Abstract
A balanced deoxyribonucleotide (dNTP) supply is essential for DNA repair. Here, we found that ribonucleotide reductase (RNR) subunits RRM1 and RRM2 accumulated very rapidly at damage sites. RRM1 bound physically to Tip60. Chromatin immunoprecipitation analyses of cells with an I-SceI cassette revealed that RRM1 bound to a damage site in a Tip60-dependent manner. Active RRM1 mutants lacking Tip60 binding failed to rescue an impaired DNA repair in RRM1-depleted G1-phase cells. Inhibition of RNR recruitment by an RRM1 C-terminal fragment sensitized cells to DNA damage. We propose that Tip60-dependent recruitment of RNR plays an essential role in dNTP supply for DNA repair.
Keywords: DNA repair, ribonucleotide reductase, Tip60, dNTPs, genomic instability, DNA double-strand breaks
Maintenance of the optimal intracellular concentrations of deoxyribonucleotides (dNTPs) is critical not only for faithful DNA synthesis during DNA replication and repair, but also for the survival of all organisms. Ribonucleotide reductase (RNR), composed of a tetrameric complex of two large catalytic (RRM1) subunits and two small subunits (RRM2 or 53R2), catalyzes de novo synthesis of dNTPs from the corresponding ribonucleotides (Reichard 1993). This reaction is the rate-limiting process in DNA precursor synthesis and is regulated by multiple complex mechanisms, including transcriptional and subcellular localization regulation of RNR (Nordlund and Reichard 2006). In order to duplicate their chromosomal DNA, mammalian S-phase cells possess 15–20 times more dNTP pools than resting quiescent cells, whereas whole dNTP pools were almost unchanged after DNA damage, suggesting the presence of a unique mechanism that supplies a sufficient quantity of dNTPs at repair sites (Hakansson et al. 2006). DNA synthesis must function properly in both repair and replication (dNTP concentrations in fibroblasts were estimated to be as follows: ∼0.5 μM in G0/G1-phase cells, and ∼10 μM in S-phase cells, given that the average volume of a fibroblast is 3.4 pL) (Imaizumi et al. 1996). Although the amount of dNTPs required for DNA repair is small, their concentration during DNA synthesis is critical because DNA polymerase involved in DNA repair (Kraynov et al. 2000; Johnson et al. 2003) has similar kinetic affinities for dNTPs (∼10 μM) to those involved in DNA replication (∼10 μM) (Dong and Wang 1995). Therefore, the dNTPs might be compartmentalized close to the damage sites during the DNA repair process. In this study, we show that, in mammals, both RRM1 and RRM2 rapidly accumulated at double-strand break (DSB) sites in a Tip60-binding-dependent manner.
Results and Discussion
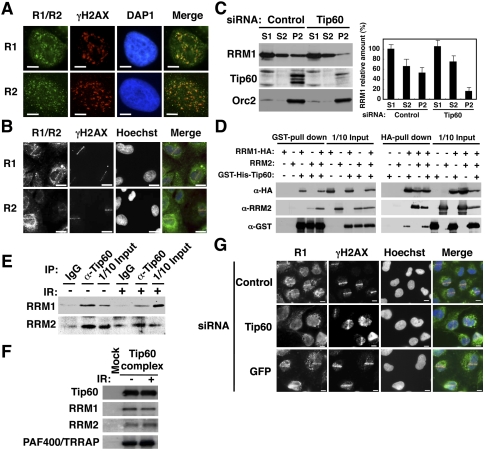
In order to understand the mechanisms by which dNTPs are sufficiently supplied at DNA damage sites in mammals, we first examined changes in the subcellular localization of RRM1 and RRM2 subunits after ionizing irradiation (IR) irradiation. Although both RRM1 and RRM2 predominantly localized in the cytoplasm as reported previously (Pontarin et al. 2008), we also detected trace, but significant, signals of both proteins in chromatin fraction (see Fig. 1C; Supplemental Fig. S4A–D). After removing soluble RNR proteins by detergent extraction, we found that RRM1 and RRM2 proteins formed nuclear foci that colocalized with γH2AX (Fig. 1A). RRM1 nuclear foci were not evident without DNA damage (Supplemental Fig. S1A) or after RRM1 depletion by siRNA (Supplemental Fig. S1B). Ultravioulet A (UVA) microirradiation resulted in the accumulation of RRM1 and RRM2 along microirradiated lines as early as 5 min after treatment (Fig. 1B). These accumulations were also observed when cells were not subjected to detergent extraction or preincubation with BrdU (Supplemental Fig. S2A,B), but were significantly compromised when R1 expression was knockdown by siRNA (Supplemental Figs. S2C, S4B), excluding the possibility that accumulated signals at DSB sites were artifacts during cell-staining processes. These results indicated that RNR, at least in part, was rapidly recruited to DSB sites.
