Abstract
The Polycomb group proteins foster gene repression profiles required for proper development and unimpaired adulthood, and comprise the components of the Polycomb-Repressive Complex 2 (PRC2) including the histone H3 Lys 27 (H3K27) methyltransferase Ezh2. How mammalian PRC2 accesses chromatin is unclear. We found that Jarid2 associates with PRC2 and stimulates its enzymatic activity in vitro. Jarid2 contains a Jumonji C domain, but is devoid of detectable histone demethylase activity. Instead, its artificial recruitment to a promoter in vivo resulted in corecruitment of PRC2 with resultant increased levels of di- and trimethylation of H3K27 (H3K27me2/3). Jarid2 colocalizes with Ezh2 and MTF2, a homolog of Drosophila Pcl, at endogenous genes in embryonic stem (ES) cells. Jarid2 can bind DNA and its recruitment in ES cells is interdependent with that of PRC2, as Jarid2 knockdown reduced PRC2 at its target promoters, and ES cells devoid of the PRC2 component EED are deficient in Jarid2 promoter access. In addition to the well-documented defects in embryonic viability upon down-regulation of Jarid2, ES cell differentiation is impaired, as is Oct4 silencing.
Keywords: Polycomb, chromatin, Histone methylation, Jarid2
Polycomb group (PcG) proteins ensure that the gene expression pattern that came about during cell fate decisions in the developmental stage are maintained in the adult (Schuettengruber et al. 2009). They are involved in various cellular processes other than cellular identity, such as proliferation and pluripotency (Schuettengruber et al. 2009). PcG proteins form several complexes that function mostly through the regulation of chromatin structure. Polycomb-Repressive Complex 2 (PRC2) is composed of four core components: Ezh2, Eed, Suz12, and RbAp46/48. Ezh2 catalyzes di- and trimethylation of the Lys 27 residue of histone H3 (H3K27me2/3), and this activity requires that Ezh2 be in complex with the other PRC2 components (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). H3K27me3 is associated with repressed chromatin states, and genome-wide approaches revealed that H3K27me3 is widely distributed among genes encoding developmental regulators (Bernstein et al. 2006; Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006; Schwartz et al. 2006; Squazzo et al. 2006).
Although H3K27me3 is found at up to 10% of the annotated genes in embryonic stem (ES) cells (Lee et al. 2006), only a small subset exhibits activation upon disruption of PRC2 through targeting of one of its core components using knockdown or knockout strategies (Chamberlain et al. 2008). Of the promoters that are enriched for H3K27me3 in wild-type ES cells, 27% are transcriptionaly up-regulated more than twofold in Eed−/− ES cells (Chamberlain et al. 2008). Moreover, deletion of Ezh2 (conditional knockout) promotes differentiation of epidermal progenitor cells through the derepression of genes that are normally expressed at later stages of differentiation (Ezhkova et al. 2009). In the case of mouse ES cells, PRC2 is dispensable for maintaining pluripotency; however, Eed−/− ES cells were more prone to differentiation (Boyer et al. 2006). The absence of Suz12 prevented differentiation, and Suz12−/− ES cells formed disorganized embryoid bodies (EBs) that lacked a proper endodermal layer (Pasini et al. 2007). In contrast, another study failed to observe spontaneous differentiation of Eed−/− ES cells, and these cells were able to contribute to all tissue lineages (Chamberlain et al. 2008). It is clear, however, that PRC2 is required for proper reprogramming of gene expression during differentiation. Whether it promotes or prevents this process might depend on the type of cells studied and/or the target and type of strategy employed for disrupting PRC2. The outcome might reflect a balance between abnormally expressed transcription factors that maintain pluripotency and transcription factors that drive ES cells toward a defined lineage.
Despite extensive studies, the means by which PRC2 can access its targets in mammals is still undetermined. None of the PRC2 core components are able to recognize a specific DNA sequence. Only AEBP2, a protein that was reported to copurify with PRC2 (Cao et al. 2002), has a Zinc finger that could contribute to PRC2 recruitment (Kim et al. 2009). We found that the PRC2 component Eed recognizes several different histone marks, all of which contain trimethylated lysines and are associated with repression, including H3K27me3 (Margueron et al. 2009). Yet, it is when Eed binds to H3K27me3 that Ezh2 enzymatic activity is up-regulated through an allosteric effect (Margueron et al. 2009). The ability of Eed to recognize trimethylated lysine is important for the transmission of the H3K27me3 mark, yet it appears dispensable for PRC2 recruitment to its target genes. While we and others have shown that PHF1 or its Drosophila homolog, Pcl, are important for the enzymatic activity of PRC2, these factors did not seem to be determinants for PRC2 recruitment (Nekrasov et al. 2007; Cao et al. 2008; Sarma et al. 2008).
In this study we sought such a factor or factors that might associate with PRC2 and facilitate its access to chromatin. We identified Jarid2 as an associating partner of PRC2. Jarid2 encodes a nuclear protein essential for mouse embryogenesis, including neural tube formation (Takeuchi et al. 1995) and heart development (Takeuchi et al. 1999; Lee et al. 2000; Toyoda et al. 2003). Its homolog in Drosophila is dJMJ (CG3654), mutation of which suppresses position effect variegation of the T(2;3)SbV rearrangement (Sasai et al. 2007).
Jarid2 contains a DNA-binding domain, denoted as the AT-rich interaction domain (ARID); a zinc finger domain; a jumonji N (JmjN) domain; and a JmjC domain (Jung et al. 2005b; Takeuchi et al. 2006). Unlike the other members of the Jarid family of proteins, Jarid2 has a long N-terminal moiety devoid of characterized domains. Many JmjC domain-containing proteins have been shown to catalyze lysine demethylation, but the residues required for iron and ascorbate binding, essential for demethylation activity, are not conserved in the JmjC domain of Jarid2 (Takeuchi et al. 2006). Jarid2 was shown to bind to the promoter and repress the expression of cyclin D1, and its overexpression negatively regulates cell proliferation (Toyoda et al. 2003; Shirato et al. 2009). Jarid2 was also reported to interact physically with Nkx2.5, GATA4, ZF496, Rb, and the myocyte enhancer factor 2 (MEF2) (Kim et al. 2004, 2005; Jung et al. 2005a; Mysliwiec et al. 2007), and its repressive activity was suggested as being mediated by the H3K9 methyltransferase G9a (Shirato et al. 2009). We found, however, that Jarid2 interacts with PRC2, stimulates its H3K27 methylation activity, has a DNA-binding activity that is not dependent on the ARID domain only, and facilitates PRC2 recruitment to its target genes. Moreover, Jarid2 is required for PRC2-repressive activity in ES cells, as evidenced by their impaired differentiation in its absence.
