Abstract
Transcriptional feedback loops are central to the generation and maintenance of circadian rhythms. In animal systems as well as Neurospora, transcriptional repression is believed to occur by catalytic post-translational events. We report here in the Drosophila model two different mechanisms by which the circadian repressor PERIOD (PER) inhibits CLOCK/CYCLE (CLK/CYC)-mediated transcription. First, PER is recruited to circadian promoters, which leads to the nighttime decrease of CLK/CYC activity. This decrease is proportional to PER levels on DNA, and PER recruitment probably occurs via CLK. Then CLK is released from DNA and sequestered in a strong, ∼1:1 PER–CLK off-DNA complex. The data indicate that the PER levels bound to CLK change dynamically and are important for repression, first on-DNA and then off-DNA. They also suggest that these mechanisms occur upstream of post-translational events, and that elements of this two-step mechanism likely apply to mammals.
Keywords: Chromatin immunoprecipitation, circadian, negative feedback loop, tiling array, transcriptional oscillations
Circadian rhythms are generated by cell-autonomous oscillators and allow biological functions to adapt to and anticipate daily variations in the environment. In eukaryotes, the molecular machinery underlying these rhythms relies on transcriptional activators and repressors, which are organized into negative feedback loops (Allada et al. 2001; Takahashi 2004). In Drosophila, two basic helix–loop–helix (bHLH) PAS domain transcription factors encoded by the genes clock (clk) and cycle (cyc) form the heterodimer CLK/CYC, which rhythmically binds to E-boxes and activates the transcription of the key clock genes period (per) and timeless (tim). The proteins PER and TIM then interact and inhibit CLK/CYC activity, which decreases and ultimately leads to a new transcriptional cycle. Similar molecular oscillations occur in mammals and involve the CLK/CYC orthologs CLOCK/BMAL1, which also bind to E-boxes and rhythmically activate the transcription of their repressor genes, Period and Cryptochrome (Hastings et al. 2003; Hardin 2005).
A dominant paradigm for circadian transcriptional repression has emerged over the last several years and involves post-translational mechanisms; i.e., the repressor protein decreases the function of an activator by mediating its post-translational modifications. For example, Drosophila PER has been proposed to inhibit circadian transcription by recruiting kinases to the CLK/CYC complex (Yu et al. 2006; Kim et al. 2007). Consistent with this view, a recent failure to detect an association of wild-type PER with DNA by chromatin immunoprecipitation (ChIP) was interpreted to suggest that PER normally promotes phosphorylation by interacting only transiently with chromatin-bound CLK (Yu et al. 2009). In mammals, a similar view prevails; namely, post-translational modifications of CLK and BMAL1 play a prominent role in the repression of circadian transcription (Kondratov et al. 2003, 2006; Tamaru et al. 2003, 2009; Dardente et al. 2007; Hirayama et al. 2007; Yoshitane et al. 2009). A phosphorylation-based catalytic mechanism is also the dominant repression paradigm in cyanobacteria (Murayama et al. 2008), and several studies in the Neurospora system have emphasized the importance of phosphorylation to the inhibition of White Collar (WC)-mediated circadian transcription by the repressor FRQ (He and Liu 2005; He et al. 2005; Schafmeier et al. 2005, 2006, 2008; Hong et al. 2008; Sancar et al. 2009).
To understand further how PER inhibits CLK transcriptional activity, we used a newly described Drosophila strain containing a V5-tagged Clk transgene (Kadener et al. 2008) for immunoprecipitation (IP) and ChIP experiments. The data provide a new perspective on the mechanisms by which PER mediates transcriptional repression. We first show that the beginning of the repression phase is associated with the binding of PER to circadian promoters, probably via a PER–CLK interaction. The PER–DNA interaction likely affects RNA polymerase II (Pol II) occupancy within the ORF despite persistent CLK DNA binding. This “on-DNA” PER–CLK interaction is followed by removal of CLK from DNA and concomitant formation of a strong 1:1 “off-DNA” PER–CLK complex with low DNA-binding affinity. Our results suggest that these events occur upstream of the post-translational modifications that influence CLK/CYC activity.
Results
CLK occupancy does not determine all aspects of circadian transcription
To address the relationship between CLK DNA binding and transcription of the important circadian genes per and tim, we took advantage of our transgenic strain containing a genomic version of Clk tagged with a V5 epitope (Kadener et al. 2008). This strain carries two Clk-V5 transgenes in addition to the two endogenous Clk genes present in the yw background. It has a circadian period of 23.28 ± 0.06 h, as increasing the number of Clk gene copies decreases circadian period (Kadener et al. 2008). CLK-mediated transcription is otherwise unaffected by Clk-V5 transgenes, and the tim pre-mRNA profile is essentially indistinguishable from that generated more than a decade ago by nuclear run-on assays in fly head extracts (Fig. 1A; So and Rosbash 1997).