Figure 1.
Tip60-dependent recruitment of RNR at DSB sites. (A) HeLa cells were exposed to IR at 1 Gy, subjected to in situ detergent extraction after 5 min, and immunostained with the indicated antibodies. Bars, 5 μm. (B) GM02063 cells were subjected to UVA microirradiation and immunostained with the indicated antibodies after 5 min. RRM1 or RRM2 and γH2AX signals are shown in green and red, respectively, in merged images. Bars, 10 μm. (C) IR-irradiated HeLa cell lysates treated with the indicated siRNAs were fractionated as described in the Materials and Methods. (Left panels) The fractions were subjected to immunoblotting using the indicated antibodies. (Right panel) The RRM1 bands were quantitated, and the results are presented as percentages of S1 fraction. Data are mean ± standard deviation (n = 3). (D) Sf9 lysates expressing RRM1-HA, RRM2, or GST-His-Tip60 were subjected to GST pull-down or HA pull-down assays using the indicated antibodies. (E) Chromatin fractions from IR- or mock-treated HeLa cells (after 5 min) were solubilized with micrococcal nuclease. The solubilized extracts were immunoprecipitated with anti-Tip60 antibodies or control IgG. The resulting precipitates and a 10% input (1/10 Input) were immunoblotted with the indicated antibodies. (F) The affinity-purified Tip60 complexes, as described in the Materials and Methods, were subjected to immunoblotting using the indicated antibodies. (G) GM02063 cells were treated with control, Tip60, or GFP siRNAs and then subjected to UVA microirradiation as in B.
In order to determine the molecular basis underlying RNR recruitment at the sites of DSBs, we performed yeast two-hybrid screening using RRM1 as a bait. Of a total of 5 × 106 transformants from a HeLa cell cDNA library, 45 positive colonies were confirmed to be lacZ-positive. They contained overlapping cDNAs derived from three genes: RRM2 and 53R2 (both encoding a small subunit of RNR), and another encoding Tip60 histone acetyltransferase (Tip60). Small C-terminal RRM1 deletion mutants (Δ761-C and Δ781-C) failed to bind Tip60, but retained the ability to bind to RRM2 (Supplemental Fig. S3A). In contrast, the N-terminal truncation mutant of Tip60 (TC2) could interact with RRM1, but no mutant with any additional truncation of TC2 was able to do so (Supplemental Fig. S3B). Full-length Tip60 failed to bind full-length RRM2 (Supplemental Fig. S3C). We generated the C-terminal fragment of RRM1 (amino acids 701–792) with a SV40 nuclear localization signal (NLS-RC1-HA) and examined its ability to bind Tip60 in vivo and in vitro. NLS-RC1-HA, but not a control NL-GFP-HA fragment, was detected in the anti-Myc immunoprecipitates when transiently coexpressed with Tip60-Myc (Supplemental Fig. S3D). Purified MBP-fused RC1 produced in Escherichia coli was capable of binding to GST-Tip60 expressed in insect cells (Supplemental Fig. S3E). Both Δ761-C and Δ781-C failed to bind chromatin, further confirming that the binding of RRM1 to chromatin required its interaction with Tip60 (Supplemental Fig. S3F).
