Abstract
Because understanding amyloid fibrillation in molecular detail is essential for development of strategies to control amyloid formation and overcome neurodegenerative disorders, increased understanding of present molecular probes as well as development of new probes are of utmost importance. To date, the binding modes of these molecular probes to amyloid fibrils are by no means adequately described or understood, and the large number of studies on Thioflavin T (ThT) and Congo Red (CR) binding have resulted in models that are incomplete and conflicting. Different types of binding sites are likely to be present in amyloid fibrils with differences in binding modes. ThT may bind in channels running parallel to the long axis of the fibril. In the channels, ThT may bind in either a monomeric or dimeric form of which the molecular conformation is likely to be planar. CR may bind in grooves formed along the β-sheets as a planar molecule in either a monomeric or supramolecular form.
Keywords: Amyloid, Binding mode, Congo Red, Fibrillation, Molecular probes, Thioflavin T
Introduction
In 1854, the term “amyloid” was used for the first time by Virchow [1] to describe an aggregated substance found in the liver of a deceased patient. Despite the misleading association to starch, the term is still used and currently 27 diseases are associated with amyloid fibril deposits of normally soluble proteins [2]. No curative treatment is yet available for these amyloid diseases, which include Alzheimer’s disease and type II diabetes [3]. Also, in the pharmaceutical development of stable protein drugs against life-threatening and seriously debilitating diseases such as diabetes, cancer, and rheumatoid arthritis [4], fibrillation is an undesirable phenomenon. Fibril formation is unwanted both from the point of view of adequate shelf life of the pharmaceutical and from the point of view of patient safety [5–9]. In recent years, more and more studies have shown that the ability to form amyloid fibrils is a generic feature of all polypeptide sequences [10], and focus has also been placed on functional use of amyloid, e.g., using amyloid as a depot for controlled continued release of peptide- or protein-based drugs [11] or functional amyloids, i.e., naturally occurring protein fibrils which play a biological role [12].
A few characteristics of the amyloid fibril structure and morphology are widely accepted, e.g., that mature fibrils are typically about 100 Å in diameter, have highly variable lengths up to several micrometers, and are often twisted but generally unbranched [13–18]. Next to the typical fibrillar appearance, one of the most prominent physico-chemical characteristics of amyloid fibrils is the cross-β X-ray fiber diffraction pattern (Fig. 1). Furthermore, two classic markers for the detection of amyloid fibrils are Thioflavin T (ThT) and Congo Red (CR). Most of the abovementioned characteristics are included in the formal definition of amyloid fibrils as “in vivo deposited material, which can be distinguished from non-amyloid deposits by characteristic fibrillar electron microscopic appearance, typical X-ray diffraction pattern and histological staining reactions, particularly affinity for the dye CR with resulting green birefringence” [19].
Fig. 1.
Schematic illustration of the cross-β structure of amyloid fibrils. a Experimental setup for X-ray fiber diffraction. The collimators filter the stream of X-rays so that only those traveling parallel to the specified direction are allowed through. When the fibril sample is exposed to the X-ray beam with the long axis of the fibrils more or less perpendicular to the direction of the beam, the meridional reflections are defined as those lying parallel to the fibril axis, and the equatorial reflections are those positioned at right angles to the fibril axis. b The X-ray fiber diffraction pattern is often interpreted as shown in the schematic illustration of stacked β-sheets with an interstrand distance of 4.8 Å and an intersheet distance of ~11 Å. c, d The strongest repeating feature of the fibril structure is β-strands running perpendicular to the fibril axis with an interstrand distance of 4.8 Å in the fibril direction [96, 97]. The adjacent chains forming the β-sheet have been proposed to be aligned as either parallel (c) or antiparallel (d) protein strands [97, 164–168]. The secondary structures are primarily stabilized by hydrogen bonding between the amino- and carbonyl groups of the main chain(s) (···) [169]. In β-sheets, the amino acid side groups (R) are directed orthogonal to the sheet plane and systematically decorate both surfaces [165]
Molecular probes for detection of amyloid fibrils and prefibrillar oligomeric species are important for diagnosis of amyloid disease as well as for increasing knowledge of the mechanism of amyloid formation. The latter is still not understood, and many different mechanisms have been proposed [20–24]. A full understanding would require structural elucidation of every species and determination of the kinetics of interconversion of all species on the reaction pathway. For examining the kinetics, extrinsic molecular probes can be very useful for high-throughput screening. An example is the fluorescent dye, ThT, which is most often used for in situ amyloid fibril detection. CR staining is not suitable for in situ detection but is the diagnostic test of choice for routine identification and screening of amyloid deposits [25].
The binding mode(s) of the molecular probes for amyloid fibrils is not fully understood. For ThT and CR, a large number of studies have examined the binding mode, but a high-resolution characterization has so far not been possible because of the insolubility and often heterogeneous nature of the amyloid fibrils. Even co-crystallization of the dyes with one of the steric zippers proposed to resemble the structure of the cross-β spine [26, 27] has not succeeded yet. The lack of high-resolution models complicates the examination of the binding mode to amyloid fibrils, and existing models are both incomplete and conflicting. In addition, the binding modes of ThT and CR analogs have hardly been reported.
Further knowledge of the binding modes is essential for a number of reasons, such as better interpretation of the characteristic amyloid signature, improved instructive guides for design of novel molecular probes, and increased understanding of the various plausible binding modes of dye binding to amyloid fibrils. Also, from a therapeutic point of view, the binding modes may be of utmost interest, since CR and some of the small organic molecules related to CR and ThT structures have been shown to inhibit fibril formation [28–31]. This review will give an overview of the classic as well as new molecular probes for amyloid fibril detection and a deeper insight into the recent understanding of their binding modes. The main focus will be on the binding mode of ThT and to a minor extent on the binding mode of CR.