Results
Jarid2 is a component of PRC2
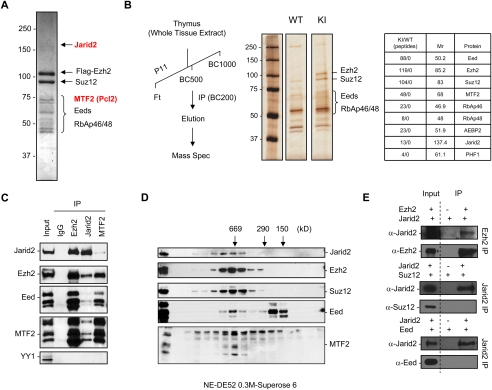
As a first step toward understanding how PRC2 accesses its target genes in mammalian cells, we sought the identities of proteins with which it might associate. The PRC2 complex was purified from 293F cells that overexpress the PRC2 component Ezh2 containing an N-terminal Flag tag. Ezh2 was immunoprecipitated from nuclear extract using M2 beads and was eluted with Flag peptides after extensive washes in a stringent buffer (500 mM KCl). Coomassie blue-stained SDS-PAGE revealed the presence of two proteins that do not correspond to the core PRC2 components (Fig. 1A). Mass spectrometric analysis allowed the identification of Jarid2 and MTF2. The interactions between PRC2 and Jarid2 or MTF2 were confirmed using cells lines that overexpress Flag-tagged versions of each of these additional PRC2-associated components (Supplemental Fig. S1).
Figure 1.
Jarid2 interacts with the PRC2 complex. (A) Flag tag purification of Ezh2 from 293F cells. After immunoprecipitation (IP), Ezh2 and associated polypeptides were eluted with Flag peptide, loaded onto SDS-PAGE, and stained with Coomassie blue. The proteins identified by mass spectrometric analyses are indicated on the right. (B) Flag tag purification of Ezh2 from mouse thymus. (Left) Purification scheme. (Center) Silver-stained SDS-PAGE of Flag-IP eluate. Control wild-type and knock-in samples were subjected to mass spectrometry. Results are presented on the right, after subtraction of peptides that were present in both control and knock-in samples. (C) Immunoprecipitation from ES cell nuclear extract using antibodies against Ezh2, Jarid2, or MTF2 and probed with the antibodies indicated on the left. IgG served as control. (D) Nuclear extracts of ES cells were fractionated on a DE52 column followed by a Superose 6 gel filtration column. The fractions were analyzed by Western blot. Molecular weight standards are shown on top. (E) Western analysis using the antibodies shown on the left of immunoprecipitations performed with antibodies shown on the right after recombinant Jarid2 was mixed with recombinant Ezh2, Suz12, or Eed protein.
With respect to MTF2, there are three paralogs in mammals that exhibit conservation with Drosophila Pcl: PHF1, MTF2, and PHF19. All three contain two PHD domains and one tudor domain (Supplemental Fig. S2A). We reported previously that PHF1 interacts with PRC2 and is required for efficient trimethylation of H3K27 (Sarma et al. 2008). Given its conservation with PHF1, we expected that MTF2 would exhibit a similar role, and thus its function was not analyzed further. Instead, we focused on the function of Jarid2 that contains a Jmjc domain and an ARID domain (Supplemental Fig. S2B). However, while Jmjc domain-containing proteins were reported to catalyze histone demethylation, Jarid2 is devoid of the conserved amino acids required for such activity (Supplemental Fig. S2C; Takeuchi et al. 2006). Moreover, attempts to demonstrate that Jarid2 has histone demethylase activity failed, and it does not exhibit dominant-negative activity against the H3K4 demethylase SMCX (data not shown).
To ensure that the proteins identified as being associated with PRC2 were not specific to conditions of Ezh2 overexpression or a particular cell line, we immunopurified PRC2 from the tissue of a whole organism. We employed mice containing a knock-in of the Ezh2 gene encoding a Flag tag fused to the C terminus of the protein (Supplemental Fig. S3A) and focused on the thymus of 1-mo-old mice, as adult tissues with a low replication rate barely express Ezh2 (Margueron et al. 2008). Whole-tissue extracts were prepared and first loaded onto a phosphocellulose column (P11) (Fig. 1B; Supplemental Fig. S3B) followed by Flag tag affinity purification. Eluates were analyzed by SDS-PAGE using silver staining and mass spectrometry (Fig. 1B), and by Western blotting (Supplemental Fig. S3C). As expected, we identified the four core PRC2 components (Ezh2, Suz12, Eed, and RbAp46/48) in the affinity-purified sample, but, more importantly, we found MTF2 and Jarid2. We also observed AEBP2 and PHF1, two proteins described previously as associating with the core PRC2 complex (Cao et al. 2002, 2008; Nekrasov et al. 2007; Sarma et al. 2008). Thus, the association of Jarid2 and MTF2 (and of AEBP2 and PHF1) with PRC2 is not due to the transformed phenotype of tissue culture cells and is evident within a developing organism.
Considering that Jarid2 and MTF2 are both highly expressed in ES cells (Supplemental Fig. S4), we next tested if their interaction with PRC2 occurs endogenously using reciprocal immunoprecipitation (Fig. 1C). While an antibody specific to Jarid2 pulled down Ezh2, Eed, and Suz12, an antibody specific to Ezh2 pulled down Jarid2, further confirming the interaction between Jarid2 and PRC2. Similarly, an antibody specific to MTF2 coprecipitated the entire PRC2 complex. Of note, in neither case was YY1 detected, indicating that, in contrast to the case in Drosophila, mammalian PRC2 is not targeted to chromatin via interaction with this sequence-specific transcription factor.
To ascertain if the association between Jarid2 and PRC2 is stable, nuclear extracts derived from ES cells naturally expressing relatively high levels of Jarid2 were fractionated by DE52, followed by Superose 6 gel filtration chromatography. Western blotting revealed a major peak at ∼650 kDa for Ezh2 and Suz12, and also for Jarid2 and MTF2 (Fig. 1D). Thus, both MTF2 and Jarid2 copurified with the Ezh2 and Suz12 core components of PRC2, indicating that each is a hitherto unrecognized component of the PRC2 complex.
To determine which of the PRC2 components is responsible for interaction with Jarid2, we performed immunoprecipitation experiments using purified recombinant proteins expressed from baculoviruses. The results showed that it is the Ezh2 component of PRC2 that interacts with Jarid2 (Fig. 1E). However, it should be noted that, under less stringent conditions, we also observed interactions between Suz12, Eed, and Jarid2 (data not shown).