Similarly to Chk1 (Niida et al. 2007; Shimada et al. 2008), endogenous RRM1 was present in cytosolic (S1), nucleoplasmic (S2), and chromatin-bound (P2) fractions (Supplemental Fig. S4A). Tip60 existed predominantly in the chromatin-bound fraction (P2). Both RRM1 and Tip60 proteins in this fraction were partly solubilized by treatment with micrococcal nuclease (Mnase), suggesting that they associated with chromatin. RRM1 knockdown showed a significant decrease of RRM1 protein levels in both soluble and chromatin-bound fractions (Supplemental Fig. S4B). IKKα and Orc2 were detected predominantly in soluble and chromatin fractions, respectively, indicating that cell fractionation was done successfully. Ectopic RRM1-HA present in the chromatin fraction was increased when Tip60-Myc-His was coexpressed, although a low level of RRM1-HA was detected in the absence of Tip60-Myc-His, presumably due to the presence of endogenous Tip60 (Supplemental Fig. S4C). The amounts of RRM1 and Tip60 bound to the chromatin were not affected by DNA damage (Supplemental Fig. S4D). However, depletion of Tip60 resulted in a reduction in the amount of RRM1 on chromatin (Fig. 1C). Taken together, chromatin binding of RRM1 appeared to be Tip60-dependent. RRM1-HA, but not the RRM2 subunit alone, formed a complex with GST-His-Tip60 in insect cells (Fig. 1D, left panels). RRM2 also formed a complex with GST-His-Tip60 in a manner dependent on the presence of RRM1-HA. Consistently, accumulation of RRM2 at DSB sites was compromised when RRM1 was depleted (Supplemental Fig. S2D). Immunoprecipitations using anti-HA antibodies demonstrated that RRM1-HA bound to both RRM2 and GST-His-Tip60 (Fig. 1D, right panels). RRM1 and RRM2 were detected in the precipitates of anti-Tip60 antibodies from the solubilized chromatin, even in the absence of DNA damage (Fig. 1E). To further confirm the interaction between RNR and Tip60, we purified the Tip60 complex from HeLa cell nuclear extracts expressing Flag-HA Tip60 as reported previously (Ikura et al. 2000, 2007). RRM1 and RRM2, as well as PAF400/TRRAP as a positive control (Murr et al. 2006), were detected in Tip60 complex from extracts with or without DNA damage (Fig. 1F). Tip60 knockdown by siRNA or shRNA abrogated accumulation of RRM1 along with microirradiated lines (Fig. 1G; Supplemental Fig. S2E). These results suggested that RRM1 recruitment at DSB sites was Tip60-dependent.
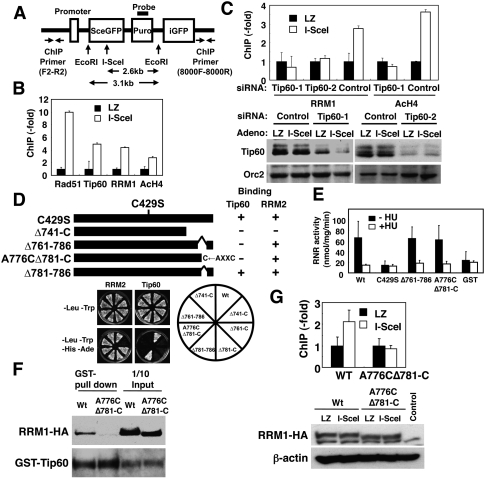
To determine precisely whether RRM1 was recruited at the site of DNA damage, we generated Ku-deficient mouse embryonic fibroblasts (MEFs) in which a single DSB was introduced after infection with adenoviruses expressing I-SceI. This DSB was not rapidly repaired by nonhomologous end-joining, making it easy to detect proteins accumulating at this DSB site by chromatin immunoprecipitation (ChIP) analysis (STEFKu70−/−phprt-DR-GFP) (Fig. 2A; Pierce et al. 2001). Introduction of the DSB was confirmed by Southern blotting (Supplemental Fig. S5). ChIP analyses revealed a substantial increase in the binding of RRM1 as well as Rad51 and Tip60 to a DNA break site. An increase in acetylation of histone H4 was also observed at the damage site (Fig. 2B). These were not seen on infection with control LacZ. Tip60 depletion by two independent siRNAs resulted in a loss of RRM1 binding to a DSB site, as well as a reduction in acetylation of histone H4 (Fig. 2C). A mutant Tip60 lacking histone-acetylating activity could recruit RRM1 to the DSB site similarly to wild-type RRM1 (Supplemental Fig. S6A). Inhibition of ATM, ATR, and DNA-PK by caffeine did not affect RRM1 recruitment (Supplemental Fig. S6B). These results further supported the notion that complex formation between RNR and Tip60 is required for recruitment of RNR to sites of DNA damage.
Figure 2.