Overview of molecular probes for amyloid fibril detection
Staining with the dye CR has been used for identification of amyloid fibrils since the beginning of the 1920s [32]. Upon binding to amyloid fibrils, CR gives a characteristic apple-green birefringence (or perhaps more correctly, shows anomalous colors) when examined between crossed polarizer and analyzer, and a characteristic shift in absorbance maximum from about 490 nm to about 540 nm is seen [32–36]. CR is most often applied in diagnosis of amyloid in ex vivo tissue sections using polarization microscopy [2], but light microscopy [37] and fluorescence microscopy [25, 37] have also been used. In vitro quantification of insulin or amyloid β (Aβ) amyloid upon CR binding can be performed using absorption spectroscopy [38, 39], although CR lacks sensitivity at low amyloid fibril concentration compared to ThT [40]. Furthermore, CR is not well suited for in situ detection, since CR interferes with processes of protein misfolding and aggregation, and is reported either to inhibit or enhance amyloid fibril formation for several proteins [29, 35, 36, 41–43].
In 1959, ThT was introduced as a dye that shows enhanced fluorescence upon binding to amyloid in tissue sections [44] and has since become a standard dye for amyloid detection. Detection is based on the fluorescence characteristics of ThT, revealing a considerably enhanced ThT fluorescence upon interaction with amyloid fibrils with excitation and emission maxima at about 450 and 480 nm, respectively [45, 46]. ThT has many applications such as diagnosis of amyloid in tissue sections using fluorescence microscopy [44, 47], direct observation of amyloid fibril growth using total internal reflection fluorescence microscopy [33, 48], monitoring of the amyloid fibril formation using fluorescence anisotropy [49], and monitoring of extracted amyloid and in vitro amyloid fibril formation using fluorescence spectroscopy [45, 46]. The latter is often monitored either as in situ measurements in a fluorescence plate reader or as ex situ measurements in a fluorescence cuvette [50]. In general, ThT does not affect, or only to a limited extent affects, the fibrillation kinetics [24, 43, 50, 51]. The fluorescence intensity of ThT has been shown to be proportional to the weight concentration of fibrils for a certain type of amyloid fibril formed from a specific protein under given conditions [52, 53] and to be independent of the number or length distribution of fibrils [52]. However, extreme care should be taken when comparing fibril formation under different conditions, since a number of factors may affect the fluorescence intensity, such as the specific protein forming the fibril [49, 54], fibril morphology [40, 45, 55–57], ThT concentration [24, 53, 58, 59], pH [46, 54, 60], and ionic strength [40].
Besides CR and ThT, few compounds have been used extensively for the detection of amyloid fibrils. Thioflavin S (ThS) has been applied to some extent, but mainly for diagnosis of amyloid fibril deposits in tissue sections using fluorescence microscopy [61, 62]. ThS is less suitable for kinetics analysis than ThT since free ThS fluorescence interferes significantly with the determination of the fibril-bound species [40, 46]. However, in the last decade, various molecular probes have been developed for detection of amyloid fibrils (Table 1). In many cases, various derivatives or related structures of both CR and ThT have been shown to be useful for in vitro and ex vivo monitoring of amyloid fibrils [63–68]. In addition, development of non-invasive methods to quantitate amyloid deposits is of great use in the diagnosis of amyloid diseases. A variety of radioactively labeled analogs of CR, ThT, and other molecular probes have been evaluated as potential in vivo position emission tomography imaging probes of amyloid deposition in the brain of Alzheimer’s patients [69–73]. A good review is already published on these radioactively labeled analogs [73], and the present review will only briefly point out one of the more successful radioactively labeled probes, namely the derivative of ThT, Pittsburgh compound-B (PiB). PiB has been modified to a neutral benzothiazole with a high affinity towards amyloid, and which efficiently crosses the blood–brain barrier [70, 74, 75]. This probe has already been applied successfully in living humans and can provide quantitative information on amyloid deposits [70, 74].
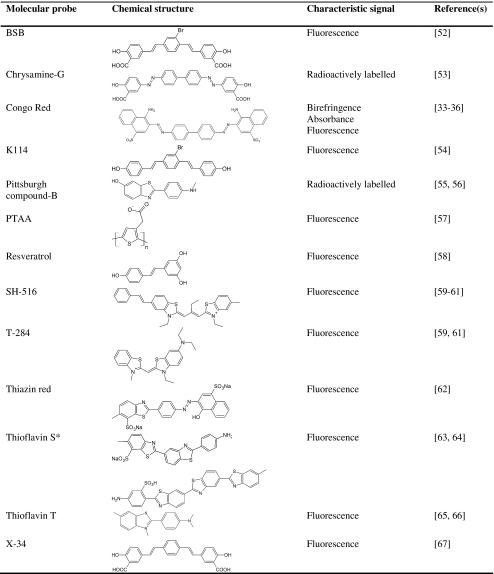
Table 1.
Some molecular probes applied for amyloid fibril detection
*Mixture of planar asymmetric dyes with charged group at the end of the molecule.
Design of new extrinsic fluorescent dyes for characterization of the fibrillation process using novel approaches is also ongoing. Perhaps most interesting is a class of luminescent conjugated polymers (LCPs), which has been shown to specifically stain amyloid aggregates in tissue samples [76–79]. In recent reviews by Nilsson et al. [80] and Åslund et al. [81], the LCPs are extensively discussed and this review will therefore only briefly point out some important facts on the use of LCPs. A great advantage of using these LCPs compared to the classic dyes and their derivatives is their specific spectroscopic signal for individual protein aggregates, which enables the LCPs to reveal different subpopulations of plaques in more detail, e.g., in Alzheimer’s and prion diseases [77, 78, 82]. To date, the polymer of polythiophene acetic acid (Table 1) is the most extensively used LCP probe [77–79, 83].
Purity and concentration determination of Thioflavin T and Congo Red
Before going into detail with the binding modes of ThT and CR in the context of amyloid fibrils, focus will be put on the purity of these probes. Both purity and an accurate determination of the concentration are of utmost importance when studying the binding mode of these molecular probes.
