Abstract
Trimethylation of histone H3 on Lys 27 (H3K27me3) is key for cell fate regulation. The H3K27me3 demethylase UTX functions in development and tumor suppression with undefined mechanisms. Here, genome-wide chromatin occupancy analysis of UTX and associated histone modifications reveals distinct classes of UTX target genes, including genes encoding Retinoblastoma (RB)-binding proteins. UTX removes H3K27me3 and maintains expression of several RB-binding proteins, enabling cell cycle arrest. Genetic interactions in mammalian cells and Caenorhabditis elegans show that UTX regulates cell fates via RB-dependent pathways. Thus, UTX defines an evolutionarily conserved mechanism to enable coordinate transcription of a RB network in cell fate control.
Keywords: Gene regulation, epigenetics, chromatin, histone demethylation, cancer
The N termini of histone tails are subject to a wide array of post-translational modifications that determine chromatin state and transcriptional activity (for review, see Rando and Chang 2009). Trimethylation of histone H3 at Lys 27 (H3K27me3) is associated with epigenetic gene silencing by the Polycomb group (PcG) proteins, which play key roles in developmental fate decisions and cell growth control (Sparmann and van Lohuizen 2006). H3K27me3 has been shown to inhibit the transcription of tumor suppressor genes (Bracken et al. 2007; Kotake et al. 2007). The H3K27 methylase EZH2 is amplified or overexpressed in human breast, prostate, and melanocytic tumors, and is a strong predictor of cancer progression (Varambally et al. 2002; Bracken et al. 2003; Kleer et al. 2003). The recent discovery of two H3K27me3 demethylases, UTX and JMJD3, revealed that removal of H3K27me3 is also under active control. UTX is required for proper activation of HOX genes and anterior–posterior identity (Agger et al. 2007; Lan et al. 2007; Lee et al. 2007), while JMJD3 can drive post-mitotic differentiation and oncogene-induced senescence (Jepsen et al. 2007; Sen et al. 2008; Agger et al. 2009; Barradas et al. 2009). Biallelic somatic mutations of UTX in human cancers suggest UTX as a tumor suppressor gene (van Haaften et al. 2009), but the mechanism of UTX-mediated tumor suppression is still unclear.
UTX is associated with the Mixed Lineage Leukemia 2–4 (MLL2–4) H3K4 methylase complexes, members of trithorax group proteins involved in gene activation (Issaeva et al. 2007; Lee et al. 2007). UTX occupancy in the HOX loci is concentrated near promoters, and can occur focally within either H3K4me2 or H3K27me3 chromatin domains (Lan et al. 2007). Thus, UTX may not simply associate with all sites of H3K4 or H3K27 methylation, but instead constitutes a dynamic and independent layer of epigenetic regulation. The full set of genes targeted by UTX is not known, nor has the relationship of UTX occupancy to underlying histone methylation states been fully explored. Here we describe the chromatin occupancy of UTX genome-wide, and reveal an unanticipated role for UTX to prevent the epigenetic silencing of the Retinoblastoma (RB) pathway and mediate cell fate control that is conserved from humans to Caenorhabditis elegans.