RRM1 is recruited at DSB sites in a Tip60-dependent manner. (A) Map of the I-SceI cassette construct containing the I-SceI site, the probe for Southern blotting, and a set of primers for the ChIP assay. (B) STEFKu70−/−phprt-DR-GFP cells infected with I-SceI adenoviruses were subjected to ChIP analysis using the indicated antibodies as described in the supporting Materials and Methods. Data are shown as percentages of increases in PCR products from cells expressing I-SceI (I-SceI) relative to those from cells expressing Lac Z (LZ). Data are mean ± standard deviation (n = 3). (C) STEFKu70−/−phprt-DR-GFP cells were transfected with two independent Tip60 siRNAs (Tip60-1 and Tip60-2) or control siRNA. ChIP analysis was performed as in B. (Bottom panels) Aliquots of cell lysates were subjected to immunoblotting using anti-Tip60 antibodies. (D) The constructs used are schematically represented, and the specific interaction between RRM1 mutants and Tip60 was assayed using yeast two-hybrid screening. (E) An in vitro RNR assay of complexes containing wild-type or various RRM1 mutants was performed as described in the Materials and Methods. (Black bars) −HU; (white bars) +HU (10 mM). Data are mean ± standard deviation (n = 3). (F) Sf9 lysates expressing GST-His-Tip60 and the indicated RRM1-HA were subjected to GST pull-down assay using the indicated antibodies. (G) Knockout–knock-in STEFKu70−/−phprt-DR-GFP cells expressing wild-type or A776CΔ781-C RRM1-HA were generated by transfection with vectors for either wild-type or A776CΔ781-C RRM1 and then with RRM1 siRNA. Expression vectors of wild type and A776CΔ781-C contain mutations in a specific sequence targeted by siRNA. (Top panel) Cells were subjected to ChIP analysis using anti-HA antibodies as in B. (Bottom panels) Aliquots of cell lysates were subjected to immunoblotting using the indicated antibodies.
We then examined if RNR recruitment at damage sites was required for effective DNA repair. We first generated RRM1 mutants that lack the ability to bind Tip60 but retain RNR activity. Given that the C-terminal CXXC motif of RRM1 is important for RNR function (Zhang et al. 2007), we constructed RRM1 mutants containing the CXXC motif but lacking Tip60-binding ability (Δ761–786 and A776CΔ781-C) (Fig. 2D). Wild-type RRM1 or its mutants were coexpressed with RRM2 in insect cells, and the resultant complexes were subjected to an in vitro RNR assay (Fukushima et al. 2001). RNR complexes containing wild-type, Δ761–786, and A776CΔ781-C RRM1 retained hydroxyurea (HU)-sensitive RNR activity (HU is a specific RNR inhibitor), whereas an inactive C429S mutant or GST protein as a negative control did not show RNR activity (Fig. 2E). The specific activity of RNR containing wild-type, Δ761–787, and A776CΔ781-C RRM1 (∼50 nmol/mg per minute) was similar to that reported previously (Guittet et al. 2001), confirming the reliability of our results. The A776CΔ781-C mutant failed to form a complex with GST-Tip60 (Fig. 2F). ChIP analysis using RRM1 knockout–knock-in STEFKu70−/−phprt-DR-GFP cells revealed that the A776CΔ781-C mutant failed to accumulate at the DSB site (Fig. 2G). These results indicated that direct interaction of RRM1 to Tip60 is required for triggering its accumulation at the DSB site.
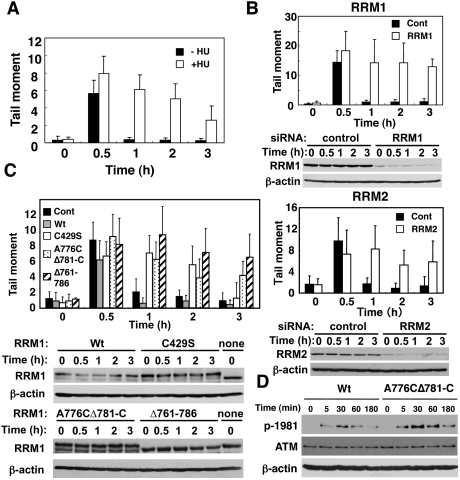
A comet assay revealed that DNA damage in cells was repaired efficiently within 1 h in the absence of HU. However, treatment with HU, and RRM1 or RRM2 depletion, resulted in an impairment of DNA repair (Fig. 3A,B). RNR activity was thus essential for effective repair. Ectopic expression of wild-type RRM1 with mutations in a specific sequence targeted by siRNA effectively rescued the impaired DNA repair in cells depleted of endogenous RRM1 (Fig. 3C). In contrast, ectopic expression of C429S, Δ761–786, and A776CΔ781-C RRM1 failed to do so. ATM was activated independently of Tip60 binding to RNR, but this activation was enhanced and prolonged in cells expressing A776CΔ781-C, presumably due to impaired DNA repair (Fig. 3D). It is therefore conceivable that recruitment of active RNR at DNA damage sites is a prerequisite for effective DSB repair, but not for activation of checkpoint signaling. Tip60 is also known to participate in transcriptional regulation of several genes. Neither RRM1 nor RRM2 proteins were affected by Tip60 depletion or overexpression (Supplemental Fig. S7), indicating that the effect of Tip60 did not result from changes in RRM1 and RRM2 expression.