Commercial ThT has a reported purity of only 65–75% (example from Sigma product information). The main impurities are salts containing sodium, chloride, and sulfur [84]. Recrystallization of ThT removes most of the impurities present in the commercial ThT, leaving not more than 0.5% proton-containing organic impurities [84]. ThT can be recrystallized from water [85–87] or from a mixture of, for example, toluene (30%) and ethanol (70%) [88]. The molar extinction coefficient at 412 nm increases from 23,800 M−1 cm−1 (Sigma product information) to 36,000 M−1 cm−1 in water for the recrystallized ThT [85–87]. Thus, for a more accurate concentration determination, the latter extinction coefficient should be used, also when applying the commercial ThT, for example, for kinetic studies. Unfortunately, many authors seem to be unaware of this [50, 58, 89] and many studies do not state the molar extinction coefficient used [54, 90–92].
As for ThT, it is important to have a high purity and an accurately determined concentration of CR when studying its binding mode in the context of amyloid fibrils. Commercial CR is contaminated with NaCl and water, making quantification by weight inaccurate [39]. Recrystallization is likely to improve the purity and make quantification by weight possible. However, quantification can also be done using the molar absorptivity value for CR in 40% ethanol (in 1 mM NaH2PO4, pH 7.0) of 5.93 × 104 AU/(cm M) at 505 nm, determined after quantification of the concentration of a CR solution in d6-dimethylsulfoxide with an internal standard by proton nuclear magnetic resonance (NMR) [39].
Binding mode of Thioflavin T
The binding mode of ThT can be defined as the orientation of ThT relative to the amyloid fibril as well as the conformation of ThT when bound to the fibril structure. A deeper insight into the binding mode of ThT in the context of amyloid fibrils, of which no high-resolution models are available, must therefore include many aspects. Of importance is the binding site which is the region or location on the fibril to which ThT binds. The binding site can, for example, be characterized by the ThT accessibility, binding stoichiometry, and affinity. Besides the conformation of ThT, the molecular form is crucial for understanding the binding mode and thus the molecular mechanism behind the characteristic ThT fluorescence. The molecular form refers to whether ThT binds as a monomer or as a supramolecular assembly in the binding site of the fibril.
Thioflavin T has easy accessibility to its binding site
Generally, ThT has easy accessibility to the binding site inducing the characteristic fluorescence in the amyloid fibrils. This is suggested by the apparent fast kinetics of ThT binding to formed fibrils (completed within 30 s) [40]. The kinetics of ThT binding has been shown to follow the Arrhenius law and may also depend on the fibril morphology, since slower binding may result from reduced accessibility in large fibrils assemblies [24, 51, 90]. In addition, conflicting reports claim that the accessibility is either dependent or independent on in situ presence of ThT [53, 58].
Orientation of Thioflavin T in its binding site
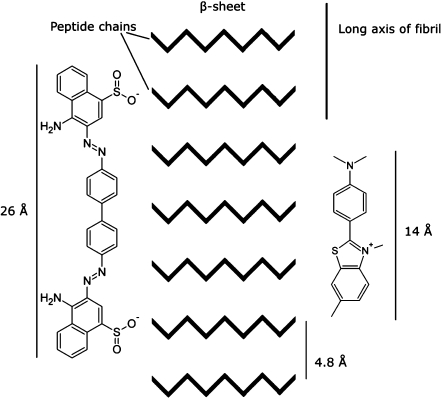
The orientation of ThT with respect to the amyloid fibrils has been suggested to be regular and specific [93]. These findings are based on amyloid spherulites, where ThT has been shown to bind with the long axis parallel to the long axis of the fibrils (Fig. 2) [93]. This orientation of ThT is consistent with a recent X-ray crystal structure of ThT bound to a “peptide self-assembly mimic” (PSAM) scaffold [94]. Also, a molecular dynamics simulation of ThT binding to two β-sheets reveals ThT binding perpendicular to the β-strands in the β-sheet [95]. However, the simulations also suggested ThT binding parallel to the β-strand at the end of the β-sheets [95]. The latter seems unlikely, since ThT has not been shown to interact with the formation and elongation of the β-sheet [40, 50]. Furthermore, the interstrand spacing along the fibril axis of 4.8 Å is unchanged in insulin fibrils upon ThT binding (Fig. 3) [53]. An increased β-strand distance would have been evident in X-ray fiber diffraction data, if ThT was bound between the β-strands.
Fig. 2.
Schematic illustration of Thioflavin T and Congo Red oriented with their long axis parallel to the long axis of the fibril. If the probes bind in an extended conformation, CR binding requires at least six continuous β-strands in a β-sheet, while ThT only needs around three to four. The distance between two sulfonic acid groups in CR is approximately 19 Å [69]. The structures are generated, and the dimensions are measured using ACD/Labs 8.00 Release (product version 8.17)
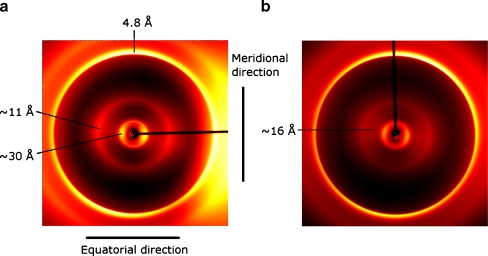
Fig. 3.