Jarid2 regulates PRC2 activity
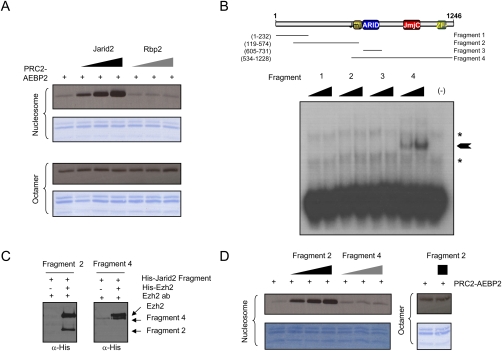
As PRC2 presumably functions through the HKMT activity associated with its Ezh2 component, we analyzed whether Jarid2 could modulate this activity. We first incubated PRC2 with increasing amounts of recombinant Jarid2 (shown in Supplemental Fig. S5A) and observed a dose-dependent increase in the methylation of histone H3 (Fig. 2A), although Jarid2 by itself did not exhibit HKMT activity (Supplemental Fig. S5B). Rbp2 (Jarid1A/KDM5A) is another member of the Jarid family of proteins that has been reported previously to interact with PRC2 (Pasini et al. 2008). It shares some homology with the C-terminal part of Jarid2, but lacks homology with the Jarid2 N terminus (Supplemental Fig. S2B). Unlike Jarid2, Rbp2 was ineffectual in regulating the HKMT activity of Ezh2 comprising PRC2 (Fig. 2A). We also analyzed whether Jarid2 could regulate other HKMTs, and observed no effect on the enzymatic activity of the nucleosomal-specific PR-Set7, or of Suv4-20h1 or Suv39h1 (Supplemental Fig. S5C). Importantly, Jarid2 enhanced PRC2 enzymatic activity toward H3K27 using oligonucleosomes, but not octamers, as substrates (Fig. 2A). HKMT assays were then probed using specific antibodies for H3K27me1/2/3 (Supplemental Fig. S5D), and we observed that Jarid2 stimulated PRC2-mediated mono- and dimethylation of H3K27 in vitro. Although we assessed the contribution of Jarid2 toward H3K27me3, we failed to detect this modification in vitro. This phenomenon—whereby the activity of PRC2 catalyzes H3K27me1/2, but not detectable H3K27me3, in vitro, while PRC2 results in the formation of H3K27me2/3, but not detectable me1, in vivo—is consistent with our previous findings (Sarma et al. 2008).
Figure 2.
Jarid2 regulates PRC2-Ezh2 enzymatic activity. (A) HKMT assay with PRC2-Ezh2 alone or in the presence of increasing amounts of Jarid2 or Rbp2. Recombinant oligonucleosomes (top) or octamers (bottom) were used as substrate. (B, top) Schematic representing the known Jarid2 domains. Also shown are the regions of Jarid2 protein that were expressed in bacteria. (Bottom) Band shift assay in the presence of increasing amounts of each of the Jarid2 fragments using the radiolabeled DNA probes selected by SELEX. The asterisks indicate nonspecific signals present in the free probes. The arrow indicates the specific band shift. (C) Coimmunoprecipitations of purified Ezh2 protein and Jarid2 fragments, as indicated. Immunoprecipitation was performed using the Ezh2 antibody. (D) HKMT assay with PRC2-Ezh2 alone or in the presence of increasing amounts of fragment 2 or fragment 4 of Jarid2 using oligonucleosomes (left) or octamers (right) as substrate.
Considering that Jarid2 modulates PRC2 activity only on nucleosomal substrates, and that two domains of Jarid2 can potentially bind DNA, we next investigated whether Jarid2 itself could bind directly to DNA. We analyzed Jarid2 DNA-binding properties by the SELEX method (systematic evolution of ligands by exponential enrichment) using full-length Jarid2, followed by band shift assays. This procedure indicated that, after six rounds of selection, Jarid2 shifted the selected DNA sequences more efficiently (data not shown). Sequencing of the selected DNA did not reveal any consensus other than a slight bias toward GC-rich sequences, suggesting that Jarid2 binds DNA without clear specificity. We next analyzed which domain of Jarid2 binds to DNA through the generation of deletion mutants, as shown in Figure 2B, and found that the C terminus of Jarid2 (mutant 4), but not the ARID domain alone (mutant 3), could shift the SELEX selected probes (Fig. 2B). This result suggests that the Jarid2 C5HC2 zinc finger, either alone or in combination with other motifs (including the ARID domain), mediates the DNA-binding properties of Jarid2.
We then employed these deletion mutants to ascertain the Jarid2 domains that interact with PRC2 and the domains that stimulate its HKMT activity. We found that fragments 2 and 4 interact with Ezh2 (Fig. 2C), and that fragment 2 containing the Jarid2 N terminus fully recapitulated the ability of full-length Jarid2 to stimulate Ezh2 activity associated with PRC2. Altogether, we conclude that Jarid2 regulates PRC2-Ezh2 activity independently of its ability to bind DNA.
Artificial recruitment of Jarid2 results in Ezh2 recruitment and increased levels of H3K27me2/3
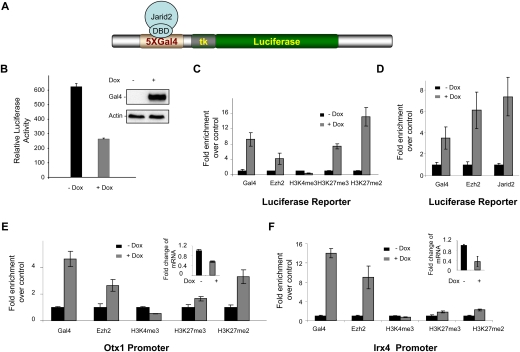
To gauge the functional import of Jarid2/PRC2 interaction, we assayed for the consequences of artificially recruiting Jarid2 to chromatin. 293 T-Rex cells containing the luciferase reporter gene expressed under the control of the TK promoter with five Gal4 DNA-binding sites were stably transfected with plasmid that expresses Gal4-Jarid2 as a function of doxycyclin induction (Fig. 3A). Gal4-Jarid2 expression led to decreased reporter gene expression (Fig. 3B). Chromatin immunoprecipitation (ChIP) results with respect to the luciferase promoter showed that this artificial recruitment of Jarid2 resulted in Ezh2 recruitment and increased H3K27me2/3 levels, while levels of H3K4me3 were only slightly diminished (Figs. 3C). Moreover, when cells were instead stably transfected with plasmid encoding Gal4-Ezh2, endogenous Jarid2 was recruited to the luciferase promoter along with Gal4-Ezh2 in a doxycyclin-dependent manner (Fig. 3D). This profile was not specific to the luciferase promoter, as evidenced by a similar outcome at the promoters of two endogenous genes that are known PRC2 targets, Otx1 and lrx4 (Fig. 3E,F, respectively). Upon Gal4-Jarid2 overexpression, Ezh2 and H3K27me2/3 levels were elevated at these promoters, and, consistent with repression of the luciferase reporter, RT–PCR results showed that both genes were repressed upon doxycyclin induction of Gal4-Jarid2 (Fig. 3E,F).
Figure 3.