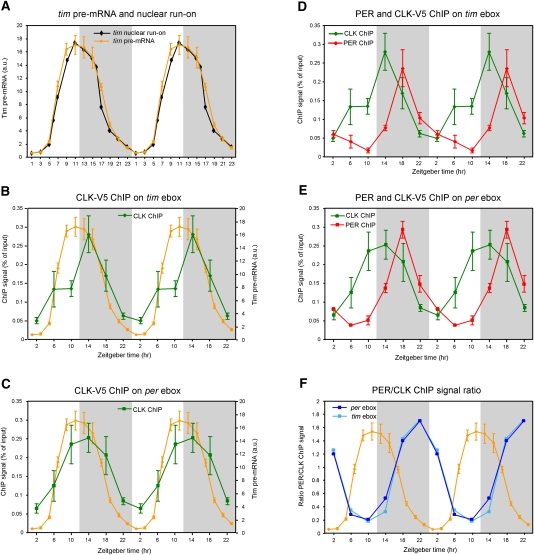
Figure 1.
CLK-V5 and PER rhythmically bind to Per and Tim promoters. (A) Circadian transcription of yw;;dClk-V5 flies. Relative levels of tim pre-mRNA were quantified by qPCR. Each time point represents the average and standard error of two independent experiments. The profile of the nuclear run-on assay (black) of tim published by So and Rosbash (1997) is displayed for comparison with tim pre-mRNA rhythms assayed by qPCR (orange). (B,C) ChIP of CLK-V5 in yw;;dClk-V5 flies. The graphs show qPCR values of CLK-V5 and PER binding to the tim promoter (B; green) and per promoter (C; green). Primers were in regions containing an ebox (see the Materials and Methods for details). Values represent the relative amount of chromatin immunoprecipitated over the input for each time point. The tim transcription curve (in orange) from A is shown for comparison. (D,E) ChIP of PER and CLK in yw;;dClk-V5 flies. The graphs show qPCR values of PER binding to the tim promoter (D; red) and the Per promoter (E; red). Graphs showing CLK binding to tim promoter (D; green) and per promoter (E; green) are identical to the graphs in B and C, respectively. (F) Quantification of the ratio of PER/CLK ChIP signal on tim (light blue) and per (dark blue) promoters. Ratios were calculated from experiments presented in B–D. The tim pre-mRNA profile from A is superimposed to the two ratio profiles.
We first assayed V5-CLK binding to the tim and per promoters by ChIP. The data show robust oscillations of CLK binding (Fig. 1B,C; Supplemental Fig. 1) and are similar to those published by Hardin and colleagues (Yu et al. 2006) using an anti-CLK antibody. Rhythmic CLK DNA binding matches most of the circadian transcription profiles (Fig. 1A–C, note that per transcription is very similar to tim transcription; So and Rosbash 1997). However, circadian transcription decreases substantially during the first half of the night and is substantially lower at Zeitgeber time 18 (ZT18) than at ZT10 despite similar CLK binding at these two times (Fig. 1; Fig. 3 of So and Rosbash 1997; Fig. 5 of Kim et al. 2007). This suggests that CLK occupancy on DNA may not fully account for transcription, and that some “on-DNA” repressive events may occur.
PER binds to DNA at the beginning of the repression phase
To determine if the transcriptional repressor PER might be relevant to on-DNA repression, we used an anti-PER polyclonal antibody (Dembinska et al. 1997) to assay by ChIP whether PER interacts with circadian promoters. The antibody generated a robust rhythmic signal on the per and tim promoters, and no rhythmic signal was apparent in the control per01-null mutant strain (Fig. 1D,E; Supplemental Fig. 1). Although the amplitude was similar to that of CLK, the PER ChIP profile peaked at ZT18 and was therefore prominently delayed relative to the CLK profile. ZT18 corresponds to the time of maximal decrease in circadian transcription, which occurs without any substantial decrease in CLK ChIP signal (Fig. 1B,C). Otherwise put, the PER/CLK ChIP signal ratio (Fig. 1F) explains much better the circadian transcription curves than CLK binding alone (Fig. 1B–E), which suggests that PER might help recruit chromatin-bound corepressors and/or decrease the binding of coactivators to circadian promoters in the first half of the night.
CLK, PER, and Pol II ChIP tiling arrays reveal new characteristics of circadian transcription
To confirm and extend those results, we assayed PER and CLK as well as Pol II DNA binding on the tim and per genes with ChIP tiling arrays (Fig. 2A,C; see the Materials and Methods for more details). The PER and CLK signals on both genes are near the start sites of transcription and manifest rhythms with time courses similar if not identical to those assayed by quantitative PCR (qPCR). However, these signals appear more spread on per than on tim at this level of resolution (Fig. 2A,C).
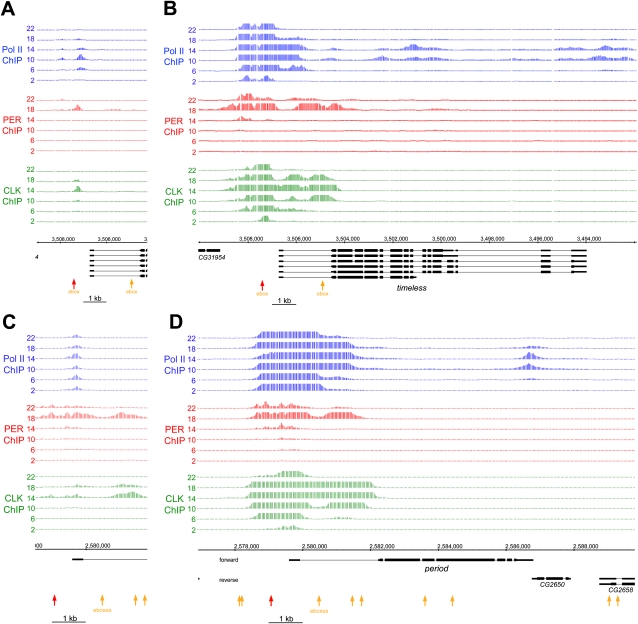
Figure 2.