Figure 3.
Recruitment of active RNR at DNA damage sites is a prerequisite for effective DNA repair. (A) HeLa cells were treated with (open bars) or without (filled bars) 2.5 mM HU, exposed to IR (4 Gy), and subjected to a comet assay as described in the Materials and Methods. The results were obtained by counting at least 50 cells per sample in three independent experiments. (B) HeLa cells were transfected with a control (filled bars) or RRM1 or RRM2 siRNA (open bars), and DNA repair was evaluated as in A. Cell lysates were subjected to immunoblotting using the indicated antibodies. (C) HeLa cells were transfected with or without (filled bars) either wild-type (gray bars), C429S (open bars), A776CΔ781-C (dotted), or Δ761–786 (hatched) RRM1. RRM1-transfected cells were then transfected with RRM1 siRNA. Expression vectors of wild type and various RRM1 mutants contain mutations in a specific sequence targeted by siRNA. DNA repair activity and expression of RRM1 were examined as in B. (D) Knockout–knock-in HeLa cells expressing wild type or A776CΔ781-C RRM1-HA were exposed to IR, and cell lysates were subjected to immunoblotting as in C.
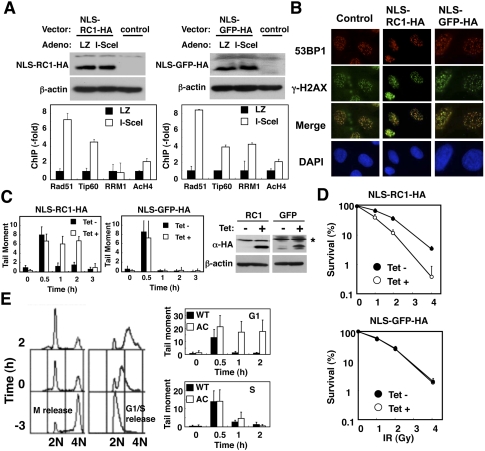
ChIP analyses revealed that NLS-RC1-HA specifically inhibited RRM1 binding, but did not affect Rad51 or Tip60 binding, or increase H4 acetylation at the DSB site in STEFKu70−/−phprt-DR-GFP cells (Fig. 4A). Expression of NLS-RC1-HA suppressed accumulation of endogenous RRM1 at DNA damage sites (Supplemental Fig. S8A,B), but did not affect the foci formation of 53BP1 at DSB sites (Fig. 4B), or complex formation and activity (Supplemental Fig. S9A,B) of endogenous RNR. However, cells expressing NLS-RC1-HA, but not NLS-GFP-HA, had unrepaired DNA in the tail at 2 h (Fig. 4C). A quantitative colony formation assay was used to examine the DNA damage sensitivity of cells expressing NLS-RC1-HA. Induction of NLS-RC1-HA sensitized cells to IR (Fig. 4D).
Figure 4.
Inhibition of recruitment of RNR at DSB sites by ectopic expression of NLS-RC1-HA abrogates DNA repair and sensitizes cells to DNA damage. (A) STEFKu70−/−phprt-DR-GFP cells expressing NLS-RC1-HA (SV40 NLS-RC1 fragment, 701–792 amino acids) or NLS-GFP-HA (GFP fragment, 1–93 amino acids) were subjected to ChIP analysis as in Figure 2B. (Top panels) Cell lysates were subjected to immunoblotting using the indicated antibodies. (B, left panels) Tet-on HeLa cells expressing NLS-RC1-HA or NLS-GFP-HA were treated with or without tetracycline (1 μg/mL), exposed to IR (4 Gy), and subjected to immunostaining with the indicated antibodies and a comet assay as in Figure 3A. (Right panels) IR-untreated lysates were subjected to immunoblotting using the indicated antibodies. (C) Asterisk (*) represents nonspecific bands. (D) These cells were exposed to the indicated dose of IR, and a quantitative colony formation assay was performed 8 d after treatment. Data are mean ± standard deviation (n = 3). (E) Knockout–knock-in HeLa cells expressing either wild-type (filled bars) or A776CΔ781-C (open bars) RRM1-HA were synchronized as described in the Materials and Methods. Synchronized cells were then released into G1 phase or S phase (time −3) and exposed to IR (4 Gy) 3 h after release (time 0). (Right panels) DNA repair was evaluated as in A. (Left panels) Cell cycle distributions are presented.