Results and Discussion
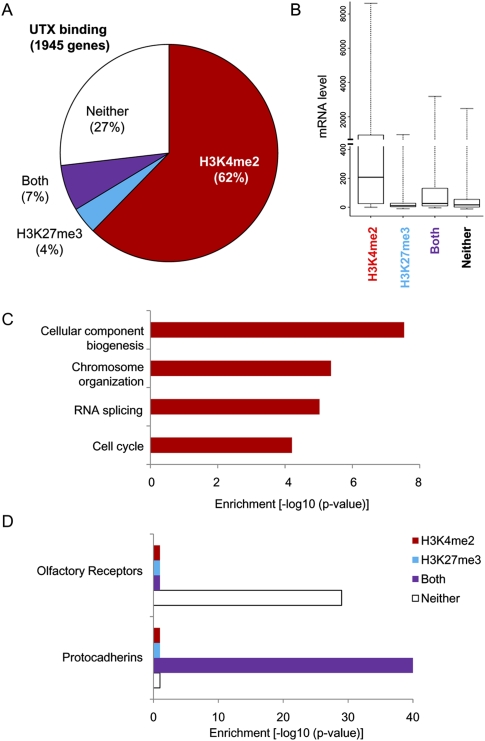
To explore the roles of UTX in transcriptional regulation, we mapped the chromatin occupancy of UTX, H3K4me2, and H3K27me3 in primary human fibroblasts using chromatin immunoprecipitation (ChIP) followed by hybridization to tiling arrays interrogating >30,000 human promoters (Materials and Methods). UTX occupied 1945 promoters, most often upstream of transcriptional start sites, and these UTX target genes can be organized into distinct functional classes based on the associated histone methylation patterns (Fig. 1A; Supplemental Table 1). The majority of UTX target genes (62%) are enriched for univalent H3K4me2, consistent with the idea that UTX is associated with the MLL2–4 complex involved in gene activation (Issaeva et al. 2007; Lee et al. 2007). UTX-occupied genes represent less than a quarter of H3K4me2-enriched genes in the genome, which is also consistent with the division of labor among more than eight known H3K4 methylases (Ruthenburg et al. 2007). UTX-occupied promoters are significantly underrepresented for co-occupancy with H3K27me3 (211 observed vs. 338 expected by chance alone, P = 6.9 × 10−17, hypergeometric distribution), suggesting that many of the UTX occupancy events lead to H3K27me3 demethylation. Twenty-seven percent of UTX target genes exhibit neither H3K4 nor H3K27 methylation, and smaller percentages of UTX targets demonstrate both histone marks (7%) or only H3K27me3 (4%). Issaeva et al. (2007) found that MLL2 is required for UTX occupancy of several, but not all, target genes, and our results expand the set of genes potentially targeted by UTX in manners independent of H3K4 methylation. These histone marks also reflect the transcriptional state of the UTX target genes as revealed by mRNA profiling: H3K4me2-occupied genes show a broad range of mRNA levels, while the three other classes of genes are largely silent (Fig. 1B).
Figure 1.
Genome-wide occupancy of UTX and histone modifications define distinct classes of UTX target genes. (A) Distribution of UTX occupancy in association with H3K27 and/or H3K4 methylation. (B) UTX target genes with H3K4 methylation tend to be expressed, whereas other classes are silent. Box plot showing 25th, 50th, and 75th percentile expression levels for different target gene categories. Whiskers show the 2.5 and 97.5 percentiles. (C) Gene Ontology terms enriched in genes with occupancy of UTX and H3K4me2. (D) Distinct gene cluster families are enriched for UTX occupancy and bivalent histone H3 modifications (protocadherins) or neither histone modification (olfactory receptors).
Gene Ontology analysis of UTX target genes revealed striking enrichment of distinct functional classes based on associated histone marks. UTX target genes with H3K4me2 demonstrate enrichment in broad classes of genes representing diverse cellular function, including chromosome organization and biogenesis, RNA splicing, and cell cycle (all reported enrichments have Benjamini-Hochberg false discovery rate [FDR] < 0.05) (Fig 1C; Supplemental Table 2). In contrast, protocadherin genes are strikingly overrepresented among UTX target genes with both H3K4me2 and H3K27me3 (33 genes observed vs. one expected by chance alone, P = 3.5 × 10−40), while genes encoding olfactory receptors are very significantly overrepresented among UTX target genes with neither H3K4 nor H3K27 methylation (59 genes observed vs. 10 expected by chance alone, P = 7.3 × 10−29) (Fig. 1D; Supplemental Fig. 1). Both protocadherin and olfactory receptor genes are organized into clusters where dozens of family members are grouped, and intricate regulatory mechanisms ensure that only one or a few select family members are expressed in each cell (Morishita and Yagi 2007; Olender et al. 2008). Finally, UTX target genes with only H3K27me3 are enriched for genes involved in developmental processes (including HOX genes), consistent with the functional class of genes known to be targeted by H3K27me3 (Bracken et al. 2006). Together, these data suggest that the majority of UTX target genes are transcriptionally active, as evidenced by H3K4 methylation on their chromatin and mRNA accumulation. The main exception to this rule is gene clusters such as HOX genes, protocadherins, and olfactory receptors, which are marked by UTX but also by silencing marks such as H3K27me3 or absence of H3K4 methylation.