CLK-V5 and PER rhythmic DNA binding occurs in the same promoter region and determines the timing of Pol II elongation. Tiling array data from Pol II (blue), PER (red), and V5 (green) ChIP experiments are displayed for tim (A,B) and per (C,D) genes. Each line illustrates one of the six time points (hours are indicated as ZT), and the data shown are the result of two independent ChIP tiling experiments. Data were analyzed using the model-based analysis of tiling arrays (MAT) algorithm (Johnson et al. 2006) and were converted to a linear scale for easier viewing using the Integrated Genome Browser from Affymetrix. The genomic location and the different isoforms for both genes are displayed in black, and the locations of the perfect consensus ebox (CACGTG) are represented by orange and red arrows (the red arrows indicate the ebox-containing region amplified by qPCR in Fig. 1). A and C represent a low magnification of the binding sites, whereas B and D represent a higher magnification of the binding sites. It is important to note that the Y-axes represent probabilities of DNA binding (MAT score) and not signal intensities (see the Materials and Methods for details). The scales of the Y-axes are set differently to allow for a spatial comparison of the signals. For example, in A the maximum values of the Y-axes are 5000 for Pol II, 500 for PER, and 80,000 for CLK.
The Pol II signals also come predominantly from the promoter regions, presumably reflecting Pol II pausing (for review, see Nechaev and Adelman 2008; Wu and Snyder 2008). Interestingly, the Pol II signal appears highly rhythmic only for tim, indicating mechanistic differences between the two genes (Fig. 2, cf. A and C; see also Taylor and Hardin 2008). This tim Pol II signal coincides with the single peak of cycling CLK signal as well as an underlying E-box, suggesting that CLK recruits Pol II rhythmically to this promoter. The per Pol II signal, in contrast, is present at or near the per transcription start site at all circadian times and does not depend on the CLK levels bound to per DNA, consistent with a previous study (Taylor and Hardin 2008). Cases of different Pol II recruitment by a transcription factor have been observed in other systems. For example, the bacterial activator CAP (catabolite activator protein) can promote RNA polymerase binding, isomerization, and promoter escape, depending on the promoter context of the target gene (Busby and Ebright 1999).
A higher-resolution view shows that cycling Pol II is also detectable within the tim ORF; it peaks at ZT10–ZT14 as expected (Fig. 2B). Although no comparable per ORF signal is apparent (Fig. 2D), the Pol II profile extends further from the promoter region at these times (Fig. 2D), consistent with the temporal profile of per transcription. This suggests that Pol II promoter escape is increased by CLK binding for per as well as for tim (Fig. 2B,D). Another difference between the genes is that cycling Pol II is apparent at the 3′ end of per, perhaps reflecting pausing at the 3′ end as well as more Pol II elongation at ZT10 and ZT14 than at other times.
The higher-resolution view also reveals several additional features of the CLK and PER profiles (Fig. 2B,D). First, some CLK signal well above background is present on both promoters, even when DNA binding is at a minimum. This is also evident from the qPCR assay (Supplemental Figs. 1, 2), and suggests that the per and tim chromatin is never completely closed. Second, the CLK ChIP signal extends across ∼4 kb of DNA at times of maximal interaction. This is probably not due to a technical artifact such as chromatin fragment size, which is 0.5–1 kb (data not shown); there are also reproducible subpatterns within the 4-kb signal region. Moreover, the signal is comparably broad, and in some cases even broader over the promoter regions of many other direct target clock genes (data not shown). Third, the PER DNA-binding profiles match very closely those of CLK, indicating that PER is also present throughout this region. As PER lacks a DNA-binding domain, it is probably recruited to chromatin via one or more DNA-binding partner proteins. The similar profiles of PER and CLK make CLK the prime candidate, but they do not exclude PER recruitment by other proteins or even by other recruitment mechanisms.
PER sequesters CLK at the end of the night/beginning of the day
Recruitment of PER to DNA by CLK suggests that we might be able to detect PER–CLK interactions without cross-linking. To this end, we assayed protein–protein interactions by standard IP procedures from fly head extracts in the CLK-V5 strain (Fig. 3).
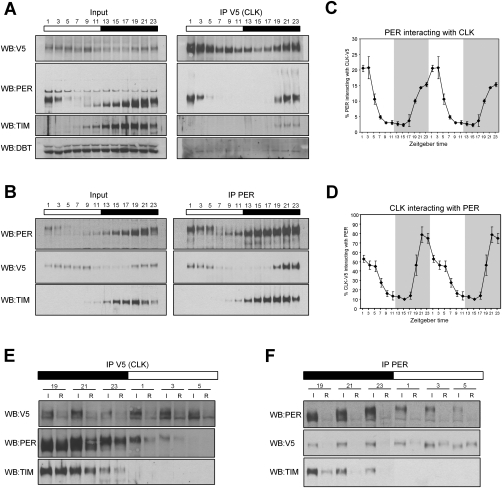
Figure 3.
