRCA1 is a transcriptional repressor of the AREG promoter. Depletion of BRCA1 by RNAi resulted in the increased AREG transcript level. According to the pathway analysis (Figure 1C), the function of the AREG protein can cause the up-regulation of the EGR1 gene that we observed (Figure 1B). Thus, we tested whether BRCA1 control of AREG transcription could affect protein levels. Western blot analysis of BRCA1-depleted HeLa cell lysates revealed that RNAi targeting BRCA1 leads to the increase of AREG protein level (Figure 4A). AREG has been reported to be present in cells as 16, 21, 25, 28 kDa proteins (21). Our data reveal that a protein with migration consistent with a 25 kDa polypeptide and reactive with the AREG-specific antibody is induced following depletion of BRCA1 (Figure 4A). This result supports the concept that the BRCA1 protein represses AREG protein expression.
Figure 4. BRCA1 controls AREG protein level.
A. Western blot analysis of samples derived from HeLa cell line. Using anti-AREG antibodies (top panel), the control sample (lane 1) was compared to the sample where BRCA1 was depleted by RNAi (lane 2). The BRCA1 depletion was confirmed using anti-BRCA1 antibodies (middle panel). The loading control was done using anti-RNA polymerase II large subunit antibody (Rpb1; lower panel).
B. Western blot analysis of samples derived from SUM149PT cell line. Exogenous expression of BRCA1 in the BRCA1-mutant SUM149PT cell line (lane 2) resulted in a decrease in AREG protein levels (top panel, lane 2). The BRCA1 expression was confirmed using anti-BRCA1 antibodies (middle panel). The loading control was done using anti-TFIIH antibody specific for the 89 kDa subunit (lower panel).
AREG was reported to be up-regulated in breast cancer (21), moreover it is known that loss of BRCA1 function can lead to breast cancer development. We asked whether loss of BRCA1 can be connected to breast cancer pathology through the AREG up-regulation. As a model to study how BRCA1 silencing can affect AREG protein level in breast tissue we have chosen SUM149PT invasive breast carcinoma cell line which was reported to have low BRCA1 transcript level due to an inactivating mutation (27). Interestingly, published reports have shown that SUM149PT cell line has increased AREG protein level in comparison to MCF10A cell line (21), indicating that BRCA1 loss and AREG up-regulation could be related.
In order to test whether BRCA1 regulates AREG protein levels, we have performed an experiment wherein BRCA1 was re-expressed in SUM149PT cells and compared the AREG level to untreated samples. The data shown in the Figure 4B illustrated that BRCA1 expression leads to the moderate, yet significant decrease of the AREG protein level (25 kDa band). The fact that the effect was moderate was not surprising and could be explained by the fact that the transfection efficiency in SUM149PT cell line was about 40% (data not shown).
Taken together, our data from two different cell lines show that BRCA1 regulates AREG expression. From this finding, we suggest a model in which loss of BRCA1 often observed in breast cancer patients results in AREG up-regulation which was shown to stimulate proliferation and migration (21).
Discussion
The BRCA1 tumor suppressor has been shown to have various cellular roles including transcription regulation, DNA damage repair, and cell cycle regulation (28). Loss of BRCA1 due to inactivation or mutation leads to cancer. Despite intensive study for several years, the question how BRCA1 loss of function leads to cancer development and progression remains unresolved. In this study, we have found that: 1) depletion of BRCA1 protein results in de-repression of a number of genes including AREG and EGR1; 2) BRCA1 binds to the AREG promoter but not the EGR1 promoter; 3) the AREG promoter contains two independent DNA sites that are responsive to BRCA1; and 4) AREG protein levels are controlled by BRCA1 protein. Taken together, these results suggest that a part of the tumorigenic phenotype of breast cancer might be explained via loss of BRCA1 repression of AREG gene expression.
In order to address the role of BRCA1 repression of transcription in tumor suppression, we first have identified the genes that significantly change their expression level in response to BRCA1 depletion in HeLa cells. We have chosen HeLa cell line for our experiments since it is a well characterized cell line which has endogenously expressed BRCA1, moreover in HeLa BRCA1 can be efficiently depleted. These data are in agreement with previously published results showing that BRCA1 can act both as transcriptional repressor and as a transcriptional activator depending on the promoter context (12, 13). Among the genes which showed high fold change we have selected those which were shown to play a role in cancer, among them AREG and EGR1 and after confirming our Affymetrix results by real-time PCR, we have focused our further study on these two genes.