Jarid2 recruits Ezh2 to an artificial promoter. (A) Schematic of the luciferase reporter under the control of the TK promoter, with Gal4 DNA-binding sites upstream. (B) Luciferase activity was measured 24 h after doxycyclin induction of the stably integrated transgene encoding Jarid2 fused to the Gal4 DNA-binding domain (Gal4-Jarid2). The values were normalized to the amount of protein. Error bars represent SD of three independent experiments. (Inset) Gal4-Jarid2 expression was checked by Western blot. Actin was used as a loading control. (C) ChIP experiments were performed before (−Dox) and after (+Dox) Gal4-Jarid2 induction. Chromatin was immunoprecipitated with antibodies against the proteins indicated at the bottom. DNA enrichment was analyzed at the luciferase transgene. Results are presented as fold of enrichment over control. Error bars represent SD of three independent experiments. (D) ChIP experiments were performed with or without induction of the stably integrated transgene encoding Gal4-Ezh2. DNA enrichment was analyzed at the luciferase transgene. (E,F) Same as C, but DNA enrichments were analyzed at the endogenous Otx1 (E) and Irx4 (F) promoters. Insets show the levels of Otx1 and Irx4 mRNAs, normalized to those of GADPH.
Jarid2, MTF2, and Ezh2 bind a common set of target genes in ES cells
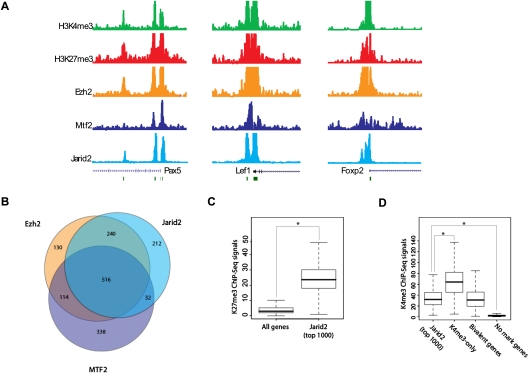
Given that Jarid2 is highly expressed in ES cells, as is also the case for some other components of PRC2, and that Jarid2 and Ezh2 are interdependent for their respective recruitment, we next compared their endogenous target genes in ES cells using ChIP coupled to high-throughput sequencing (ChIP-seq) (Fig. 4A). As we expected, there was considerable overlap, as evidenced by >50% of the 1000 genes scored having Jarid2/Ezh2/MTF2 in common and >75% when considering just Ezh2 and Jarid2 (Fig. 4B). Moreover, comparison with ChIP-seq data for the PRC2-mediated H3K27me3 modification exhibited a considerable enrichment for this mark among Jarid2 targets (Fig. 4C).
Figure 4.
Jarid2 binds to PRC2 target cenes in ES cells. (A) ChIP-seq tracks showing H3K4me3, H3K27me3, Ezh2, Mtf2, and Jarid2 at three representative genes in mouse ES cells. (B) Venn diagram showing overlap among the top 1000 gene promoters occupied by Ezh2, Mtf2, or Jarid2, respectively. (C) Box plot comparing H3K27me3 ChIP-seq signals for all genes and for the top 1000 Jarid2 targets in mouse ES cells. (*) P < 0.001. (D) Box plot showing H3K4me3 ChIP-seq signals for the top 1000 Jarid2 targets, for genes enriched for H3K4me3 but not H3K27me3, for “bivalent” genes (enriched for both H3K4me3 and H3K27me3), and for those genes that do not show a significant signal for either H3K4me3 or H3K27me3 in mouse ES cells. (*) P < 0.001. Significance was scored using the Student t-test.
Genes in ES cells contain so-called bivalent domains that comprise both transcriptionally repressive (H3K27me3) and transcriptionally permissive (H3K4me3) modifications (Bernstein et al. 2006). These domains are presumably primed for appropriate signals that are determinants to the requisite transcriptional outcome. Interestingly, on average, H3K4me3 enrichment among Jarid2 target genes is comparable with that observed for bivalent domains and lower than that reported for genes targeted by only H3K4me3 (Fig. 4D), as expected given their high enrichment for H3K27me3.
Depletion of Jarid2 partially impairs PRC2 recruitment
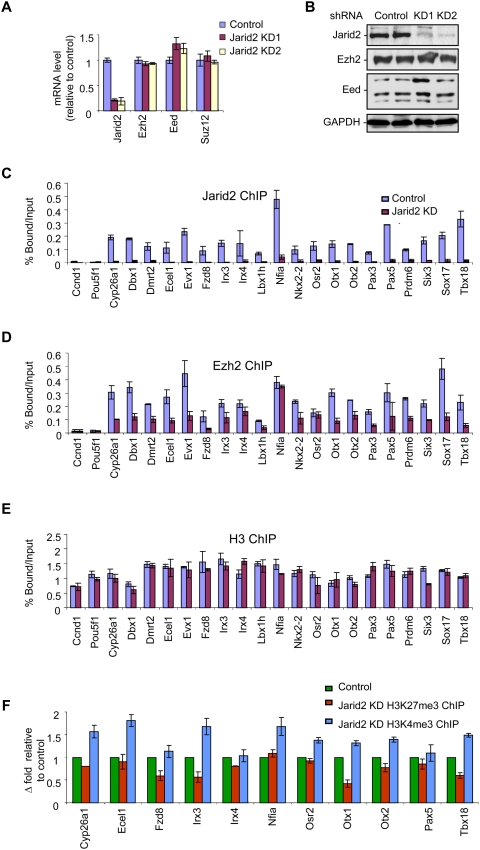
We next investigated the effects, if any, of Jarid2 depletion in vivo with respect to Ezh2 recruitment to endogenous loci. To this end, we established stable ES cell lines in which Jarid2 expression was markedly decreased by shRNA, as evidenced at the mRNA and the protein levels (Fig. 5A,B, respectively), while that of other PRC2 components, Ezh2 and Eed, or the control GADPH was unaffected. We performed manual ChIP analyses and confirmed that Jarid2 and Ezh2 do co-occupy all of the PRC2 target genes that we analyzed (Fig. 5C,D, respectively). Overall, we observed a good correlation between Ezh2 and Jarid2 enrichment at the genes analyzed. As expected, Jarid2 was barely detectable at these genes in the case of cells expressing Jarid2-specific shRNA (Fig. 5C). Of note, ChIP data derived instead using an antibody against Ezh2 showed that Jarid2 depletion resulted in a marked decrease (on average, 50%) in Ezh2 enrichment (Fig. 5D), while a similar analysis for histone H3 showed its presence was unperturbed (Fig. 5E). Interestingly, although there was a tendency for H3K27me3 levels to be decreased and H3K4me3 to be increased at some target genes, these effects appeared to be gene-specific (Fig. 5F). This observation likely illustrates that PRC2 activity is regulated in multiple ways, such that interference with only one such regulatory mode results in moderate and gene-specific changes in H3K27me3. In agreement with this result, knockdown of Jarid2 did not significantly affect the global levels of H3K27me3 (Supplemental Fig. S6).