Given that levels of dNTP pools are higher during S phase than during G1 phase (Hakansson et al. 2006), recruitment of RNR at damage sites may function at a specific phase of the cell cycle where dNTP pools are low. To address this issue, we synchronized cells at S phase or G1 phase by arrest and release of thymidine or nocodazole, respectively. Recruitment of wild-type RRM1 at a DSB site was observed at both G1 and S phase (Supplemental Fig. S10). However, a comet assay revealed that A776CΔ781-C failed to rescue the impaired DNA repair in RRM1-depleted cells at G1 phase, but not at S phase (Fig. 4E). Consistently, RRM1 mutation of Tip60 binding slightly sensitizes cells to Zeocin (Supplemental Fig. S11A), which causes DNA strand breaks, but not to MMC (Supplemental Fig. S11B), which can cause interstrand cross-linking repaired mainly at S–G2 phase. Intriguingly, this G1-phase-specific impairment of DNA repair was restored when excess amounts of dADP, dGDP, dCDP, and dUMP (250 μM) were supplied in the culture medium (Supplemental Fig. S12). These results suggested that recruitment of RNR was required specifically for effective DNA repair in cells with low levels of dNTPs.
The present study suggests that the RNR recruitment to DSB sites likely provides mechanistic insights into the regulatory events that ensure a balanced supply of dNTPs during mammalian DNA repair. RNR appears to form a complex with Tip60 independently of DNA damage. Thus, it is possible that the RNR–Tip60 complex might have an alternative function, such as regulation of transcription. In response to DNA damage, regulation of the RNR subunit by Wtm1 and Dif1 in budding yeast is radically different in terms of cellular localization (Lee and Elledge 2006; Lee et al. 2008) from that observed in the present study; however, important changes in the subcellular localization of RNR might be conserved. Given that Tip60 is a key regulator of DNA damage responses, the concomitant recruitment of RNR at damage sites suggests the presence of a synthetic regulatory mechanism for DNA repair in mammals.
Materials and methods
Antibodies
Antibodies used were as follows: α-Rad51 (Ab-1, Oncogene Research Products), α-RRM1 (sc-11733 and sc-11731, Santa Cruz Biotechnologies), α-HA (11 666 606 001, Roche Applied Sciences; and PM002, MBL), α-Myc (sc-40 and sc-789, Santa Cruz Biotechnologies), α-RRM2(sc-10844, Santa Cruz Biotechnologies), α-GST (sc-459, Santa Cruz Biotechnologies), α-Chk1 (sc-8408, Santa Cruz Biotechnologies), α-IKKα (sc-7182, Santa Cruz Biotechnologies), α-Orc2 (sc-13238, Santa Cruz Biotechnologies), α-ATM (sc-23921, Santa Cruz Biotechnologies), α-ATMp1981 (no. 4526, Cell Signaling), α-acetylated histone H4 (no. 06-866, Upstate Biotechnologies), and α-phospho-histone H2AX (411-pc-020, TREVIGEN; and 05-636, Upstate Biotechnologies). Anti-Tip60 rabbit polyclonal antibodies were generated by immunization with recombinant GST-His-Tip60 produced in insect cells, and the serum obtained was affinity-purified using a GST-His-Tip60 column.
Two-hybrid interaction assays
The pGBKT7-RRM1 plasmid was generated by insertion of the full-length human RRM1-encoding sequence. pGBKT7-RRM1 was transformed into the yeast strain AH101 and mated with yeast Y187 pretransformed with a HeLa cell cDNA library (BD Biosciences). The deletion mutants of RRM1 and Tip60 were amplified by PCR using specific sets of primers. Primer sequences are supplied in the Supplemental Material.
Affinity purification of Tip60 complex
Affinity purification of Tip60 complex was performed as described previously (Ikura et al. 2000, 2007). For the induction of DNA damage, cells were γ-irradiated (12 Gy) after centrifugation.