The comparison of our data in HeLa cells with published results from MCF10A mammary epithelial cells (MECs) in 3D culture following depletion of the BRCA1/CtIP/ZBRK1 repressor complex (29) revealed similar effect for AREG and EGR1. In that study, depletion of BRCA1 resulted in a 50% increase in expression of AREG and EGR1. Though the effect on these genes after BRCA1 depletion had a lower magnitude, the trend is in the same direction, suggesting that BRCA1 control of AREG is not limited to HeLa cells.
AREG and EGR1 up-regulation of transcription as a response to BRCA1 depletion illustrates BRCA1 involvement in the regulation of these genes, however the microarray results, alone, do not answer the question whether BRCA1 regulates AREG and EGR1 transcription directly. ChIP data showed the presence of BRCA1 on AREG promoter but not on the EGR1 promoter. Moreover, EGR1 is a downstream effector in a gene activation cascade initiated by AREG (30). Taken together, our data suggest that BRCA1 is present on AREG promoter and acts as a transcriptional repressor, and the EGR1 up-regulation is secondary to AREG de-repression when BRCA1 is depleted. Moreover, EGR1 leads to direct activation of GDF15, which is up-regulated in breast cancer (31, 32) (Figure 1C), therefore our data suggest how BRCA1, AREG, EGR1, and GDF15 can be connected together.
Since our data revealed that BRCA1 silencing is involved into EGR1 transcription up-regulation indirectly, we have focused our further study on AREG. AREG is a member of the epidermal growth factor family and a ligand of the epidermal growth factor receptor (EGFR). It is expressed as a transmembrane precursor that is cleaved by ADAM17 protease and released to activate its receptor (23). Analysis of AREG depleted mice revealed an essential role of AREG in mammary epithelial development (23, 33). AREG overexpression has been frequently observed in colon, breast, prostate, pancreas, lung, ovary cancer as well as in squamous cell carcinomas and myeloma cells (34-40). AREG can promote its own expression through the activation of EGFR in a positive feedback AREG autocrine loop (21). Overexpression of AREG in MCF10A cell line leads to the increased cell motility and invasion (21).
Even though AREG function has been well described (21, 23, 41) and the AREG promoter was characterized (15), a full understanding of AREG transcriptional control remains undetermined. Only a few transcription factors have been shown to regulate AREG transcription, among them Sp1 binding at the positions −3/−8 (15), CREB binding at the positions −57/−64 (42), WT1 zinc finger transcription factor binding at the positions −68/−84 of the AREG promoter and activating transcription (43). 17β-estradiol has been shown to induce AREG mRNA expression through activation of estrogen receptor (44) though no estrogen response element is characterized on this promoter. None of these known enhancers correlates with the BRCA1 elements that we mapped, using the reporter assay, on the AREG promoter.
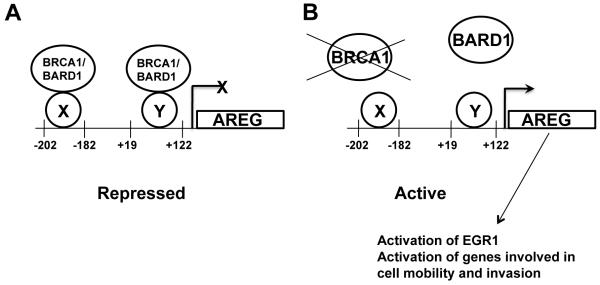
BRCA1 has been reported to regulate transcription on a variety of promoters, however on its own binds DNA only in a sequence non-specific manner (25, 26). BRCA1 can regulate transcription by interacting with other transcription factors which can bind to specific sites on DNA and in this way BRCA1 alters the function of these transcription factors (28, 45). We have identified two independent BRCA1-response elements on the AREG promoter located at −202/−182 and +19/+122 positions. We hypothesize that since BRCA1 is a transcriptional repressor of the AREG promoter, the BRCA1 protein, in a complex with BARD1, binds to a sequence-specific transcription factor/activator bound to either of the identified DNA elements, and represses AREG expression (Figure 5). Identification of the BRCA1 binding sequence-specific transcription factor(s) is the subject of continuing investigation.
Figure 5. A model suggesting how BRCA1 loss of function might lead to AREG up-regulation and contribute to cancer development and progression.