Figure 5.
Jarid2 contributes to PRC2 recruitment to its target genes. (A,B) Two clones in which Jarid2 was stably knocked down were compared with the parental cell line with respect to the levels of mRNA (A) and protein (B) of PRC2 components. mRNA levels were normalized to those of GADPH (A) and GADPH served as a loading control for the Western blot (B). (C–F) ChIP experiments comparing the control parental ES cells versus ES cells in which Jarid2 was stably knocked down. Chromatin was immunoprecipitated with either Jarid2 (C), Ezh2 (D), H3 (E), or H3K27me3 and H3K4me3 (F) antibodies. (C–E) Results are represented as percent of input. Error bars represent SD of triplicates. (F) Results are represented as fold change over control. Error bars represent SD of triplicates.
Jarid2 recruitment in vivo is dependent on the integrity of PRC2
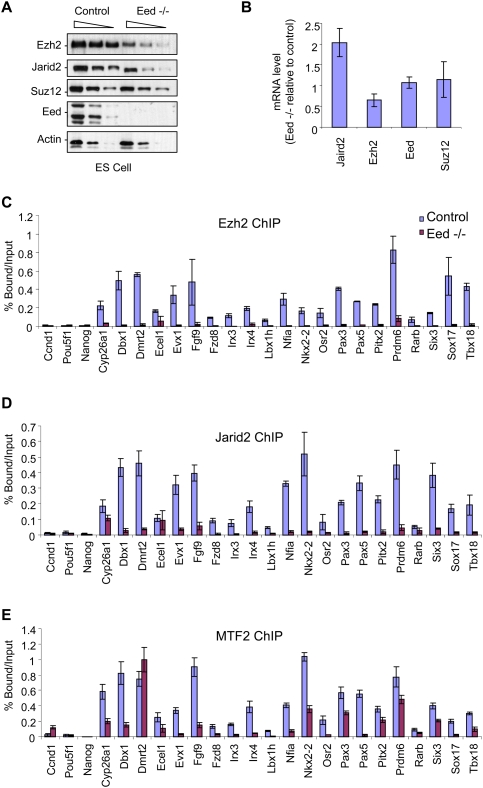
As our results thus far indicated that Jarid2 and the core PRC2 component Ezh2 interact, and that each is effective in recruiting the other, we next investigated their ability to access their common endogenous target genes when another core component of PRC2 is depleted in vivo. We used previously characterized EED-null ES cells (L196P mutation, null mutation) (Morin-Kensicki et al. 2001). In this model, the absence of Eed protein is associated with reduced levels of Ezh2 protein (around sixfold), without a significant change in Suz12 protein accumulation (Fig. 6A). Interestingly, we also observed a decrease in Jarid2 protein (twofold), but not at the mRNA level (Fig. 6A,B, respectively), suggesting that, similar to the case of Ezh2, the integrity of the PRC2 complex might be important in stabilizing the Jarid2 protein.
Figure 6.
PRC2 is required for Jarid2 targeting. (A,B) The levels of protein (A) and mRNA (B) with respect to PRC2 components present in wild-type versus Eed−/− ES cells are shown. (A) Actin served as a loading control. (B) mRNA levels are presented relative to the wild type, and were normalized to those of GADPH. (C–E) ChIP experiments comparing wild-type with Eed−/− ES cells. Chromatin was immunoprecipitated with Ezh2 (C), Jarid2 (D), or MTF2 (E) antibodies. Results are represented as percent of input. Error bars represent SD of triplicates.
ChIP experiments revealed that Ezh2 (Fig. 6C) and Jarid2 (Fig. 6D) were each severely impaired in their ability to occupy their natural target genes in Eed−/− cells. In the case of MTF2, the deficiency in its recruitment was, in general, more moderate (Fig. 6E). These results indicate that, at least in ES cells, Ezh2 and Jarid2 are partially interdependent.
Jarid2 is required for proper ES cell differentiation
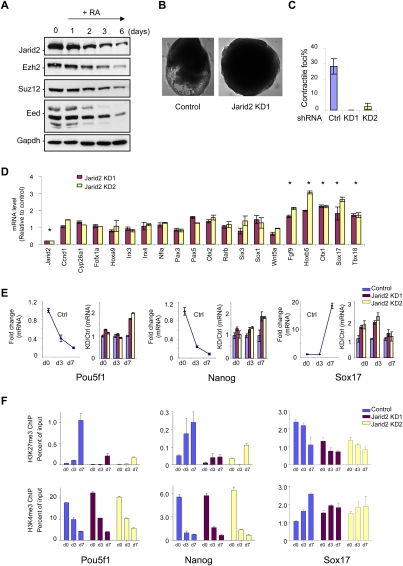
We next tested if Jarid2 can impact the role of PRC2 in a biologically relevant context. Since we found Jarid2 expression to be elevated in ES cells, and ES cells devoid of the PRC2 core component Suz12 exhibit defects in the differentiation process in vitro (Pasini et al. 2007), we focused on ES cell differentiation as a function of the presence of Jarid2. Similar to the core PRC2 components, Jarid2 expression decreases over time of exposure of ES cells to retinoic acid (RA) treatment, during which the cells undergo differentiation (Fig. 7A).
Figure 7.
Jarid2 is required for proper ES cell differentiation. (A) Western blot of PRC components present in ES cells as a function of time of RA-induced differentiation, as shown at the top. The antibodies used are indicated on the left. (B) Control parental ES or Jarid2 knockdown stable clones were induced for differentiation under conditions conducive to the formation of EBs. Bright-field images of representative EBs at day 8 after induction of differentiation are shown. (C) EBs at day 7 after induction of differentiation were plated on regular cell culture dishes to promote cardiomyocyte differentiation. After 4 d, spontaneous contractions of the EBs were quantified for control versus Jarid2 knockdown ES cells. (D) mRNA levels of a representative set of genes (noted at the bottom) that are targeted by PRC2 and Jarid2, in the case of undifferentiated ES cells stably knocked down for Jarid2, shown relative to the control parental cell line and normalized against those of GADPH. Error bars represent SD of triplicates. (*) P < 0.05 indicates statistically significant changes (Student's t-test). (E) mRNA levels of three markers of ES cell differentiation (Pou5f1, Nanog, and Sox17) as a function of time of induction of ES cell differentiation into EBs. mRNA levels are represented relative to undifferentiated ES cells (left panel), or relative to the control parental cell line (right panel) at each time point. Error bars represent SD of triplicates. (F) Enrichment of H3K27me3 (top panel) or H3K4me3 (bottom panel) was analyzed by ChIP at different times during differentiation, as indicated, in the case of the Pou5f1, Nanog, and Sox17 promoters. Results are expressed as percent of input. Error bars represent SD of triplicates.