In situ detergent extraction and immunofluorescence analysis
Immunofluorescence on paraformaldehyde-fixed cells was performed according to a previous report (Green and Almouzni 2003), using the indicated antibodies.
Microirradiation
Microirradiation was performed as described previously (Ikura et al. 2007). In brief, GM02063 cells were maintained on the microscope stage in a Chamlide TC live-cell chamber system (Live Cell Instrument) at 37°C. Microirradiation was performed using an LSM510 confocal microscope (Carl Zeiss). Sensitization of cells was performed by incubating the cells for 20 h in medium containing 2.5 μM deoxyribosylthymine and 0.3 μM bromodeoxyuridine (Sigma), and then staining with 2 μg/mL Hoechst 33258 (Sigma) for 10 min before UVA microirradiation. The 364-nm line of the UVA laser was used for microirradiation (three pulses at 30 μW). Samples were examined with a Zeiss Axioplan 2 equipped with a charge-coupled device camera AxioCam MRm controlled by Axiovision software (Zeiss).
Knockdown experiments
HeLa cells or STEFKu70−/−phprt-DR-GFP cells were transfected with either control siRNA (Silencer Negative Control #1, Ambion 4611), siRNAs for human Tip60 (sc-37966, Santa Cruz Biotechnologies), mouse Tip60-1 (sc-37967, Santa Cruz Biotechnologies), mouse Tip60-2 (D-057795-02-0010, Dharmacon), or RRM1 (GGAUCGCUGUCUCUAACUUtt) using Lipofectamine 2000 reagent (Invitrogen).
Subcellular fractionation and Mnase treatment
Subcellular fractionation was performed according to a previous report (Mendez and Stillman 2000). The isolated chromatin fraction (1 × 106 cells) was treated with Mnase (15 U) for 30 min at 37°C.
Establishment of STEFKu70−/− cells containing a phprt-DR-GFP cassette
The phprt-DR-GFP vector (10 μg) was linearized with PvuI and transfected into STEFKu70−/− cells. Cells were selected with 1.25 μg/mL puromycin for 12 d, and single colonies were screened by Southern blotting using puromycin cDNA as a probe. Clones having only one copy of the phprt-DR-GFP cassette were used for experiments.
Establishment of Tet-on HeLa cells expressing NLS-RC1
pcDNA4/TO-NLS-RC1 (10 μg) was linearized with XhoI and transfected into HeLa T-Rex cells (Invitrogen). Positive clones were selected with Zeocin (250 μg/mL) and Blastcidin (5 μg/mL) for 12 d and screened by immunoblotting using anti-HA antibodies for the detection of NLS-RC1 induction in the presence of tetracycline (1 μg/mL).
Generation of adenoviruses expressing I-SceI endonuclease
The full-length I-SceI fragment harboring the CAG promoter and poly A signal was subcloned into pAd/PL-DEST (Invitrogen). Adenoviruses expressing I-SceI were generated according to the manufacturer's protocol (Invitrogen).
ChIP assay
A population of STEFKu70−/−cells (1 × 107) containing phprt-DR-GFP cells infected with adenoviruses expressing I-SceI was cross-linked with 1% formaldehyde for 10 min at 37°C. ChIP assays were performed essentially as described (Shimada et al. 2008). Precipitated DNA was resuspended in 50 μL of water and analyzed by quantitative real-time PCR with the ABI PRISM7000 system using Power SYBR Green PCR Master Mix (Applied Biosystems) as described (Katsuno et al. 2009). Primers used for detection of the I-SceI break site were indicated in Figure 2A. As an internal control for normalization of the specific fragments amplified, mouse GAPDH locus was amplified using whole genomic DNAs with mGAPDH-F and mGAPDH-R. Primer sequences are supplied in the Supplemental Material.
Comet assay
Alkaline comet assays were performed using a Trevigen's Comet Assay kit (4250-050-k) according to the manufacturer's instructions. DNA was stained with SYBR Green, and slides were photographed digitally (Nikon Eclipse E800 lens and Fuji CCD camera). Tail moments were analyzed as reported previously (Park et al. 2006) using TriTek Comet Score Freeware.
Measurement of DNA damage sensitivity
Tet-on HeLa cells expressing NLS-RC1-HA or NLS-GFP-HA were irradiated with varying doses of IR in the presence or absence of doxycycline (1 μg/mL), and then washed with PBS. Eight days after an additional incubation, surviving colonies were counted, and their relative numbers were expressed as percentages of the untreated cells (n = 3).