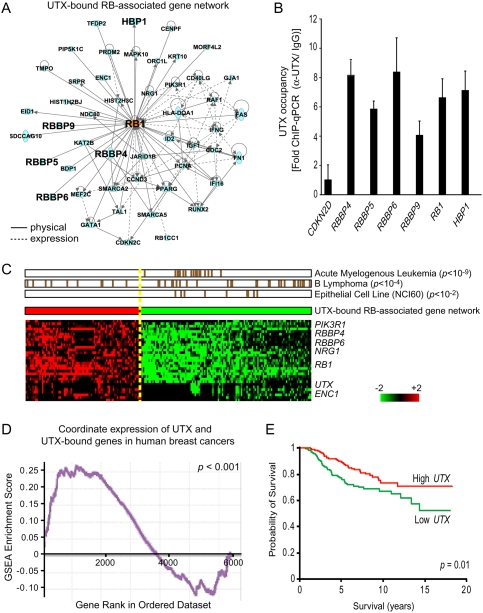
To better understand potential functional roles of UTX, UTX-bound genes were organized by Ingenuity Pathway Analysis, a bibliomic approach that organizes sets of genes into structured networks and nominates biological contexts for gene function (Calvano et al. 2005). Notably, and consistent with the independent Gene Ontology analysis, the most significant gene network among UTX-bound genes is involved in cell cycle, and this network of 49 genes is centered on the tumor suppressor gene RB, a key inhibitor of G1–S transition and enforcer of terminal differentiation (Fig. 2A; for review, see Burkhart and Sage 2008). UTX bound directly to multiple genes that encode proteins that interact physically with pRB and function in the RB pathway, including RB1 itself, HBP1, and RB-binding proteins (RBBP) 4, 5, 6, and 9; all of these genes were also enriched for H3K4me2 occupancy (Supplemental Table 1). ChIP followed by quantitative PCR (qPCR) confirmed UTX occupancy of these RB pathway genes (Fig. 2B). Several of these pRB-binding proteins are individually required for RB function, including HBP1, a high-mobility group protein required for RB-dependent cell cycle arrest (Tevosian et al. 1997). These data suggest the hypothesis that UTX may enable the coordinate transcription of genes encoding the pRB complex.
Figure 2.
UTX regulates many RB-binding proteins, which are coordinately decreased in expression in human cancers. (A) Top network of genes occupied by UTX organized by Ingenuity Pathway Analysis. (Center) Forty-nine UTX-occupied genes are known to be associated with RB. The solid line indicates known physical interaction, while the dashed line indicates known interaction by regulation of expression. Genes of special interest are in large font. (B) Validation of UTX occupancy of RB pathway genes by ChIP-qPCR. CDKN2D is not a UTX target gene and served as negative control. (C) The UTX–RB gene network is coordinately repressed in human cancers. We interrogated a compendium of 1973 microarrays representing 22 human tumor types and diverse normal controls for coordinate regulation of the genes occupied by UTX. In the bottom matrix, each column is a sample showing significant coordinate induction (red) or repression (green) of UTX network genes (P < 0.05, FDR < 0.05); each row is a UTX network gene. The middle bar shows the average level of activity of the UTX–RB network in each sample. The top matrix displays whether the coordinate induction or repression of UTX–RB network is enriched for specific clinical annotations. Each brown hatch mark indicates a sample with the indicated annotation; each row is an enriched annotation (P < 0.05, FDR < 0.05, hypergeometric distribution) that shows selective deactivation of the network in cancers relative to their normal counterparts. (D) RB pathway gene expression is correlated with UTX. Genes are sorted and ranked based on Pearson correlation to UTX. Gene set enrichment analysis of the Pearson correlation of UTX-bound genes in 295 human breast cancer patients shows that UTX-bound genes are significantly positively correlated with UTX expression (P < 0.001). (E) UTX expression level is a predictor of patient survival. Kaplan-Meier analysis of patients with high versus low UTX expression is shown for the 295 breast cancer patients (160 patients have “high” UTX expression; 135 patients have “low” UTX expression).