A. A model illustrating BRCA1-dependent AREG regulation with intact BRCA1.
B. A model illustrating BRCA1-dependent AREG regulation when BRCA1 protein is depleted.
AREG is often overexpressed in breast cancer (46), moreover AREG expression in breast cancer samples does not correlate with ERα (46, 47) and AREG and EGFR expression in invasive carcinomas is correlated with the absence of estrogen receptor (48). Tumors resulting from BRCA1 mutations frequently are ER-independent. The ER-independent invasive ductal carcinoma cell line SUM149PT has been characterized by low BRCA1 transcript due to the 2288delT BRCA1 inactivating mutation (27) and has an increased AREG mRNA and protein level (21). Our results suggest that there is a direct link to explain this prior observation. Our experiments show for the first time that BRCA1 depletion leads to AREG up-regulation in the HeLa cell line, and BRCA1 expression in SUM149PT cell line leads to reduction in the amount of AREG protein, supporting the idea that BRCA1 controls AREG expression. Moreover, AREG overexpression in MCF10A leads to activation of the genes involved in cell mobility and invasion (21) which allows us to suggest that BRCA1 loss of function can contribute to cancer development and progression through AREG up-regulation (Figure 5). Taken together, our data suggest that a part of the tumorigenic phenotype of breast cancer might be explained via loss of BRCA1 repression of AREG gene expression.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant CA90281 (to J.D.P.) and by start up funds from the Ohio State University Comprehensive Cancer Center and by CRUK Grant C423/A7640 (to Prof. Sibylle Mittnacht, Institute of Cancer Research, London, UK). We thank Alice Smith for help with qPCR (Institute of Cancer Research, London, UK).
Literature
- 1.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Couch FJ. Genetic epidemiology of BRCA1. Cancer Biol Ther. 2004;3:509–14. doi: 10.4161/cbt.3.6.840. [DOI] [PubMed] [Google Scholar]
- 3.Magdinier F, Ribieras S, Lenoir GM, Frappart L, Dante R. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene. 1998;17:3169–76. doi: 10.1038/sj.onc.1202248. [DOI] [PubMed] [Google Scholar]
- 4.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 5.Starita LM, Parvin JD. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr Opin Cell Biol. 2003;15:345–50. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 6.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 7.Parvin JD, Sankaran S. The BRCA1 E3 ubiquitin ligase controls centrosome dynamics. Cell Cycle. 2006;5:1946–50. doi: 10.4161/cc.5.17.3208. [DOI] [PubMed] [Google Scholar]
- 8.Chapman MS, Verma IM. Transcriptional activation by BRCA1. Nature. 1996;382:678–9. doi: 10.1038/382678a0. [DOI] [PubMed] [Google Scholar]
- 9.Scully R, Anderson SF, Chao DM, et al. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci U S A. 1997;94:5605–10. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–6. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 11.Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–53. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwitz AA, Sankaran S, Parvin JD. Direct stimulation of transcription initiation by BRCA1 requires both its amino and carboxyl termini. J Biol Chem. 2006;281:8317–20. doi: 10.1074/jbc.C500475200. [DOI] [PubMed] [Google Scholar]
- 13.Horwitz AA, Affar el B, Heine GF, Shi Y, Parvin JD. A mechanism for transcriptional repression dependent on the BRCA1 E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2007;104:6614–9. doi: 10.1073/pnas.0610481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sui G, Soohoo C, Affar el B, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:5515–20. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plowman GD, Green JM, McDonald VL, et al. The amphiregulin gene encodes a novel epidermal growth factor-related protein with tumor-inhibitory activity. Mol Cell Biol. 1990;10:1969–81. doi: 10.1128/mcb.10.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–91. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, Weaver Z, Linke SP, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–95. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 19.Scully R, Ganesan S, Brown M, et al. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–6. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- 20.Starita LM, Machida Y, Sankaran S, et al. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol Cell Biol. 2004;24:8457–66. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281:37728–37. doi: 10.1074/jbc.M606532200. [DOI] [PubMed] [Google Scholar]
- 22.Bieche I, Lerebours F, Tozlu S, Espie M, Marty M, Lidereau R. Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature. Clin Cancer Res. 2004;10:6789–95. doi: 10.1158/1078-0432.CCR-04-0306. [DOI] [PubMed] [Google Scholar]
- 23.Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z. Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development. 2005;132:3923–33. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigismund S, Woelk T, Puri C, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A. 2005;102:2760–5. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Direct DNA binding by Brca1. Proc Natl Acad Sci U S A. 2001;98:6086–91. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simons AM, Horwitz AA, Starita LM, et al. BRCA1 DNA-binding activity is stimulated by BARD1. Cancer Res. 2006;66:2012–8. doi: 10.1158/0008-5472.CAN-05-3296. [DOI] [PubMed] [Google Scholar]