PER sequesters CLK in a stochiometric complex at the end of the night/beginning of the day. (A,B) IP of CLK-V5 and PER in yw;;dClk-V5 flies. Input samples correspond to ∼15% of the amount of the IP sample. Membranes were analyzed by immunoblotting using antibodies directed against V5, PER, TIM, and DBT. (C,D) Quantification of the PER immunoprecipitated by CLK-V5 (from A) and CLK-V5 immunoprecipitated by PER (from B), respectively. Two independent experiments for each IP were quantified using Image J software (NIH). The ratio of the IP/input signals was calculated and the relative amount of protein immunoprecipitated was plotted as the percentage of PER immunoprecipitated by CLK-V5 (C) or CLK-V5 immunoprecipitated by PER (D). Each time point corresponds to the average ± standard error. (E,F) Effect of CLK-V5 (E) and PER (F) immunodepletion on CLK-V5, PER, and TIM levels. Levels of CLK-V5, PER, and TIM were compared before ([I] input) and after ([R] remaining) the CLK-V5 IP. Similar amounts of total proteins are loaded in every well.
The anti-V5 antibody efficiently immunoprecipitated CLK-V5 at all time points. PER was also immunoprecipitated with this antibody, but in a highly time-dependent manner, indicating a robust rhythm of PER–CLK interaction. PER was first detectable at ZT19, which is much later than the appearance of PER on the circadian promoters by ChIP. The interaction then increases, with the strongest signals present from ZT21 to ZT3. The complex appears to preferentially contain hyperphosphorylated PER (Fig. 3A), suggesting, perhaps, that the CLK–PER complex is a preferred substrate for PER phosphorylation. The PER signal decreases thereafter, in parallel with the degradation of PER in the early daytime (Fig. 3A).
To confirm these results, we performed the reciprocal IP with the anti-PER polyclonal antibody. PER immunoprecipitated CLK-V5 only from ZT19 to ZT5, with little or no interaction between ZT7 and ZT17 (Fig. 3B), similar if not identical to the IP assay with anti-V5. Since we do not detect a significant PER–CLK interaction by IP before ZT19, PER binding to DNA-bound CLK is only visible with formaldehyde cross-linking, and therefore is probably much weaker than at later times. Quantification of the CLK-V5 and PER IPs (Fig. 3C,D) as well as a comparison of CLK levels before and after PER immunodepletion (Fig. 3F) showed that ∼80% of CLK-V5 was immunoprecipitated with anti-PER at peak interaction times; i.e., ZT21 and ZT23. Since the complex survives the overnight incubation and washing steps used for IP, it is remarkably stable at these times and may even contain all of CLK in vivo.
Other proteins are also present in this complex. Indeed, the two known core clock components TIM and DBT were also immunoprecipitated by CLK-V5. Although the signals were weaker than the PER signal (Fig. 3A, both input and IP membranes were processed together and are displayed with same exposure), they progressively increased from ZT19 to ZT23 like the PER signal (Fig. 3A). This strongly suggests that at least a portion of the CLK/PER-repressive complex also contains TIM and DBT. PER IP also indicated a strong TIM–PER interaction, as most if not all of TIM was associated with PER at all times of day (Fig. 3B,F). The time-independent association of PER with TIM reflects the previously described PER–TIM complex (Zeng et al. 1996), which is presumably distinct from the PER–CLK complex.
PER sequesters CLK in a close to 1:1 stochiometric complex
To further characterize the PER–CLK complex, we measured the stochiometric relationship between these two proteins at peak interaction times. We used a dilution series of V5-tagged PER from S2 cell extracts to characterize and quantitatively compare signals between different antibodies; e.g., the polyclonal anti-PER antibody and the anti-V5 antibody. Signals are directly comparable, since the V5 antibody generates the same signal for the same molar amounts of every protein tagged with a single V5 epitope.
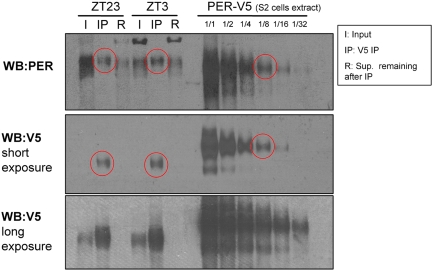
A dilution series of V5-tagged PER on the same gel as samples from a CLK-V5 IP performed at ZT23 and ZT3 showed that PER and CLK-V5 are present in this complex at a 1:1 ratio or nearly so (Fig. 4, circled bands have similar intensities). This suggests that PER sequesters nearly all of CLK off-DNA in a remarkably stable 1:1 complex. The comparisons also indicate that PER is present in fly head extracts at approximately eightfold excess over CLK-V5 at ZT23 and at about fourfold excess at ZT3 (Fig. 4).
Figure 4.