X-ray fiber diffraction images of: a insulin fibrils formed in 25 mM HCl (pH 1.6) without ThT and b with ThT bound (mixed in a molar ratio of 1:2). Modified from Groenning et al. [53] with permission from Elsevier
Structural changes in the binding site upon Thioflavin T binding
Prior to ThT binding, it is believed that no rate-limiting conformational change occurs either in the amyloid fibril or of ThT. Instead, tertiary and quaternary conformational changes in the fibril may occur after dye binding [40]. An X-ray fiber diffraction study on insulin fibrils with and without ThT also showed some structural changes, especially among the equatorial reflections (Fig. 3) [53]. A decrease in peak intensity for the reflection at 11 Å, often interpreted as an intersheet spacing [96, 97], relative to a peak at around 16 Å was observed. Importantly, the reflection at 11 Å still remains [53]. Several explanations can be proposed for this phenomenon such as ThT binding between some β-sheets in the fibrils increasing the distance from 11 to 16 Å, ThT association in a regular manner in the fibril structure yielding the 16 Å reflection, or other changes in the protofilament structure or its association into protofibrils and fibrils [53]. Morphological changes of the fibrils are also indicated by the presence of low-resolution changes upon ThT binding, but in the example of insulin fibrils under the given conditions, the 30 Å reflection, which may be interpreted as a center-to-center distance of the elongating units in the protofilaments, is unchanged (Fig. 3) [53].
Binding affinity and stoichiometry of Thioflavin T in amyloid fibrils
Most studies performed on the binding affinity and stoichiometry of ThT focus on the binding mode of the fluorescent ThT as this is often the binding mode of interest (Table 2). However, more than one binding site population for ThT has been shown to be present in insulin, Aβ(1–40) and HET-s fibrils [53, 90, 91], and a pronounced difference between the total amount of ThT bound to the amyloid fibrils and the amount of bound ThT inducing the characteristic ThT fluorescence has been observed (Table 2) [53, 90]. Hence, all bound ThT does not result in the characteristic fluorescence. Interestingly, a ThT binding process prior to the one providing the characteristic fluorescence may also occur [53]. This suggests that the binding process that induces the characteristic ThT fluorescence may involve binding of two ThT ions, or the binding process may involve a fast dye binding event followed by rate-limiting tertiary or quaternary conformational changes of the protein or the probe within the fibril. The former is in agreement with the proposed occurrence of the characteristic fluorescence by binding of a ThT dimer [53, 87], while the latter has previously been proposed for ThT binding to Aβ(1–40) fibrils [40].
Table 2.
Thioflavin T binding to various amyloid fibril
| Type of fibrils | n | Kd (μM) | Measurement | Reference |
|---|---|---|---|---|
| AA fibrils (pH 9.0) | 1.4 AU/μg/ml | 0.760 | Fluorescence | [66] |
| AA fibrils (pH 7.5) | – | 0.033 | Fluorescence | [76] |
| Aβ(1–40) fibrils (pH 6.0) | – | 20 | Stopped-flow fluorescence | [40] |
| – | 11 | Fluorescence | [76] | |
| – | 2 | Fluorescence | [65] | |
| Aβ(1–40) fibrils (pH 7.4) | 0.025 mol/mola | 0.75 | Fluorescence | [116] |
| Aβ(1–40) fibrils (pH 8.5) | 2.2 AU/μg/ml | 0.86 | Fluorescence | [121] |
| Aβ(1–28) fibrils (pH 6.0) | – | 0.54 | Fluorescence | [65] |
| ApoAII fibrils (pH 7.5) | – | 0.53 | Fluorescence | [76] |
| ASSAM fibrils (pH 9.0) | 4.4 AU/μg/ml | 0.035 | Fluorescence | [66] |
| HET-s fibrils (pH 2.0) | 0.01 mol/mol | 23 (0 M NaCl) | Fluorescence | [115] |
| 0.11 mol/mol | 3.4 (2.4 M NaCl) | |||
| HET-s fibrils (pH 7.0) | 0.11 mol/mol | 1.5 (0 M NaCl) | Fluorescence | [115] |
| 0.13 mol/mol | 1.7 (2.4 M NaCl) | |||
| Insulin fibrils (pH 1.6) | 0.66 mol/mol | 64 | UV absorbance | [75] |
| 0.11 mol/mol | – | Fluorescence | [75] | |
| Insulin fibrils (pH 7.5) | – | 0.5 | Fluorescence | [76] |
| 0.09 mol/mol | – | Fluorescence | [75] | |
| 0.14 mol/mol | 0.059 (Kd,1) | ITC | [75] | |
| 0.59 mol/mol | 23 (Kd,2) | |||
| 0.89 mol/mol | 10 | UV absorbance | [75] | |
| TTR fibrils (pH 7.5) | – | 5.2 | Fluorescence | [76] |
n is number of binding sites, while Kd is dissociation constant. – no data. Protein nomenclature is AA (Apo)serum AA, Aβ amyloid β, ApoAII apolipoprotein AII, ASSAM murine senile amyloid protein, TTR transthyretin
aMoles of ThT per mole of protein in fibril form
Whether the characteristic fluorescence is induced by only a single binding site population or multiple populations is still being debated [40, 46, 53, 90, 91]. Notably, if several binding site populations for ThT are present, they are likely to have relatively similar affinities [40]. The affinity of ThT towards various amyloid fibrils, based on the characteristic fluorescence, is 0.033 to 23 μM (Table 2). Generally, the affinity decreases at acidic pH, probably due to electrostatic repulsion below the isoelectric point of the protein [53, 54, 90]. This is supported by a dramatic increase in affinity at highly acidic pH when the ionic strength is increased, probably due to shielding of like charges (Table 2) [90]. At neutral pH, the affinity is almost independent of ionic strength [90]. Finally, a neutral charge of ThT favors the ThT interaction by increasing the affinity of various neutral, but non-fluorescent, ThT analogs [71, 73]. Importantly, the increased affinity may be ascribed to the binding of some neutral ThT analogs in a class of binding site different from that of ThT [91].