- 27.Elstrodt F, Hollestelle A, Nagel JH, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–5. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 28.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–63. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 29.Furuta S, Wang JM, Wei S, et al. Removal of BRCA1/CtIP/ZBRK1 repressor complex on ANG1 promoter leads to accelerated mammary tumor growth contributed by prominent vasculature. Cancer Cell. 2006;10:13–24. doi: 10.1016/j.ccr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Kimura H. Schwannoma-derived growth factor must be transported into the nucleus to exert its mitogenic activity. Proc Natl Acad Sci U S A. 1993;90:2165–9. doi: 10.1073/pnas.90.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wollmann W, Goodman ML, Bhat-Nakshatri P, et al. The macrophage inhibitory cytokine integrates AKT/PKB and MAP kinase signaling pathways in breast cancer cells. Carcinogenesis. 2005;26:900–7. doi: 10.1093/carcin/bgi031. [DOI] [PubMed] [Google Scholar]
- 32.Welsh JB, Sapinoso LM, Kern SG, et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci U S A. 2003;100:3410–5. doi: 10.1073/pnas.0530278100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luetteke NC, Qiu TH, Fenton SE, et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–50. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 34.Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Salomon DS, Normanno N, Ciardiello F, Brandt R, Shoyab M, Todaro GJ. The role of amphiregulin in breast cancer. Breast Cancer Res Treat. 1995;33:103–14. doi: 10.1007/BF00682718. [DOI] [PubMed] [Google Scholar]
- 36.Bostwick DG, Qian J, Maihle NJ. Amphiregulin expression in prostatic intraepithelial neoplasia and adenocarcinoma: a study of 93 cases. Prostate. 2004;58:164–8. doi: 10.1002/pros.10322. [DOI] [PubMed] [Google Scholar]
- 37.Ebert M, Yokoyama M, Kobrin MS, et al. Induction and expression of amphiregulin in human pancreatic cancer. Cancer Res. 1994;54:3959–62. [PubMed] [Google Scholar]
- 38.Fontanini G, De Laurentiis M, Vignati S, et al. Evaluation of epidermal growth factor-related growth factors and receptors and of neoangiogenesis in completely resected stage I-IIIA non-small-cell lung cancer: amphiregulin and microvessel count are independent prognostic indicators of survival. Clin Cancer Res. 1998;4:241–9. [PubMed] [Google Scholar]
- 39.Tsai ST, Yang KY, Jin YT, Lin YC, Chang MT, Wu LW. Amphiregulin as a tumor promoter for oral squamous cell carcinoma: involvement of cyclooxygenase 2. Oral Oncol. 2006;42:381–90. doi: 10.1016/j.oraloncology.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 40.D’Antonio A, Losito S, Pignata S, et al. Transforming growth factor alpha, amphiregulin and cripto-1 are frequently expressed in advanced human ovarian carcinomas. Int J Oncol. 2002;21:941–8. [PubMed] [Google Scholar]
- 41.Berasain C, Castillo J, Perugorria MJ, Prieto J, Avila MA. Amphiregulin: a new growth factor in hepatocarcinogenesis. Cancer Lett. 2007;254:30–41. doi: 10.1016/j.canlet.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 42.O’Reilly SM, Leonard MO, Kieran N, et al. Hypoxia induces epithelial amphiregulin gene expression in a CREB-dependent manner. Am J Physiol Cell Physiol. 2006;290:C592–600. doi: 10.1152/ajpcell.00278.2005. [DOI] [PubMed] [Google Scholar]
- 43.Lee SB, Huang K, Palmer R, et al. The Wilms tumor suppressor WT1 encodes a transcriptional activator of amphiregulin. Cell. 1999;98:663–73. doi: 10.1016/s0092-8674(00)80053-7. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Lacaci I, Saceda M, Plowman GD, et al. Estrogen and phorbol esters regulate amphiregulin expression by two separate mechanisms in human breast cancer cell lines. Endocrinology. 1995;136:3983–92. doi: 10.1210/endo.136.9.7649107. [DOI] [PubMed] [Google Scholar]
- 45.Benezra M, Chevallier N, Morrison DJ, MacLachlan TK, El-Deiry WS, Licht JD. BRCA1 augments transcription by the NF-kappaB transcription factor by binding to the Rel domain of the p65/RelA subunit. J Biol Chem. 2003;278:26333–41. doi: 10.1074/jbc.M303076200. [DOI] [PubMed] [Google Scholar]
- 46.Qi CF, Liscia DS, Normanno N, et al. Expression of transforming growth factor alpha, amphiregulin and cripto-1 in human breast carcinomas. Br J Cancer. 1994;69:903–10. doi: 10.1038/bjc.1994.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LeJeune S, Leek R, Horak E, Plowman G, Greenall M, Harris AL. Amphiregulin, epidermal growth factor receptor, and estrogen receptor expression in human primary breast cancer. Cancer Res. 1993;53:3597–602. [PubMed] [Google Scholar]
- 48.Ma L, de Roquancourt A, Bertheau P, et al. Expression of amphiregulin and epidermal growth factor receptor in human breast cancer: analysis of autocriny and stromal-epithelial interactions. J Pathol. 2001;194:413–9. doi: 10.1002/path.902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.