As a first test of this idea, we examined the coordinate activation or deactivation of UTX and the 49 UTX-occupied RB gene networks in a compendium of 1973 microarrays, comprised of 22 human tumor types and their normal counterparts, using the gene module map method (Segal et al. 2004). For each microarray, we determined whether these 50 genes were significantly coinduced or corepressed (P < 0.05, FDR < 0.05), and next examined whether those microarrays demonstrating coordinate UTX target gene regulation were systematically enriched for specific clinical annotations (P < 0.05, FDR < 0.05). As shown in Figure 2C, UTX and its targets were indeed coordinately expressed in many instances. Notably, while coordinate expression of UTX and its targets occurred in diverse instances, and therefore were not enriched for any specific clinical annotation, coordinate repression of UTX and its targets were enriched in cancers relative to their normal counterparts, including human acute myelogenous leukemia (P < 10−9) and human B-cell lymphomas (P < 10−4), and among epithelial cell lines in the NCI60 collection of tumor cell lines (P < 10−2). In human primary breast cancer where high EZH2 expression is a known poor prognosis factor (Kleer et al. 2003), UTX target genes including RB1, HBP1, and several RBBPs are significantly and positively correlated with UTX expression (P < 0.001) (Fig. 2D), and low UTX expression is a significant predictor of subsequent patient death (P = 0.01) (Fig. 2E). Thus, loss of UTX expression may be a rather common occurrence in human cancers, is linked to transcriptional silencing of the RB pathway, and has prognostic significance for cancer progression.
Interestingly, the prognostic impact of the UTX level is conditional on EZH2 status: In tumors with a low EZH2 level, low UTX has a substantially larger impact on subsequent patient survival and metastasis, and, conversely, the EZH2 level has a large impact on patient survival and metastasis in tumors with high UTX expression. But in tumors that have EZH2 overexpression, the UTX level is not a significant predictor of patient survival, nor is the EZH2 level significant in tumors with low UTX expression (Supplemental Fig. 2). These data are consistent with the idea that excess EZH2 or lack of UTX comprises redundant mechanisms to dysregulate H3K27me3 levels in cancer.
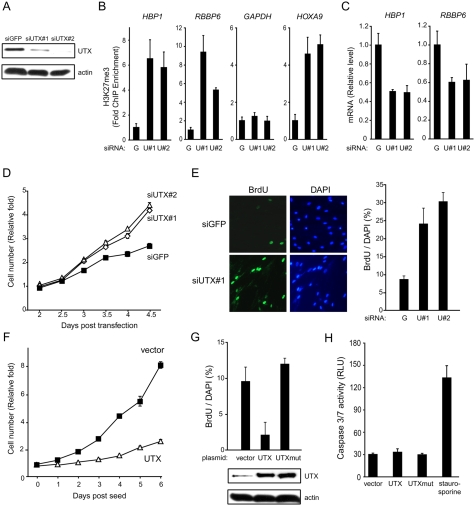
Next, we directly tested the effect of UTX on the chromatin states and transcription of RB pathway genes and cell cycle progression. Acute depletion of UTX by two independent siRNAs in primary human fibroblasts led to increased H3K27me3 level on HBP1 and RBBP6 promoters (Fig. 3A,B), and their cognate mRNA levels were decreased correspondingly (Fig. 3C). The gain of H3K27me3 on pRB pathway promoters was as strong as or stronger than that seen on HOXA9, a known UTX target gene (Lan et al. 2007), but H3K27me3 did not increase on GAPDH, a gene not targeted by UTX (Fig. 3B). Rb1 also showed little change in H3K27me3 or mRNA levels in the same experiments. Thus, UTX is continually required to prevent H3K27me3 accumulation on select genes in the pRB network. Moreover, acute depletion of UTX caused significantly increased cell proliferation and S-phase entry, as measured by serial cell counts and bromodeoxyuridine (BrdU) incorporation (Fig. 3D,E). Fluorescence-activated cell sorting confirmed a decrease in G1 fraction and corresponding increase in S and G2 fractions of the cell cycle (8% vs. 17% S-phase fraction in GFP vs. UTX-depleted cells, respectively). Conversely, enforced expression of UTX in primary human fibroblasts by retroviral transduction led to cell cycle arrest but not apoptosis, as measured by serial cell counts, BrdU incorporation, and apoptotic caspase activity (Fig. 3F–H). Importantly, the catalytic-inactive mutant UTXH1126A was unable to inhibit BrdU incorporation, indicating that H3K27me3 demethylase activity is required to induce cell cycle arrest (Fig. 3G). Collectively, these data suggest that UTX demethylates H3K27me3 at the promoters of genes encoding pRB complex subunits to enable their coordinate transcription, and UTX is necessary and sufficient to mediate cell cycle arrest of primary human cells.
Figure 3.