For ThT ions inducing the characteristic fluorescence, the binding stoichiometry to amyloid fibrils varies from about 0.01 to 0.1 mol of ThT per mole of protein in fibril form (Table 2) [53, 90, 91]. The binding stoichiometry is similar for insulin fibrils at low and neutral pH [53]. The same has been observed for fibrils of the prion protein fragment HET-s(218–289) at high ionic strength, but without salt a 10-fold decrease in the binding stoichiometry was detected at acidic pH compared to neutral pH [90]. This suggests that the poor ThT binding may be due to charge repulsion [90]. Although charge repulsion can affect the binding stoichiometry, specific electrostatic interactions may be of only minor importance for the binding [53].
Thioflavin T conformation upon fibril binding
The ThT ion in solution consists of a benzothiazole and a benzamine ring freely rotating around a shared C―C bond. Non-bound ThT prefers a non-planar conformation with a torsional angle between the two aromatic systems of ~35° according to quantum mechanics calculations [87, 98, 99]. For interpretation of the occurrence of the characteristic ThT fluorescence and the location of the binding site, the torsional angle of the amyloid bound ThT is likely to be important. Two conflicting conformations have been proposed with ThT either being bound in a planar (or near-planar) conformation [60, 93, 99, 100], or in a significantly twisted conformation [90, 94, 101]. For example, an X-ray crystal structure of ThT bound to the active site gorge in acetylcholinesterase (AChE) has revealed a planar conformation of ThT, where ThT binds with high affinity and has a characteristic fluorescence similar to that in amyloid fibrils [100]. If ThT binds in a similar binding mode to the amyloid fibrils, the binding may be induced by a channel of similar size rather than specific interaction with the secondary structure. This conformation is in agreement with the model proposed by Krebs et al. where ThT is suggested to bind as a planar monomer in the regular grooves formed by the side chains of the residues of the β-strands on the surface of the β-sheet in the fibril [93]. On the other hand, a significantly twisted conformation has been suggested due to a strong Cotton effect detected as a negative circular dichroism (CD) signal around 450 nm. This may mean that the otherwise achiral ThT acquires chiral properties when bound in a twisted conformation in amyloid fibrils [90, 101]. However, recently the chirality was shown not to be a prerequisite for the occurrence of the characteristic ThT fluorescence and the chiral moieties binding ThT have been attributed to the structure of the high-order fibril assemblies [102].
Molecular form of Thioflavin T bound to amyloid fibrils: monomer, dimer, or micelle?
The molecular form of ThT upon binding in amyloid fibrils has been suggested to be either monomeric [93, 98, 99, 103, 104], dimeric [87, 105], or micellar [89]. The monomer hypothesis is based on ThT behaving as a molecular rotor [60, 93, 98, 99, 103, 104], while the dimer hypothesis is based on ThT forming an excimer in cavities of appropriate size [53]. These two hypotheses will be discussed later.
ThT has also been proposed to bind to amyloid fibrils as a preformed micelle of about 3 nm in diameter. The micelles have been suggested to have a hydrophobic interior formed by the benzamine rings, while the positively charged region of the benzothiazole ring would point towards the solvent [89]. In several cases, the ability of ThT to form micelles has been shown, revealing a critical micelle concentration (cmc) in the range of 4.0–31 μM in water [49, 89, 90]. Thus, electrostatic repulsion of the positively charged ThT does not hinder micelle formation, in similarity to the proposed ability of ThT to form dimers upon binding in cavities [87, 105]. However, recent evidence against the micelle theory has been put forward, for instance its inconsistency with ThT preferring to bind parallel with the long axis of the fibril [93]. Another argument against this hypothesis is the occurrence of characteristic ThT fluorescence even well below cmc, i.e., in the range 0.5 to 2.5 μM ThT [24, 59, 90], although it cannot be ruled out that micelles can form on surfaces below cmc. Atomic force microscopy (AFM) images have been interpreted as ThT micelles bound to amyloid fibrils [89], but may alternatively be interpreted as stacking of ThT rotated along its lateral axis [95].
Spectral properties of Thioflavin T in solution
The spectral properties of ThT in aqueous solution, and thus the size of the red shift accompanying ThT binding to amyloid fibrils, have been questioned. The emission at around 445 nm, when excited at 350 nm, for free ThT in aqueous solution has been suggested to originate from impurities [99, 103, 106]. This hypothesis deals with the fact that the absorbance spectrum of ThT has a maximum at 412 nm, which does not correspond with the fluorescence excitation spectrum [106]. A weak point in this hypothesis is the continuous presence of the excitation band at 350 nm with an emission maximum at 445 nm after recrystallization of ThT [87]. Recently, an alternative explanation has been suggested where the excitation at 350 nm may be due to the emission of an isolated ThT chromophore, for example, due to the fluorescence of a benzothiazole ring [106]. This may be a more plausible explanation for the ThT fluorescence at 450 nm when ThT is free in aqueous solution.
Effect of primary and secondary protein structures on Thioflavin T fluorescence
The characteristic ThT fluorescence is observed upon binding of ThT to amyloid fibrils formed from proteins with a wide variety of size and primary, secondary, and tertiary structures [54]. Thus, the primary structure of the proteins is not considered important for obtaining a proper binding between ThT and amyloid fibrils, although it may account for some of the observed differences in fluorescence intensity among different amyloid fibrils [54]. Minor differences in excitation and emission maxima of ThT are also observed upon binding to amyloid fibrils formed by various proteins [22, 107].
The β-pleated sheet secondary structure of the amyloid fibrils may be important in binding of ThT. However, the β-pleated sheet peptide backbone alone is suggested to be insufficient to form a proper ThT binding site where the characteristic ThT fluorescence is induced. This statement is based on the fact that not all β-sheet fibrils induce the characteristic change in ThT fluorescence; for example, β-sheet fibrils of a peptide of the central strand of islet amyloid polypeptide (SNNFGAILSS), poly-l-lysine, and poly-l-serine do not [54]. In addition, many β-sheet-rich proteins, such as chymotrypsin, bovine IgG, transthyretin, or concanavalin A, have failed to induce the characteristic ThT fluorescence [54, 60, 87]. The latter may be due to the often heavily twisted conformation of larger β-sheets in water-soluble proteins, since flatness of the β-sheet has been proposed to be critical for high-affinity ThT binding [94]. It may also be due to the β-sheets of the native proteins being shorter and less regular than the sheets in amyloid fibrils [93]. Notably, binding of ThT in for example grooves at the surface of β-sheets is not likely to be compatible with β-sheets alone being insufficient to induce the characteristic fluorescence.