Stable ES cells expressing Jarid2-specific shRNA were next inspected for their differentiation potential. The formation of multicellular aggregates, called embryoid bodies (EBs), in vitro can recapitulate in a time-dependent manner many aspects of early embryogenesis. While EBs are characterized by a central cavity (cystic EB) after 4–5 d of differentiation in normal ES cells, such structures were not apparent until day 7 in the case of the Jarid2 knockdown cells. At day 8, we detected EBs with yolk sac in normal ES cells, but, again, such levels of differentiation were rarely detected in the Jarid2 knockdown case, even though the size of the EBs were similar or slightly larger (Fig. 7B).
Jarid2 mutant embryos have been reported to display a range of cardiac defects, including ventricular septal defects, noncompaction of the ventricular wall, and a double-outlet right ventricle (Takeuchi et al. 1999; Lee et al. 2000; Toyoda et al. 2003). In view of this phenotype, and to substantiate the importance of Jarid2-mediated regulation of gene expression and associated histone methylation, we analyzed cardiac myocyte formation by plating the EBs at day 7 of differentiation. While 25% of control EBs exhibited spontaneous contraction after 4 d of growing the EBs on plates, this property was barely detectable in Jarid2 knockdown cells, confirming their aberrancy in cardiac lineage development (Fig. 7C).
Our initial comparison of parental ES cells versus their derivatives expressing Jarid2-specific shRNA revealed that only a subset of Ezh2/Jarid2 targets was inappropriately up-regulated in the Jarid2 knockdown case (Fig. 7D). However, when we performed a similar investigation of genes encoding key factors in the differentiation process, we observed that Jarid2 knockdown in ES cells led to a delay in gene repression after differentiation, as in the case of Pou5f1 that encodes the transcription factor Oct4 and of the gene encoding the transcription factor Nanog, or inappropriate elevated levels before and after differentiation, as in the case of Otx1 and Sox17 (Fig. 7E). This correlated with marked deficiencies in PRC2-mediated H3K27 methylation, relative to the control, as evidenced by ChIP results (Fig. 7F). This deficiency was specific to the marks catalyzed by PRC2, as the levels of H3K4 methylation were unaffected (Fig. 7F). Thus, the depletion of Jarid2 hampers the requisite access of PRC2 to chromatin, and therefore the PRC2-mediated chromatin modifications that specify gene repression. This then results in the inappropriate delay in the gene repression program that by default impairs the process of ES cell differentiation.
Discussion
Since the first characterization of the PRC2 core complex, the subsequent, persuasive evidence supports that PRC2 is actually a family of complexes whose composition varies during development, as a function of cell type, or even from one promoter to another. In this study, we identified two new components that interact with PRC2: MTF2 and Jarid2. Our analyses of the proteins that interact with the PRC2 complex initiated with transformed cells. Yet it has become clear that interactions observed using transformed cells might be specific to such cells, and not a determinant to the integrity of a normal organism. Thus, we incorporated studies of a developmentally relevant process and confirmed that the interactions observed between PRC2 and Jarid2 were of consequence to the developmental program.
MTF2 is a paralog of Drosophila Pcl. We reported previously that PHF1, another mammalian paralog of Pcl, was required for efficient H3K27me3 and gene silencing in HeLa cells (Sarma et al. 2008). Although PHF1 appears dispensable for PRC2 recruitment in HeLa cells, work in Drosophila has suggested that the absence of Pcl could impair PRC2 gene targeting (Savla et al. 2008). It is possible that the other paralogs of Pcl (MTF2 and PHF19) exhibit a role that is partially redundant with PHF1 function and thereby maintain PRC2 recruitment upon its knockdown. Pcl and its mammalian paralogs contain two PHD domains and a tudor domain, domains reported to potentially recognize methylated histones. Although the ability of Pcl to specifically bind modified histone has not been elucidated to date, it is tempting to speculate that the PHD and tudor domains could target Pcl to specific chromatin regions. Its presence would then stabilize PRC2 recruitment and promote its enzymatic activity. In support of this hypothesis, we observed that, whereas Ezh2 targeting is severely impaired in Eed−/− ES cells, MTF2 recruitment is affected in a promoter-dependent manner and to a lesser extent than that of Ezh2. This observation suggests that MTF2 gene targeting could be partially independent of PRC2.
The exact function of Jarid2 is more enigmatic. Indeed, Jarid2 is a member of a family of enzymes capable of demethylating histones (Klose et al. 2006). However, Jarid2 is devoid of the amino acids required for iron and αKG binding, and consequently is unable to catalyze this reaction. We considered that Jarid2 could act as a dominant negative and inhibit the activity of other histone demethylases; however, coexpression of Jarid2 with, for instance, SMCX did not affect H3K4me3 demethylation (data not shown). Jarid2 has two domains that could potentially bind DNA: the ARID domain and a zinc finger. Although the ARID domain of Jarid2 was reported to bind DNA (Kim et al. 2003; Patsialou et al. 2005), our band shift assay suggests that other parts of the Jarid2 C terminus (potentially a zinc finger) are also important for binding to DNA. The SELEX experiment performed with the full-length Jarid2 did not allow us to identify any sequence-specific DNA binding, but did result in a slight enrichment of GC-rich DNA sequences. Importantly, we found that the N-terminal part of Jarid2 could robustly stimulate PRC2-Ezh2 enzymatic activity on nucleosomes. We found that a knockdown of Jarid2 decreased the enrichment of PRC2 at its target genes. Conversely, overexpression of a Gal4-Jarid2 chimera recruited PRC2 at a stably integrated reporter and increased PRC2 enrichment at its target genes, supporting the hypothesis that Jarid2 contributes to PRC2 recruitment.
In the case of Drosophila, PRE (Polycomb group response element) sequences have been described, and PRC2 access to chromatin is expected to involve the concerted action of several distinct and specific DNA-binding proteins that interact directly or indirectly with PRC2 (Schuettengruber and Cavalli 2009; Simon and Kingston 2009). However, these same DNA-binding factors, or even a combination thereof, are also found at active genes devoid of PRC2 (Schuettengruber et al. 2009). What distinguishes PRE sequences harboring PRC2 from active genes is still not clear. During the evolution from Drosophila to mammals, only a few of the DNA-binding factors that bind PREs (Dsp1 and Pho) are conserved. Either PRC2 recruitment in mammals involves other mechanisms, or distinct transcription factors have emerged to stabilize PRC2 at its target genes. A recent study has identified a presumed mammalian PRE (Sing et al. 2009); however, the role of this putative PRE at the endogenous locus that is enriched for PRC2 is not reproduced when the element is integrated upstream of a transgene, as PRC2 is absent (Sing et al. 2009). Of note, whereas DNA-binding proteins are likely to play an important role for PRC2 recruitment in mammals, some studies have now suggested that long noncoding RNA could also be involved in this process (Rinn et al. 2007; Pandey et al. 2008; Zhao et al. 2008). These observations together suggest that the recruitment of PRC2 to target genes is complex and requires more than one factor. Our findings suggest that the DNA-binding activity of Jarid2 is one such factor, but its affinity for DNA is low and likely requires the help of other factors.