UTX is necessary and sufficient to mediate cell cycle arrest of primary human fibroblasts. (A) Depletion of UTX protein by siRNAs. (B) UTX depletion causes increased H3K27me3 levels at UTX-occupied sites of target genes. ChIP-qPCR of H3K27me3 at the indicated promoters is shown; HOXA9 and GAPDH served as positive and negative controls, respectively. (C) UTX depletion causes decreased mRNA levels of HBP1 and RBBP6 as assayed by qRT–PCR. (D) UTX depletion increases cell proliferation. Serial cell counts of fibroblasts transfected with one of two siRNAs targeting UTX versus control siRNA targeting GFP. (E) Increased S-phase entry of UTX-depleted cells. BrdU immunofluorescence (left) and quantification (right) showing increase percentage of S-phase cells in population versus control. n = 1000+ for each condition. (F) Overexpression of UTX results in decreased cell proliferation. Serial cell counts of fibroblasts transduced with UTX or puromycin-resistant vector alone. (G) Catalytic activity of UTX is required to induce cell cycle arrest, as measured by BrdU incorporation. (UTXmut) UTXH1126A. n = 1000+ in each condition. (H) UTX overexpression does not induce apoptosis. Staurosporine treatment served as positive control. Mean ± SD is shown in all bar graphs.
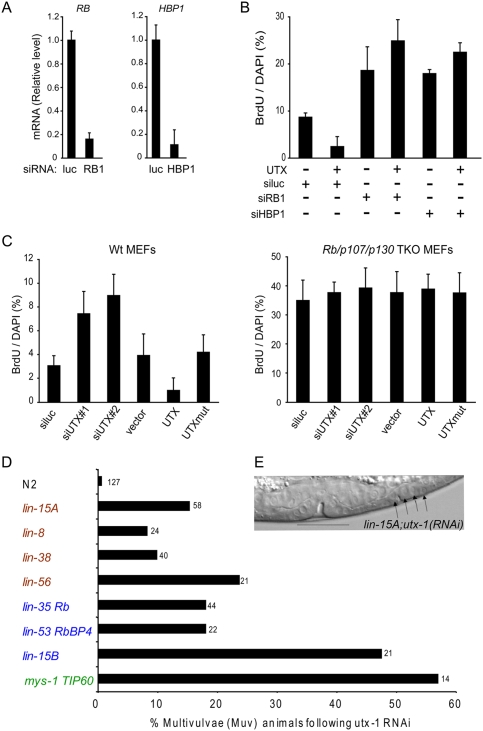
Our model predicts that RB pathway genes are required for UTX-mediated cell cycle arrest. Overexpression of UTX led to cell cycle arrest as measured by BrdU incorporation, but concomitant depletion of either RB or HBP1 reversed the cell cycle arrest (Fig. 4A,B). Indeed, RB- or HBP1-depleted cells cycle more than wild-type cells, and become insensitive to UTX overexpression. We also employed a genetic approach to examine the epistasis between the UTX and RB pathways. In the mouse, Rb and its family members p107 and p130 exhibit cross-regulation and compensation, but triple knockout (TKO) of all three Rb family members fully inactivates the RB pathway (Sage et al. 2003). In the TKO setting, we hypothesized that loss of UTX can no longer have an additive effect on relaxing the G1 checkpoint, and overexpression of UTX should no longer lead to cell cycle arrest. Indeed, in wild-type mouse embryonic fibroblasts (MEFs), two independent siRNAs targeting mouse Utx led to ectopic cell proliferation, and enforced expression of UTX, but not its catalytic mutant, led to cell cycle arrest. But in TKO MEFs, neither Utx depletion nor overexpression had an appreciable effect on the cell cycle (Fig. 4C). In sum, these data suggest that UTX controls cell cycle arrest by activating a RB-dependent pathway.
Figure 4.