Effect of solvent viscosity and polarity on Thioflavin T fluorescence
A number of studies have tried to simulate the microenvironment surrounding the bound ThT probe in search of an explanation for the occurrence of the characteristic fluorescence by focusing on the effect of solvent viscosity and polarity (dielectric constant) [60, 99, 104, 106, 108].
In several of these studies, increased viscosity or rigidity of the microenvironment has been shown to increase the quantum yield [60, 99, 103]. The hypothesis is that ThT behaves as a molecular rotor, where the viscous solvents block rotation in the dye molecule due to steric hindrance, which results in suppression of quenching and a high quantum yield of fluorescence [98, 103, 104]. Interestingly, the fluorescent quantum yield of ThT is observed only to be dependent on viscosity when a proper environment, i.e., with a lower dielectric constant, surrounds ThT [104]. However, no simple relationship between fluorescence quantum yield and dielectric constant is observed [104]. Finally, viscosity alone does not account for the characteristic ThT fluorescence, since no red shift in excitation maximum is observed [54], and since a tight packing of ThT in β-cyclodextrin (β-CD) and in the β-sheet cavity of transthyretin (TTR) did not induce the fluorescence [87]. The red shift may depend on the solvent. For example, the presence of polyol solvents such as glycerol and ethylene and solvents such as chloroform, dichlorobenzene, and dichloromethane significantly red-shifts the excitation maximum of ThT [54, 58, 99].
The importance of cavities on the characteristic Thioflavin T fluorescence
A proper binding environment for ThT to induce the characteristic ThT fluorescence is provided in cavities of an appropriate diameter and length, such as in the cavity of γ-cyclodextrin (γ-CD) and AChE [85, 87, 109]. For the fluorescence to occur, the cavity diameter should be of 8–9 Å, while the cavity needs to be long enough to cover the entire length of the ThT ions [87, 105]. Cavities with a smaller diameter have been shown to be unable to induce the characteristic ThT fluorescence such as in TTR and β-CD [87, 105]. A cavity with a diameter of 8–9 Å can indeed be present in the amyloid fibril structure, for example, as a central cavity in the protofilament [110, 111] or between the protofilaments when associating to form protofibrils and mature fibrils [17, 97, 112–115]. Binding in a cavity is in agreement with the minor influence of the primary and secondary structures on the occurrence of the characteristic fluorescence, the observation that specific electrostatic interactions were not crucial for the binding, and offers an explanation for the specificity of ThT.
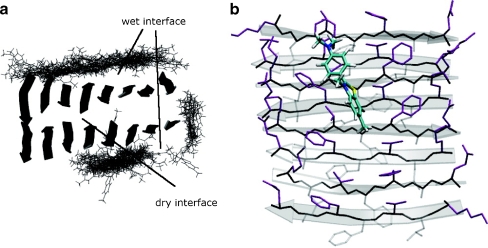
ThT binding in cavities, channels, or grooves of the amyloid fibrils may be in the form of a monomer [93] or a dimer [53]. According to a recent X-ray crystal structure of one ThT molecule bound in the active site gorge of AChE, a monomer can bind in a cavity of 8–9 Å [100]. However, a monomer has most often been proposed to bind in narrow grooves formed between the side chains of the residues of the β-strands on the surface of the β-sheet [93, 116]. Recently, more insight into the β-sheet packing of small peptides in crystals has been obtained, revealing a dry and a wet interface of the β-sheet [26, 27]. The wet interface resembles the surface of the β-sheet and molecular dynamics simulations have shown binding of ThT in a regular groove on the surface (Fig. 4) [116]. In contrast to ThT binding in the grooves between adjacent residues, ThT has also been suggested to interact with the β-sheet by docking onto its surface as revealed by the X-ray crystal structures of ThT bound to a PSAM scaffold [94]. The mechanism proposed for the interaction is suggested to involve the aromatic side chains of tyrosine as a particular favorable binding site for ThT [94]. A similar mechanism involving π-stacking and hydrophobic interactions also occurs upon ThT binding in the gorge of AChE, which is highly enriched with aromatic side chains [100]. However, to induce the characteristic ThT fluorescence, the monomer is most often suggested to behave as a molecular rotor, as described previously. For the dimer, the characteristic ThT fluorescence is proposed to occur due to an excited-state dimer (excimer) formed in cavities with a diameter of 8–9 Å [53, 87, 109]. Excimer formation is suggested to induce the observed red shift due to the formation of an increased conjugated system. The excimer formation is an excited-state reaction where a molecular process changes the structure of the excited-state fluorophore. The excited-state reaction occurs subsequent to excitation, i.e., the excited-state species is not excited directly but is formed from the initially excited species [117]. Excimer formation has previously been observed for molecules with a structure similar to ThT, e.g., 2-phenylindone with a monomer emission near 390 nm and a red-shifted excimer emission at 420 nm [117]. An excimer formation of ThT has also previously been proposed for ThT bound to deoxyribonucleic acid (DNA) [118].
Fig. 4.