PER and CLK form an apparent 1:1 stable complex. Stochiometry of the PER/CLK complex was determined by analyzing on the same gel samples from a CLK-V5 IP performed in yw;;dClk-V5 flies at ZT23 and ZT3 (input samples correspond to ∼15% of the amount of the IP sample) (as in Fig. 3) with a dilution series of extract from PER-V5-expressing S2 cells. Immunoblots were performed with either a polyclonal PER antibody or a V5 antibody. The PER-V5 samples from S2 cells allowed us to compare the levels of CLK and PER in the off-DNA complex. By comparing the polyclonal PER signals from the IPs and the S2 cell extracts, we show that the amount of PER in the complex at ZT23 and ZT3 is approximately equal to the 1/8 dilution of PER-V5 S2 cell extract (red circles). When the 1/8 dilution of PER-V5 S2 cell extract is blotted with anti-V5 antibody, this band is approximately equal to the amount of CLK-V5 in the PER/CLK complex (red circles). Since the V5 antibody generates the same signal for the same molar amounts of every protein tagged with a single V5 epitope, we conclude that PER and CLK are roughly equimolar in the “off-DNA” complex.
Discussion
Circadian transcriptional repression is believed to occur by catalytic post-translational events in animal systems as well as Neurospora. We report here in the Drosophila model two different mechanisms that occur sequentially. First, the beginning of the repression phase is associated with PER binding to circadian promoters, probably via a PER–CLK interaction. The PER–DNA interaction likely inhibits CLK-mediated transcription despite persistent CLK DNA binding. This “on-DNA” phase is followed by the release of CLK from DNA and concomitant formation of a strong, close to 1:1 “off-DNA” PER–CLK complex with low affinity for DNA and in which most of CLK is sequestered.
The interaction of PER with DNA is prominent, as CLK-mediated transcription starts to decrease at ZT14 and is then maximal at ZT18 when the decrease in transcription (slope) is approximately maximal. The increase in on-DNA PER between ZT10 and ZT18 parallels the substantial, well-established rise in PER levels during these 8 h, and indicates that mass action may be sufficient to account for this increase (Fig. 5). The data therefore suggest that this increase in on-DNA PER affects the rate of transcription, and that possible effects of post-translational mechanisms on DNA-bound CLK or of chromatin modification on transcription are downstream from this PER–CLK ratio. Although the mechanism of transcriptional inhibition is not known, PER presumably recruits or potentiates corepressors, or it inhibits the recruitment or activity of coactivators.
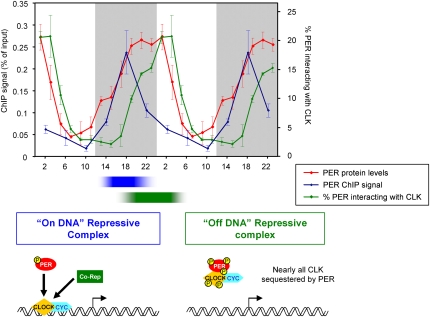
Figure 5.
PER represses CLK/CYC transcription activity by on-DNA and off-DNA mechanisms. PER represses CLK/CYC-mediated transcription by two different mechanisms that occur sequentially. First, PER represses CLK/CYC transcription activity by binding to circadian promoters (blue; PER ChIP signal on tim ebox) (Fig. 1D) and then by sequestering CLK off-DNA in a 1:1 PER:CLK complex (green; percentage of PER interacting with CLK) (Fig. 3C). These two events accommodate well the rhythmic levels of PER protein (red; levels quantified from two independent experiments). A schematic representation of these two mechanisms is depicted with the “on-DNA repressive complex” on the left and the “off-DNA repressive complex” on the right. See the text for more details.
The inability to assay a soluble PER–CLK interaction until ZT19 suggests that formaldehyde cross-linking captures earlier interactions of PER with DNA-bound CLK that are too weak to survive a standard soluble IP assay. We speculate that the in vivo stability of these early PER–CLK interactions may be enhanced by a high local concentration of CLK due to several adjacent DNA-binding sites, as indicated by the broad (∼4-kb) CLK-interacting region of DNA (Fig. 2B,D). The mixed cytoplasmic/nuclear localization of PER (Shafer et al. 2002) compared with the predominantly nuclear localization of CLK (Houl et al. 2006) also suggest a labile PER–CLK interaction before ZT18.
Between ZT18 and ZT22, there is a decrease in the association of CLK with DNA as well as a striking increase in the levels of an ∼1:1 soluble PER–CLK complex, suggesting that these two phenomena are mechanistically related (Fig. 5). This 1:1 PER–CLK complex presumably has a low DNA affinity, which largely accounts for the low transcription rates after ZT19. The formation of a stable stochiometric repression complex contrasts with the transient, phosphorylation-based repression mechanism in Neurospora. In this system, the repressor FRQ is present in nuclei at a much lower molar ratio than the activator WC complex (He and Liu 2005; Schafmeier et al. 2005; Hong et al. 2008).