Genetic interaction between UTX, RB, and RB-binding proteins. (A) Depletion of RB1 or HBP1 by siRNAs. (siluc) Control siRNA targeting luciferase. (B) RB or HBP1 depletion reverses UTX-mediated cell cycle arrest. Quantifications of percentage of cells in S phase as measured by BrdU incorporation is shown. n = 1000+. (C) UTX modulation of cell cycle progression in mouse embryonic fibroblasts (MEFs) requires the Rb family members. (Left) Wild-type MEFs. (Right) Rb, p107, and p130 triple knockout (TKO) MEFs. (D) C. elegans UTX-1 is synMuv. RNAi of utx-1 was performed on eight characteristic synMuv strains and N2 wild-type worms. Wild-type worms were exposed to utx-1 RNAi and showed a very weak Muv phenotype (<1% penetrance). Strains representing class A (lin-15A, lin-8, lin-38, and lin-56; genes labeled in red), class B (lin-35 Rb, lin-53 RbBP4, and lin-15B; genes labeled in blue), and class C (mys-1 TIP60; gene label in green) were used to test for UTX-1 synMuv activity. All of these strains present a normal vulva (Ferguson and Horvitz 1989), except for mys-1, which produces <10% Muv (Ceol and Horvitz 2004). Penetrance of the synMuv interactions vary between 8% and 24% for class A and 18% and 48% for class B, and is 57% for class C. Overall, utx-1 genetically interacts with classes A, B, and C, and behaves similarly to set-16 mll (K Fisher and GB Poulin, pers. comm.). The number of worms analyzed is indicated above each bar. (E) Photomicrograph of representative synMuv phenotype in an early L4 stage worm. Arrows indicate extra cells adopting the vulval fate. The normal vulva is underlined.
In addition to cell cycle regulation, the RB pathway also controls lineage-specific differentiation. In the classic genetic model C. elegans, genes encoding RB and RB-binding proteins were first discovered for their roles in controlling vulval cell fate (for review, see Fay and Yochem 2007). The vulva develops from six equipotent vulval precursor cells (VPCs), in which only three adopt the vulval fate. Animals in which more than three VPCs adopt the vulvlal fate are defined as Multivulvae (Muv). The synthetic Multivulvae (synMuv) genes are a group of genes that act redundantly to prevent extra adoption of the vulval fate. The synMuv genes can be classified into three classes: A, B (containing most genes encoding RB and RBBPs), and C. Mutants of each class present a normal vulva (an exception is class C) (see the legend for Fig. 4), but double mutants between classes are defined as synMuv. We reasoned that, if worm UTX-1 functions in the RB pathway, depletion of UTX-1 should cause the synMuv phenotype in appropriate genetic backgrounds.
We inactivated UTX-1 in wild-type animals and eight characteristic synMuv backgrounds representing classes A, B, and C (Fig. 4D,E). Consistent with being synMuv, UTX-1 RNAi in wild-type animals had no vulvae abnormalities, but caused Muv in all synMuv backgrounds. This promiscuous synMuv activity is anticipated because lin-53/RBBP4, a presumptive UTX target gene, is also synMuv A and B (Solari and Ahringer 2000), and set-16, encoding a C. elegans MLL homolog, is also synMuv with classes A, B, and C (K Fisher and GB Poulin, pers. comm.). The promiscuous synMuv activity of UTX-1 is consistent with UTX-1 regulation of Rb, Rb-binding proteins, and likely additional targets that are in the class A and C group of synMuv genes. Taken together, these results suggest that UTX regulation of RB and RB-binding proteins occurs in vivo, can affect developmental fate decisions, and is conserved from worms to humans.
In this study, we identify promoters occupied by UTX genome-wide, and reveal RB-binding proteins as key target genes mediating UTX-dependent cell cycle arrest and fate decision. Our results suggest that UTX demethylates H3K27me3 at the promoters of genes encoding pRB complex subunits to enable their coordinate transcription, and loss of UTX may lead to coordinate silencing of the RB network in cancer. UTX-mediated cell cycle arrest requires its H3K27me3 demethylase activity, and the prognostic impact of UTX expression is conditional on EZH2, suggesting that H3K27me3 demethylation is an important readout of UTX activity. Nonetheless, the genome-wide colocalization of UTX with H3K4 methylation and genetic interaction profiles in C. elegans paralleling SET-16/MLL strongly suggest that UTX is also linked with H3K4 methylation.
The two H3K27 demethylases UTX and JMJD3 appear to have different roles regulating cell proliferation. JMJD3, but not UTX, is transcriptionally induced by oncogenic Ras to activate expression of the CDKN2A cyclin-dependent kinase inhibitor (Agger et al. 2009; Barradas et al. 2009). Consistently, UTX depletion provides normal cells with an immediate proliferative advantage (Fig. 3), whereas JMJD3 depletion does not affect cell proliferation until a senescence checkpoint is engaged (Agger et al. 2009; Barradas et al. 2009). These findings imply a division of labor among H3K27 demethylases in cell cycle control. UTX transcript level is constant through the cell cycle (Supplemental Fig. 3A), but is induced during wound healing (Shaw and Martin 2009); in contrast, EZH2 is induced in S phase (Bracken et al. 2003). UTX occupancy of RB network genes also differs in embryonic stem cells (Supplemental Fig. 3B). While our results strongly link UTX to the RB network, how the UTX–RB network may be modulated during the cell cycle or differentiation should be investigated in future studies. In this regard, vulval fate determination in C. elegans requires the interplay of receptor tyrosine kinase signaling, Notch signaling, and chromatin regulation (Fay and Yochem 2007). Our identification of UTX as a synMuv gene identifies a genetically tractable system to dissect the regulation of UTX activity.