Molecular dynamics simulation of Thioflavin T (ThT) binding to β-sheets formed by KLVFFAE peptides having a dry and a wet interface. a ThT molecules (shown in lines) bound to the β-sheets. b A representative binding of the ThT ion (mainly green) to one of the β-sheets is shown. Reprinted with permission from Wu et al. [116]. Copyright 2007 American Chemical Society
Plausible binding modes of Thioflavin T in amyloid fibrils
In summary, it seems likely that the binding location for ThT inducing the characteristic fluorescence is in a cavity with a diameter of 8–9 Å, where ThT has relatively easy access and will be bound with its long axis parallel to the long axis of the fibril. Pinpointing the exact channels in which the binding takes place is complicated by the lack of high-resolution structural fibril models. However, the low-capacity single population of binding sites (or multiple with fairly close affinity) for ThT inducing the characteristic fluorescence shows that ThT is not covering the entire surface of the amyloid fibrils. This can be exemplified assuming ThT ions to be stacked one on top of the other, end by end, throughout the central cavity of an insulin protofilament, which will then yield a maximal binding capacity of 0.22 mol of ThT per mole of insulin in fibril form, i.e., twice the determined low capacity of the binding site [53]. Therefore, we have previously proposed a more rare but plausible binding location, where ThT is bound between the protofilaments forming protofibrils, or between the protofibrils forming the mature fibril. For insulin fibrils, 0.9–1.3 or 1.4–1.9 ThT ions were estimated to stack end to end throughout a cavity between the protofilaments or protofibrils, respectively (Fig. 5) [53]. The main population of ThT ions being bound does not induce the fluorescence, and this majority of ThT may be bound in the grooves at the surface of the β-sheet. Binding at these sites is in agreement with the maintenance of the interstrand and intersheet distance of 4.8 and 11 Å, respectively [53]. Furthermore, the structural changes in the amyloid fibril to accommodate ThT, resulting in a distance of 16 Å, may indicate binding of ThT between some β-sheets increasing the typical 11 Å intersheet distance.
Fig. 5.
Two models of the fluorescence-inducing binding of Thioflavin T (ThT) to an insulin fibril consisting of three intertwining protofibrils, where each protofibril has two protofilaments. a, b Binding of ThT as a dimer between the three protofibrils is illustrated on the contoured density cross section of the insulin fibril as red dots (a) and on the subunit repeat of the fibril obtained by small-angle X-ray scattering as red rectangles (b). c, d, e Binding of a ThT monomer between the protofilaments forming the protofibrils is illustrated on the contoured density cross section of the insulin fibril (c) and on the model of a protofibril consisting of two protofilaments formed by a helical oligomer (top view (d) and side view (e)). Eight ThT ions are shown stacking end-by-end (e). The length of the oligomer in the protofilament is 200 Å. Reprinted from Groenning et al. [53] with permission from Elsevier. The contoured density cross sections and the structural models of insulin fibrils were originally modified from Jimenez et al. [14]. Copyright 2002 National Academy of Sciences, USA and Vestergaard et al. [20], respectively
The nature of the molecular form of ThT upon binding to amyloid fibrils is likely to be planar and either monomeric or dimeric. Further examination of the occurrence of the characteristic ThT fluorescence is needed to elucidate whether ThT behaves as a molecular rotor with ThT being locked in a certain position (except from planar) or formation of a more conjugated system involving ThT (for example excimer formation) takes place.
Binding mode of Congo Red
When examining the binding mode of CR, many of the aspects to consider are similar to the ones of relevance for characterizing the binding mode of ThT. Therefore, comparison with ThT will be done throughout this chapter whenever relevant in the context of the most recent knowledge on the binding mode of CR.
Congo Red orientation in and effect on the fibril structure
The CR molecules are most likely arranged along the amyloid fibrils, all having the same orientation as suggested from the dye becoming dichroic and birefringent [17, 32, 119]. In contrast, dual binding modes of CR being bound with CR both parallel and perpendicular to the β-strands have been suggested based on molecular dynamics simulation (Fig. 6) [116]. However, the primary binding mode of CR was binding to a regular groove formed by the β-strands along the wet side of the β-sheet extension direction (Fig. 6a) [116]. The latter is in agreement with most studies, which suggest that the long axis of the CR molecule lies perpendicular to the direction of the β-strands [120–124]. In addition, CR binding by intercalation between the β-strands with the long axis of the dye parallel to the β-strands [125] of already formed fibrils is generally considered unlikely [116, 126]. Whether CR binding parallel to the β-strands at the end of the fibrils plays a role in the inhibitory effect of CR on amyloid fibrillation remains an open question.
Fig. 6.
Molecular dynamics simulation of Congo Red (CR) binding to β-sheets formed by GNNQQNY peptides revealing a dry and wet interface. The X-ray crystal structure of the peptide is determined by Eisenberg and coworkers [26]. The primary binding mode is CR binding as a monomer (a) in grooves at the surface of the β-sheets. However, CR is also shown to bind as a dimer (b), parallel to the β-strands (d) or with only one CR bound and the other CR in solution (c). Reprinted with permission from Wu et al. [116]. Copyright 2007 American Chemical Society
A limited number of studies have looked into plausible structural changes in the amyloid fibril upon CR binding [127]. Larger rearrangements are not expected, which is supported by a Raman study revealing an unaltered secondary structure of Aβ(1–40) fibril with and without CR [127].
Binding kinetics, affinity, and stoichiometry of Congo Red in amyloid fibrils
Generally, the binding kinetics of CR to amyloid fibrils is slower than the binding kinetics of ThT [128]. For example, CR binding to fibrils of poly-l-lysine reaches its equilibrium at around 60 min [121]. This suggests that the molecular form of CR that interacts with the amyloid fibrils may not have easy access to its binding site. In terms of accessibility, the larger size of CR may be critical. Despite the slower kinetics of CR binding and CR being longer than ThT (Fig. 2), the binding stoichiometry generally reveals a significantly higher capacity of the CR binding site compared to ThT inducing the characteristic fluorescence (Tables 2 and 3). The 20-fold increase in the binding stoichiometry of CR compared to ThT in insulin fibrils suggests that CR is not only bound in central channels of 8–9 Å in diameter, as is suggested for ThT. Rather, CR is likely to bind to the β-sheets surface, maybe at a similar binding location as the majority of bound ThT, which does not induce the characteristic ThT fluorescence. For CR, a single type of binding site population in amyloid fibrils is revealed using absorbance spectroscopy [121]. Thus, if more than one binding site population is present, they are of similar affinity.