As there are no striking increases in PER levels after ZT18, key qualitative changes may occur after this time; for example, the addition of other components and/or post-translational modifications. These changes presumably contribute to removing CLK from DNA and to creating the strong 1:1 PER–CLK complex in the late night–early morning with a greatly reduced affinity for circadian promoters. It has not escaped our attention that ZT18 is precisely when TIM goes from being predominantly cytoplasmic to being predominantly nuclear within l-LNvs (Shafer et al. 2002). This event may therefore contribute to the removal of CLK and PER from DNA. The post-translational modification possibility is supported by the mobility change of CLK from ZT17 to ZT19 (Supplemental Fig. 2), as well the lower mobility of both PER and CLK within the 1:1 stochiometric complex. We suggest, however, that the prior on-DNA PER–CLK complex is the substrate for these modifications, and is therefore upstream of phosphorylation events that might increase the stability of the off-DNA.
In conclusion, our data provide a new mechanistic view of PER-mediated transcriptional repression and emphasize the importance of the PER levels and the PER:CLK ratio. This includes the increases that occur on-DNA during the early night as circadian transcription is decreasing, as well as the ∼1:1 PER–CLK ratio that is found off-DNA in the late night–early morning when CLK DNA-binding affinity is at its nadir. Indeed, these two phases may be connected by the increasing ratio of PER–CLK on-DNA: It may dictate the circadian timing of the decrease in CLK DNA affinity, ultimately resulting in the departure from DNA of the stable PER–CLK complex. It is notable that a recent study in the mammalian system has described a prominent PER–CLK interaction that appears very important to repression of the CLK–BMAL1 complex (Chen et al. 2009). These new insights support the notion that PER acts as a stable complex component rather than catalytically to effect transcriptional repression in flies and, perhaps, also in mammals.
Materials and methods
Fly strains
The following flies were used: yw, per01, and yw;;WT dClk-V5 (Kadener et al. 2008).
S2 cell transfection
S2 cells were seeded in a six-well plate and maintained in 10% fetal bovine serum (Invitrogen) insect tissue culture medium (HyClone). Transfection was performed at 90% confluence according to the company's recommendations using Cellfectin (Invitrogen) and 100 ng of pAc:Per-V5 plasmid per well (McDonald et al. 2001). Two days post-transfection, S2 cells were lysed with RBS buffer and lysate was processed for Western blot analysis.
Western blotting
S2 cell lysates and fly head extracts were generated using a slightly modified RBS buffer (20 mM HEPES at pH 7.5, 50 mM KCl, 10% glycerol, 2 mM EDTA, 1% Triton X-100, 0.4% NP-40, 1 mM DTT, 1× protease inhibitor cocktail [Roche]) (Yu et al. 2006) and a homemade mix of phosphatase inhibitor consisting of 1 mM Na3VO4, 20 mM β-glycerophosphate, 0.1 μM okadaic acid, 25 mM NaF, and 10 mM sodium pyrophosphate. For S2 cell lysates, 250 μL of buffer were used for one well of a six-well plate. Fifty fly heads were homogenized on ice with 75 μL of buffer and using pellet pestle (South Jersey Precision Tool and Mold, Inc.). After high-speed centrifugation, lysates were mixed with 5× SDS buffer and boiled for 10 min. Each sample was loaded on a NuPAGE Novex 3%–8% Tris-Acetate gel (Invitrogen) and run following manufacturer's instructions. Gels were transferred on a nitrocellulose membrane using the iBlot Dry Blotting system (Invitrogen). Membranes were blocked and incubated with primary and secondary antibodies according to standard techniques. Anti-V5 (Sigma), rabbit anti-PER (Dembinska et al. 1997), rat anti-TIM (Zeng et al. 1996), guinea pig anti-DBT (gift from J.L. Price), and horseradish peroxidase-conjugated secondary antibodies (Amersham) were used. Peroxydase activity was detected with ECL reagent (Amersham) according to standard techniques.
Analysis of gene expression by real-time PCR
Total RNA was prepared from fly heads using Trizol reagent (Invitrogen) and DNase-treated using RQ1 DNase (Promega) according to the manufacturer's protocols. cDNA derived from this RNA (using Invitrogen SuperScript II and random primers) was used as a template for quantitative real-time PCR performed with the Corbett Research Rotor-Gene 3000 real-time cycler. The PCR mixture contained Platinum Taq polymerase (Invitrogen), optimized concentrations of SYBR green (Invitrogen), and the corresponding primers forward tim mRNA and pre-mRNA (CAGCACTGAAACTATAACACGATC), reverse tim mRNA (GAGGTACTTCTTAGCCATTTGC), and reverse tim pre-mRNA (GTGTTAGGTCAGGTCTAAGC). Rp49 primers have been described elsewhere (Kadener et al. 2008). Cycling parameters were 3 min at 95°C, followed by 40 cycles of 30 sec at 95°C, 45 sec at 55°C, and 45 sec at 72°C. Fluorescence intensities were plotted versus the number of cycles by using an algorithm provided by the manufacturer. mRNA levels were quantified using a calibration curve based on dilution of concentrated cDNA. mRNA values from heads were normalized to that from ribosomal protein 49 (rp49).