Among the many UTX target genes, HBP1 emerges as a key effector of UTX. HBP1 was initially discovered as a HMG-box protein that physically interacted with pRB and can enforce cell cycle arrest (Tevosian et al. 1997). Somatic mutations and loss of expression of HBP1 in human cancers indicate that HBP1 is also a tumor suppressor gene (Paulson et al. 2007). UTX occupies the HBP1 promoter and is required to prevent H3K27me3 and silencing of HBP1, allowing HBP1 to accumulate and work with pRB. Further, cell cycle arrest induced by UTX can be suppressed by concomitant inactivation of HBP1. Our data thus implicate a UTX–RB–HBP1 pathway that functionally links three human tumor suppressor genes. Drosophila Utx is also a suppressor of tissue overgrowth that modulates the fly Rb pathway (H-M Herz, LD Madden, Z Chen, C Bolduc, E Buff, R Gupta, R Davuluri, A Shilatifard, IK Hariharan, and A Bergmann, in prep.). Together with our studies in mammalian cells and C. elegans, we suggest that the UTX–RB connection is evolutionarily conserved and fundamental to metazoan growth control.
Materials and methods
ChIP–chip and data analysis
ChIP with affinity-purified UTX antiserum (Agger et al. 2007; Issaeva et al. 2007; Lan et al. 2007) and H3K27me3 and H3K4me2 (Rinn et al. 2007) from human fibroblasts were as described and were hybridized to arrays tiling human promoters (Roche Nimblegen). Peak calling (Johnson et al. 2008), Gene Ontology analysis (Lan et al. 2007), Ingenuity Pathway Analysis (Calvano et al. 2005), gene module map (Segal et al. 2004), and GSEA (Subramanian et al. 2005) were as described. qPCR primers are listed in Supplemental Table 3. Array data are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), GSE 16221.
Cell proliferation assays
Primary human or mouse fibroblasts were transfected with siRNAs or plasmids as in Lan et al. (2007), and were scored for BrdU incorporation as described (Liu et al. 2007) or for viable cells using MTT assay (Roche).
C. elegans RNAi
N2, lin-15A(n767), lin-8(n111), lin-38(n751), lin-56(n2728), lin-35(n745), lin-53(n833), lin-15B(n744), and mys-1(n4075)/nT1[qIs51] strains were fed UTX-1 bacterial RNAi clone and scored for Muv phenotype using DIC microscopy.
Detailed information is available in the Supplemental Material.
Acknowledgments
We thank E. Canaani for the gift of anti-UTX antibody; J. Sage and D. Burkhart for TKO MEFs; A. Bergmann for communicating unpublished information; and J. Sage, P. Khavari, and Seung Kim for input. This work was supported by grants from the California Institute for Regenerative Medicine (RN1-00529-1), the National Cancer Institute (R01-CA118487 to Y.S. and R01-CA118750 to H.Y.C.), and the American Cancer Society (RSG 07-084-01-MGO). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1882610.
Supplemental material is available at http://www.genesdev.org.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A–ARF locus in response to oncogene- and stress-induced senescence. Genes & Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes & Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- Fay DS, Yochem J. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol. 2007;306:1–9. doi: 10.1016/j.ydbio.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. Systematic evaluation of variability in ChIP–chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes & Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Liu H, Adler AS, Segal E, Chang HY. A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 2007;3:e91. doi: 10.1371/journal.pgen.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Yagi T. Protocadherin family: Diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Olender T, Lancet D, Nebert DW. Update on the olfactory receptor (OR) gene superfamily. Hum Genomics. 2008;3:87–97. doi: 10.1186/1479-7364-3-1-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, et al. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67:6136–6145. doi: 10.1158/0008-5472.CAN-07-0567. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes & Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: Loss of Polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F, Ahringer J. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol. 2000;10:223–226. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, Shih HH, Mendelson KG, Sheppard KA, Paulson KE, Yee AS. HBP1: A HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes & Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]