A critical issue at this juncture is whether or not the composition of PRC2 changes during development. Here, we reported that Jarid2 interacts with PRC2, but its expression, unlike the PRC2 core components, seems to be restricted to some cell lines. In agreement with previous gene expression profiles that monitored mRNA levels during the reprogramming of mouse embryonic fibroblast cells into ES cells (Mikkelsen et al. 2008), we observed that Jarid2 expression is higher in undifferentiated ES cells and decreases upon differentiation. Polycomb target genes are enriched with the H2A variant H2A.Z in undifferentiated ES cells (Creyghton et al. 2008); furthermore, H2A.Z and PRC2 targeting are interdependent in these cells. This result suggests that PRC2 recruitment might involve distinct mechanisms in ES cells and differentiated cells. It is possible that Jarid2 somehow contributes to this specificity.
Knockdown of Jarid2 in undifferentiated ES cells does not give rise to an obvious phenotype; gene expression patterns appear to be only moderately affected, and cell proliferation is unchanged (data not shown). In contrast, when cells are induced to differentiate, a process that entails dramatic changes in gene expression, we now observed impairments as a function of Jarid2 knockdown. Interference with Jarid2 resulted in a failure to accurately coordinate the expression of genes required for the differentiation process, consistent with the previous report on Suz12 knockout cells (Pasini et al. 2007). Instead of the requisite silencing of OCT4 and Nanog loci that occurs upon normal differentiation, each of which become enriched in H3K27me3, Jarid2 knockdown prevented such H3K27 methylation at these genes, and this correlated with their delayed repression. Thus, the Jumonji family of proteins that usually exhibits demethylase activity that might function in opposition to the role mediated by PRC2 contains the member Jarid2 that is devoid of such activity and instead facilitates the action of PRC2 through enabling its access to chromatin.
Materials and methods
Cloning
The IMAGE clone containing human JARID2 (IMAGE ID 4520786) was purchased from American Type Culture Collection. Full-length JARID2 was cloned into the 5′ Not1 and 3′ Kpn1 sites in pFlag-CMV4 vector (Sigma). Human MTF2 was cloned by PCR from HeLa cell cDNA. Subsequent cloning was done using standard techniques. JARID2 fragments were cloned by PCR into the PET102 TOPO plasmid (Invitrogen) for bacterial expression.
Stable cell line
A stable cell line expressing the Gal4-DBD-Jarid2 chimera was generated by transfection of pCDNA4-TO-Gal4-Jarid2 into 293 T-Rex cells containing a stably integrated 5XGal4RE-tk-Luc-neo construct (Sarma et al. 2008) and screened for expression in single colonies. The 293F cell line overexpressing Flag-Ezh2 was described previously (Kuzmichev et al. 2004). The ES Eed−/− cell was a generous gift from T. Magnuson.
Antibody
Polyclonal antibody specific to Jarid2 was raised against amino acids 1–123, expressed in bacteria using the pET102 plasmid (Invitrogen), and affinity-purified using GST-Jarid2 1–203 (pGEX 6P1). Anti-MTF2 antibody was raised against amino acids 302–593 of hMTF2 that was expressed from the pGEX-6P1 plasmid and from which GST was excised. The antibody was affinity-purified using the antigen. Antibodies against total H3 and against H3K27me3 for ChIP (ab6002) were purchased from Abcam. Antibody against Gal4-DBD for use in ChIP, and against H3K27me1 and H3K27me3 for Western blot, were purchased from Upstate Biotechnologies (Millipore). Antibodies specific to YY1 and Gal4-DBD (for Western blot) were purchased from Santa Cruz Biotechnologies. Anti-Flag was purchased from Sigma. Antibodies specific to Ezh2, Suz12, and H3K27me2 were described previously (Kuzmichev et al. 2004).
Nuclear extracts and immunoprecipitation
For nuclear extract preparation, cells were incubated with buffer A (20 mM Tris-HCl at pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1 mM PMSF) for 10 min on ice, centrifuged at 3500 rpm, resuspended in buffer C (20 mM Tris-HCl at pH 7.9, 1.5 mM MgCl2, 0.42 M NaCl, 0.5 mM DTT, 0.2 mM PMSF, 0.2 mM EDTA), sonicated, and centrifuged at 14,000 rpm. For immunoprecipitation, 1 mg of nuclear extract was incubated with 1∼3 μg of antibody bound to protein A/G beads overnight, then the beads were washed three times with BC300 buffer (50 mM Tris at pH 7.9, 300 mM KCl, 2 mM EDTA, 10% glycerol, protease inhibitors) plus 0.1% NP40, and eluted with 0.2 M glycine (pH 2.6) or 2× SDS loading buffer.
Affinity purification of PRC2
Nuclear extract from a 293F Flag-Ezh2 stable cell line was incubated with M2 beads overnight, and the beads were then washed with BC buffer containing 500 mM KCl extensively. Proteins were eluted with Flag peptide at 0.2 mg/mL concentration. Samples were concentrated with acetone before loading onto SDS-PAGE.
Recombinant protein purification and interaction
Recombinant proteins were produced in SF9 insect cells after infection with the corresponding baculovirus as described previously (Margueron et al. 2008). Proteins were purified on Ni-NTA beads in His buffer (350 mM NaCl, 0.5% NP40, 15% glycerol, 10 mM HEPES at pH 7.6, with protease and phosphatase inhibitors). Incubation and washes for the immunoprecipitations were done in His buffer. SDS-PAGE loading buffer was used for the elution. Reconstitution of PRC2 was described previously (Margueron et al. 2009). AEBP2 and Rbp2 baculoviruses were a generous gift from Dr. Y. Zhang. AEBP2 was purified by Flag affinity and mixed at an equimolar ratio with rPRC2.
HKMT assay
HKMT assays were done as described previously (Margueron et al. 2009). For titration experiments, the molar ratio between Jarid2, or Rbp2 or Jarid2 fragments, and PRC2 ranged from 1/10 to 1/1. Nucleosomes or octamers were added after a 15-min incubation of all proteins together at room temperature.
EZH2 knock-in mice
Knock-in mice were generated by homologous recombination at the Institute Clinique de la Souris (ICS). The following amino acid sequence was inserted in front of the translation stop codon: DVEAWLGARVPLVETGENLYWQGGDYKDDDDK. Targeted ES cells were screened by PCR and injected into blastocysts. After germline transmission, deletion of the neomycin cassette was achieved by breeding with a CMV-Cre transgenic line (ICS).