IP
For each IP, 100 μL of fly heads were homogenized in 3–4 vol of RBS buffer (see Western blotting method). After high-speed centrifugation, ∼75% of each extract (the other ∼25% was used as input) was incubated overnight with either 20 μL of anti-V5 agarose beads (Sigma) or 3 μL of rabbit anti-PER antibody (Dembinska et al. 1997) for 3–4 h and then overnight with protein G Plus/protein A agarose beads (Calbiochem). Beads were then centrifuged at 1000g for 20 sec and the supernatant was kept on the side. Beads were then washed three times for 10 min with RBS buffer, mixed with 5× SDS buffer, and boiled for 10 min. Samples were then processed for Western blotting as described above, side by side with the input.
ChIP
ChIP protocol was based on a previous protocol (Yu et al. 2006) with modifications. yw;;Clk-V5 flies were entrained for 3 d in LD conditions and then collected every 4 h. To provide background binding controls, ChIPs were also performed with yw (V5 antibody) and Per01 flies (per antibody). One milliliter of fly heads was homogenized in 3 mL of NEB buffer (10 mM HEPES-Na at pH 8.0, 10 mM NaCl, 0.1 mM EGTA-Na at pH 8.0, 0.5 mM EDTA-Na at pH 8.0, 1 mM DTT, 0.5% Tergitol NP-10, 0.5 mM Spermidine, 0.15 mM Spermine plus protease inhibitor tablets [Roche]) for a total of 30 min (2-min homogenization 10 times, 1 min on ice 10 times). Homogenate was dumped into a 70-μm cell strainer placed in a 50-mL falcon tube and centrifuged at 300g for 1 min. Filtered homogenate was centrifuged at 6000g for 10 min. The nuclei-containing pellets were resuspended in 1 mL of NEB and centrifuged at 20,000g for 20 min on sucrose gradient (0.65 mL of 1.6 M sucrose in NEB, 0.35 mL of 0.8 M sucrose in NEB). The pellet was resuspended in 1 mL of NEB and 11% formaldehyde (diluted in Schneider's media; Sigma) was added to a final concentration of 1%. Nuclei were cross-linked for 10 min at room temperature before cross-linking was quenched by adding 1/10 vol of 1.375 M glycine. The nuclei were collected by centrifugation at 6000g for 5 min. Nuclei were washed twice in 1 mL of NEB and resuspended in 450 μL of Sonication buffer (10 mM HEPES-Na at pH 7.5, 2 mM EDTA at pH 8.0, 1% SDS, 0.2% Trition X-100, 0.5 mM Spermidine, 0.15 mM Spermine). Nuclei were sonicated using a Fisherbrand Sonic Dismembranator at setting 2 (57 W) four times for 15 sec on and 2 min on ice. Sonicated nuclei were centrifuged at 15,000g for 10 min and frozen at −80°C in 150-μL aliquots. The majority of the sonicated chromatin was ∼500- to 1000 base pairs (bp) in length.
Twenty-five microliters of sonicated chromatin were removed for the input sample. The remaining 125 μL of chromatin were diluted in 1.1 mL of IP buffer (50 mM HEPES/KOH at pH 7.6, 2 mM EDTA, 1% Triton, 0.1% NaDeoxycholate in PBS). Samples were rotated overnight at 4°C after adding antibodies: 15 μL of anti-V5 antibody (Abcam ab9116), 15 μL of anti-PER antibody (Dembinska et al. 1997), or 1 μL of anti-Pol II antibody (gift from A. Greenleaf). Protein G-Sepharose beads (Zymed) were blocked overnight in 0.1 mg/mL yeast tRNA and 1 mg/mL BSA in IP buffer. After overnight incubation, the beads were washed once in IP buffer, added to the chromatin/antibody mixture, and then incubated for an additional 2 h at 4°C.
Beads were spun down at 10,000 rpm for 20 sec and were washed once in 1.5 mL of ChIP Wash buffer (50 mM HEPES-KOH at pH 7.6, 1 mM EDTA, 1% Triton, 0.1% NaDeoxycholate, 0.1% Sarkosyl, 0.1% BSA, 0.5 M KCl in PBS). Beads were resuspended in 1 mL of ChIP Wash buffer and rotated for 30 min at 4°C. Beads were then washed once in Li Wash Buffer (10 mM Tris-Cl at pH 8.0, 0.25 M LiCl, 0.5% NP40, 0.5% NaDeooxychoalte, 1 mM EDTA) and once in cold TE (pH 8.0) before being eluted with 150 μL of ChIP Elution buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% SDS, 1 mM DTT, 0.1 mg/mL Proteinase K). ChIP Elution buffer (150 μL) was also added to the input sample. Both IP and input samples were incubated for 2 h at 37°C. The sepharose beads were removed from the IP samples and then all samples were decross-linked overnight at 65°C. DNA was isolated from the samples using PCR purification kit (Qiagen).
Dilutions (1:10) of all samples were used in qPCR. Primers for TIM (TimE1) and PER [PerE5(CRS)] were described previously (Taylor and Hardin 2008). qPCR was repeated at least twice and the average results are presented as the percentage of the input.