RNR assay
Insect cells were coinfected with baculoviruses expressing wild-type RRM1 or its mutants, and with those expressing wild-type RRM2. RNR complexes were immunopurified, and their activities were determined according to a method reported previously (Fukushima et al. 2001). Amounts of wild-type RRM1 protein or its mutant proteins were determined by SDS-PAGE and used for calculating specific activities.
Cell cycle synchronization
For synchronization of cells at S phase, knockout–knock-in HeLa cells expressing wild-type or A776CΔ781-C RRM1-HA were first synchronized at the G1/S boundary by exposure to 2.5 mM thymidine for 16 h, and then released into S phase by wash-out of thymidine with PBS and the addition of 20% FBS containing DMEM. Cells were then exposed to IR 3 h after release. For synchronization of cells at G1 phase, knockout–knock-in HeLa cells were synchronized at M phase by exposure to 100 ng/mL nocodazole for 16 h and released into G1 phase by wash-out of nocodazole with PBS and addition of 20% FBS containing DMEM. Cells were then exposed to IR 3 h after release.
Acknowledgments
We thank M. Delhase for critical reading of the manuscript; M. Jasin for hprt-DR-GFP and pCBASce vectors; M. Fukushima for critical advice on the RNR assay; A. Kurimasa for STEFKu70−/− MEFs; K. Murata, C. Namikawa-Yamada, and H. Kojima for technical assistance; and M. Inagaki and H. Goto for fluorescence microscopy. This work was supported in part by the Ministry of Education, Science, Sports, and Culture of Japan through Grants-in-Aid for Scientific Research (B) (to M.N.) and (C) (to H.N.), the YASUDA Medical Foundation (to M.N.), and the Sagawa Cancer Foundation (to M.N.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1863810.
Supplemental material is available at http://www.genesdev.org.
References
- Dong Q, Wang TS. Mutational studies of human DNA polymerase α. Lysine 950 in the third most conserved region of α-like DNA polymerases is involved in binding the deoxynucleoside triphosphate. J Biol Chem. 1995;270:21563–21570. doi: 10.1074/jbc.270.37.21563. [DOI] [PubMed] [Google Scholar]
- Fukushima M, Fujioka A, Uchida J, Nakagawa F, Takechi T. Thymidylate synthase (TS) and ribonucleotide reductase (RNR) may be involved in acquired resistance to 5-fluorouracil (5-FU) in human cancer xenografts in vivo. Eur J Cancer. 2001;37:1681–1687. doi: 10.1016/s0959-8049(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guittet O, Hakansson P, Voevodskaya N, Fridd S, Graslund A, Arakawa H, Nakamura Y, Thelander L. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J Biol Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Jean-Louis F, Dubertret ML, Bailly C, Cicurel L, Petchot-Bacque JP, Dubertret L. Effect of human basic fibroblast growth factor on fibroblast proliferation, cell volume, collagen lattice contraction: In comparison with acidic type. J Dermatol Sci. 1996;11:134–141. doi: 10.1016/0923-1811(95)00431-9. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Trincao J, Aggarwal AK, Prakash S, Prakash L. Deoxynucleotide triphosphate binding mode conserved in Y family DNA polymerases. Mol Cell Biol. 2003;23:3008–3012. doi: 10.1128/MCB.23.8.3008-3012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A–Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci. 2009;106:3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov VS, Showalter AK, Liu J, Zhong X, Tsai MD. DNA polymerase β: Contributions of template-positioning and dNTP triphosphate-binding residues to catalysis and fidelity. Biochemistry. 2000;39:16008–16015. doi: 10.1021/bi0008480. [DOI] [PubMed] [Google Scholar]
- Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes & Dev. 2006;20:334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell. 2008;32:70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap–Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Banerjee B, Hande MP, Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol. 2007;27:2572–2581. doi: 10.1128/MCB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontarin G, Fijolek A, Pizzo P, Ferraro P, Rampazzo C, Pozzan T, Thelander L, Reichard PA, Bianchi V. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci. 2008;105:17801–17806. doi: 10.1073/pnas.0808198105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–232. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang K, Chen CC, Feser J, Huang M. Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1. Proc Natl Acad Sci. 2007;104:2217–2222. doi: 10.1073/pnas.0611095104. [DOI] [PMC free article] [PubMed] [Google Scholar]