PRC2 purification from thymus
Thymus from 1-mo-old female mice, either wild type or with an Ezh2 knock-in (inbred mixed strain), were collected, and whole-tissue extracts were prepared as described previously (Margueron et al. 2008). After dilution in 10 vol of BC100 plus protease inhibitor, samples were loaded on a P11 column pre-equilibrated with BC100 and washed extensively. The P11 column was eluted with BC500 and BC1000. BC500 elutions were dialyzed against BC200 and incubated for 2 h with M2 beads (Flag-IP). After washing with BC200, beads were eluted with glycine. Eluates were analyzed by Western blot, silver-stained SDS-PAGE, and mass spectrometry.
shRNA
shRNAs were designed to target the ORF of the mouse Jarid2 gene at sites A (344–362 nucleotides [nt]) and B (243–261 nt). The oligonucleotide sequence is available in Supplemental Table S1. Oligonucleotides were annealed and subcloned downstream from the H1 promoter in pSUPER.retro.puro (OligoEngine) using BglII and HindIII. To generate ES cell lines stably expressing Jarid2 shRNA, E14Tg2A.4 ES cells were transfected by Amaxa nucleofector (Lonza) and selected with 2 μg/mL puromycin; the clones were then screened for Jarid2 knockdown by quantitative RT–PCR.
RT–PCR
Total RNA was isolated using Trizol reagent (Invitrogen). First-strand cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) and oligo(dT)20. Quantitative PCR was performed in triplicate using SYBR green reagent (Roche) in the MxPro machine (Stratagene). At least three independent experiments were performed for each assay. Primer sequences are provided in Supplemental Table S1.
RA treatment and ES cell differentiation
Mouse ES cell line E14Tg2A.4 was cultured on gelatin-coated dishes in GMEM media supplemented with 10% FBS (Hyclone), 100 mM MEM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 1 mM L-glutamine (Invitrogen), and 103 U/mL leukemia inhibitory factor (LIF) (Chemicon). For differentiation assays, cells were plated at a density of 0.3 × 106 per 10-cm dish. Twenty-four-hour after plating (d0 time point), LIF was removed and RA (Sigma) was added at a concentration of 2 μM. For formation of EBs, the cells were seeded onto 10-cm Corning Ultra-Low-Attachment Dishes (Corning Catalog no. 3262) containing 15 mL of ES medium without LIF, and the media were changed every other day. For differentiating ES cells into cardiac myocytes, day 7 EBs were seeded onto tissue culture grade plates and cardiomyocyte beating was observed at 2 d and 4 d thereafter.
ChIP
ChIP was performed essentially as described previously (Margueron et al. 2008; Sarma et al. 2008). Quantitative PCR was performed in triplicate using SYBR green reagent (Roche) in the MxPro machine (Stratagene). At least three independent experiments were performed for each ChIP assay. Primer sequences are provided in Supplemental Table S1.
ChIP-seq
V6.5 mouse ES cells were cultured as described previously (Mikkelsen et al. 2007; Ku et al. 2008). ChIP experiments for MTF2 were performed in V6.5 ES cell nuclei prepared as described (Ku et al. 2008). ChIP experiments for Jarid2 were performed as described, with modifications (Orlando et al. 1997). ES cells were cross-linked with formaldehyde and nuclei were isolated by incubation in buffer S (20 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.1% NP-40, 1 mM DTT, protease inhibitor cocktail [Roche]). Swelled nuclei were passed through a 16-gauge needle 20 times before being centrifuged at 8000 rpm for 5 min. Nuclei were then resuspended in cold-modified RIPA buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, protease inhibitor cocktail). Chromatin was then sonicated to achieve 200–700 base pairs (bp) in fragment size range. Lysates were precleared with Protein A sepharose beads (Sigma) for 1 h before incubating with anti-Jarid2 antibodies (described above) overnight at 4°C. Antibody–Jarid2–chromatin complex was captured by Protein A sepharase beads and subjected to extensive washes with modified RIPA buffer, RIPA buffer supplemented with 500 mM NaCl, LiCl buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and TE buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA). ChIP-enriched lysate was then treated with RNase A, followed by proteinase K digestion and cross-linking reversal for 6 h at 65°C. Enriched DNA was extracted by phenol/chloroform/isoamylalcohol, followed by precipitation with glycogen in sodium acetate and ethanol. DNA pellet was resuspended in TE and quantified using Qubit (Invitrogen). Illumina-solexa library construction, sequencing, data processing, and identification of enriched levels were performed as documented previously (Mikkelsen et al. 2007; Ku et al. 2008). It should be noted that the ChIP protocol used for Jarid2 ChIP experiments was biased toward GC-rich promoters genome-wide (M Ku and BE Bernstein, unpubl.). However, Jarid2 targets remained strongly enriched when compared with non-Jarid2 targets in both ChIP-quantitative PCR validation and ChIP-seq signal intensities. The top 1000 genes targeted by Jarid2 and MTF2 are available in Supplemental Tables S2 and S3, respectively.
Gene Expression Omnibus has approved and assigned GEO accession number GSE19708 for our ChIP-seq data set.
Gel mobility shift assay
32P-labeled double-stranded oligonucleotides and proteins were incubated in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM dithiothreitol (DTT), 10% glycerol, 50 mM KCl, and 25 μg/mL poly(dA)–poly(dT) in a total volume of 12 μL for 30 min at 4°C. The products of the reaction were analyzed by 5% native polyacrylamide gel electrophoresis (37.5:1) in 0.2× TG buffer (5 mM Tris-HCl, 50 mM glycine at pH 7.6). The dsDNA was generated by PCR from the pool of probes selected for their binding to full-length His-Jarid2 by SELEX.
Acknowledgments
We are grateful to Dr. L. Vales for comments on the manuscript and active discussions. This work was supported by grants from the NIH (GM64844 and R37GM37120 to D.R.) and by HHMI (to B.B. and D.R.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1886410.
Supplemental material is available at http://www.genesdev.org.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kim TG, Lyons GE, Kim HR, Lee Y. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem. 2005a;280:30916–30923. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn. 2005b;232:21–32. doi: 10.1002/dvdy.20204. [DOI] [PubMed] [Google Scholar]
- Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278:42247–42255. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol. 2004;24:10151–10160. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG, Jung J, Mysliwiec MR, Kang S, Lee Y. Jumonji represses α-cardiac myosin heavy chain expression via inhibiting MEF2 activity. Biochem Biophys Res Commun. 2005;329:544–553. doi: 10.1016/j.bbrc.2005.01.154. [DOI] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–146. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Mysliwiec MR, Kim TG, Lee Y. Characterization of zinc finger protein 496 that interacts with Jumonji/Jarid2. FEBS Lett. 2007;581:2633–2640. doi: 10.1016/j.febslet.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsialou A, Wilsker D, Moran E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 2005;33:66–80. doi: 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M. The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J. 2007;274:6139–6151. doi: 10.1111/j.1742-4658.2007.06135.x. [DOI] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes & Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kojima M, Nakajima K, Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech Dev. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]