Drosophila tiling array
Probes for GeneChip Drosophila Tiling Array 2.0 (Affymetrix) were made according to the protocol provided by Affymetrix. Ten microliters of the IP samples and 1 μL of input samples were amplified to generate probes. qRT–PCR was used to verify that the enrichment in the IP sample was maintained through the amplification process. The arrays were hybridized, washed, and scanned according to the manufacturer's recommendations. Two independent ChIPs were hybridized to tiling arrays for each time point. In addition, two samples from a PER ChIP in a per01 background were also used as probes for tiling arrays. Data were analyzed using the model-based analysis of tiling arrays (MAT) algorithm (Johnson et al. 2006) and were converted to a linear scale for easier viewing on the Integrated Genome Browser from Affymetrix. The output values, called MAT scores, are influenced by the overall background and represent a probability of DNA binding rather than signal intensity. Because the overall background is higher in the PER ChIP, the MAT scores are lower than in the CLK-V5 ChIP despite roughly similar signal intensities.
Due to the possibility of a background signal with the polyclonal PER antibody, we subtracted the PER ChIP signal generated in a per01 flies from that generated in yw;;Clk-V5 flies.
Acknowledgments
We thank our colleagues in the Rosbash laboratory for helpful discussions and comments on the manuscript, especially Emi Nagoshi, Weifei Luo, Yue Li, and Yuhua Shang, as well as our ex-laboratory mates Sebastian Kadener and Pipat Nawathean. We also thank Jeffrey L. Price for the gift of the anti-DBT antibody, Arno L. Greenleaf for the gift of the anti-Pol II antibody, Frank Weber and Paul Hardin for sharing unpublished information, and Mike Marr for helpful discussions. We are also grateful to Danielle Saly for technical help. J.S.M was a recipient of an EMBO long-term fellowship. This work was supported in part by NIH grants NS44232 and NS45713 to M.R.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1883910.
Supplemental material is available at http://www.genesdev.org.
References
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: The genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardente H, Fortier EE, Martineau V, Cermakian N. Cryptochromes impair phosphorylation of transcriptional activators in the clock: A general mechanism for circadian repression. Biochem J. 2007;402:525–536. doi: 10.1042/BJ20060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembinska ME, Stanewsky R, Hall JC, Rosbash M. Circadian cycling of a PERIOD-β-galactosidase fusion protein in Drosophila: Evidence for cyclical degradation. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol. 2005;15:R714–R722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- He Q, Liu Y. Molecular mechanism of light responses in Neurospora: From light-induced transcription to photoadaptation. Genes & Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Shu H, Cheng P, Chen S, Wang L, Liu Y. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem. 2005;280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Hong CI, Ruoff P, Loros JJ, Dunlap JC. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes & Dev. 2008;22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houl JH, Yu W, Dudek SM, Hardin PE. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J Biol Rhythms. 2006;21:93–103. doi: 10.1177/0748730405283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol. 2007;27:5014–5028. doi: 10.1128/MCB.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes & Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Lee C, Gorbacheva VY, Chernov MV, Antoch MP. Post-translational regulation of circadian transcriptional CLOCK(NPAS2)/BMAL1 complex by CRYPTOCHROMES. Cell Cycle. 2006;5:890–895. doi: 10.4161/cc.5.8.2684. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Oyama T, Kondo T. Regulation of circadian clock gene expression by phosphorylation states of KaiC in cyanobacteria. J Bacteriol. 2008;190:1691–1698. doi: 10.1128/JB.01693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Promoter-proximal Pol II: When stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brunner M, Schafmeier T. Activity of the circadian transcription factor White Collar Complex is modulated by phosphorylation of SP-motifs. FEBS Lett. 2009;583:1833–1840. doi: 10.1016/j.febslet.2009.04.042. [DOI] [PubMed] [Google Scholar]
- Schafmeier T, Haase A, Kaldi K, Scholz J, Fuchs M, Brunner M. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Schafmeier T, Kaldi K, Diernfellner A, Mohr C, Brunner M. Phosphorylation-dependent maturation of Neurospora circadian clock protein from a nuclear repressor toward a cytoplasmic activator. Genes & Dev. 2006;20:297–306. doi: 10.1101/gad.360906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T, Diernfellner A, Schafer A, Dintsis O, Neiss A, Brunner M. Circadian activity and abundance rhythms of the Neurospora clock transcription factor WCC associated with rapid nucleo-cytoplasmic shuttling. Genes & Dev. 2008;22:3397–3402. doi: 10.1101/gad.507408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS. Finding new clock components: Past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T, Isojima Y, van der Horst GT, Takei K, Nagai K, Takamatsu K. Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells. 2003;8:973–983. doi: 10.1046/j.1365-2443.2003.00686.x. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Hirayama J, Isojima Y, Nagai K, Norioka S, Takamatsu K, Sassone-Corsi P. CK2α phosphorylates BMAL1 to regulate the mammalian clock. Nat Struct Mol Biol. 2009;16:446–448. doi: 10.1038/nsmb.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Hardin PE. Rhythmic E-box binding by CLK–CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol. 2008;28:4642–4652. doi: 10.1128/MCB.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Snyder M. RNA polymerase II stalling: Loading at the start prepares genes for a sprint. Genome Biol. 2008;9:220. doi: 10.1186/gb-2008-9-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitane H, Takao T, Satomi Y, Du NH, Okano T, Fukada Y. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol. 2009;29:3675–3686. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes & Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]