Table 3.
Congo Red binding to various amyloid fibril in vitro
| Type of fibrils | na | Kd (μM) | Measurement | Reference(s) |
|---|---|---|---|---|
| Aβ(1–40) fibrils (pH 7.4) | 0.60 | 1.1 (0 M NaCl) | Abs (centrifugation) | [148] |
| 2.6 | 1.5 (0.1 M NaCl) | Abs (centrifugation) | [148] | |
| 0.43 (0 M NaCl) | – | Abs (centrifugation) | [149] | |
| 0.89 (0.14 M NaCl) | – | Abs (spectrophotometric) | [39] | |
| α-Poly-l-lysine (pH 7.4) | 36 | – | Abs (filtration) | [140] |
| β-Poly-l-lysine (pH 11) | 42 | 0.51 | Abs (filtration) | [140] |
| Insulin fibrils (pH 7.4) | 1.8 | 0.13 | Abs (spectrophotometric) | [38, 140] |
| 2.2 | 0.10 | Abs (filtration) | [38, 140] | |
| Poly-l-serine (pH 7.4) | 0.12 | 0.42 | Abs (filtration) | [140] |
| Polyglycine (pH 7.4) | 0.33 | 0.82 | Abs (filtration) | [140] |
Abs is absorbance and centrifugation, spectrophotometric and filtration refer to different methods applied to measure the concentration of bound dye. n is number of binding sites, while Kd is dissociation constant. – no data
aMoles of CR per mole of protein in fibril form
The affinity of CR towards various amyloid fibrils varies from 0.1 to 1.5 μM (Table 3) and is thus less variable than the affinity observed for the ThT binding (Table 2). This may partly be due to the affinity being insensitive to ionic strength, at least at neutral pH [129], or because most studies are done at neutral pH (Table 3) due to the insolubility of CR at acidic pH. Despite the insensitivity of the affinity towards ionic strength, the same study reveals a dramatic reduction in the binding capacity of CR at low ionic strength, indicating that CR–fibril interactions are not predominantly ionic in nature [129]. In contrast, most studies suggest that ionic interactions are important for CR binding to amyloid fibrils. For example, the stoichiometry of CR binding was decreased as pH was increased for CR binding to β-poly-l-lysine, suggesting that protonation of the poly-l-lysine is important for the binding [121]. Furthermore, the importance of the ionizable group on CR for the binding is shown by removing one of these groups and thereby reducing the affinity dramatically [73]. Inducing structural changes at the central portion of CR does not change the binding affinity significantly [73], but changing the distance of 19 Å between the di-anionic groups affected the binding constant [73]. The spacing of 19 Å corresponds to the distance between five β-strands (Fig. 2). A binding model stressing the significance of ionic interactions between the fixed distance di-anions of CR and the repetitive layer structure of amyloid fibrils has been suggested by Klunk et al. [69, 121].
Studies have also shown that hydrophobic interactions play an important role for burial of the hydrophobic part of the CR molecule [116, 120]. In addition, an increase in hydrophobicity affects the spectrophotometric properties of CR showing an intensity increase and a slight red shift in the absorption band around 498 nm [127]. This shift is in parallel with the one observed in amyloid fibrils, although the magnitude is much smaller [127]. Based on the importance of hydrophobic interactions, a binding model is suggested where the CR molecules are stabilized by being inserted into the grooves on the β-sheet surface [93, 116, 120]. In consistency with a model based on hydrophobic interactions, the β-pleated sheet conformation does not appear to be absolutely necessary for the CR binding [121, 130]. For example, CR binds to the α-helical form of poly-l-lysine [121].
Molecular form of Congo Red bound to amyloid fibrils: monomer, dimer, or micelle?
CR has been proposed to bind to amyloid fibrils in different molecular forms, such as monomeric [35, 69, 116, 125, 126, 131, 132], dimeric [116], or micellar [133–135]. CR binding as a monomer is the most widely believed hypothesis, although it is evident that CR is able to self-assemble in solution [133, 134].
Binding of CR dimers to the grooves on the surface of a β-sheet has been observed during molecular dynamics simulations, although it does not appear to be the primary binding mode (Fig. 6b) [116]. Interestingly, a CR dimer has also been shown to bind in the central cavity of γ-CD using NMR spectroscopy and isothermal titration calorimetry (ITC) [136] (Fig. 7). Thereby, a cavity with a diameter of 8–9 Å is a plausible binding site for CR, as it also is for ThT. Evidence against this binding site being the only one for CR is that CR is also able to bind to non-β-sheet proteins where such channels are not present [130, 137] and the high capacity of the CR binding site as described previously.
Fig. 7.
Binding of Thioflavin T (ThT) and Congo Red (CR) in γ-cyclodextrin (γ-CD) forming a 2:2 complex. a The energetically most favorable binding of two ThT in complex with two γ-CDs. The molecular modeling is illustrated in two views rotated 90° where the ThT ions have a red and a green surface. Reprinted from Groenning et al. [87] with permission from Elsevier. b Proposed structure of two CR in complex with two γ-CDs. Inspired by Mourtzis et al. [136]
No evidence of circular CR micelles on the surface of amyloid fibrils has been shown, but an increase in height was observed for the CR bound fibrils using AFM [89]. However, a rod- or ribbon-like micellar species of CR has been proposed to bind as a single ligand to amyloid fibrils [133–135, 138]. The micelles have been suggested to self-associate through face-to-face stacking of their elongated, symmetric polyaromatic rings with a twist of the assembled molecules to avoid direct contact between the sulfonic groups [133, 134]. These micelles are suggested to bind to the β-sheets in amyloid fibrils [133, 134].
Congo Red conformation and mechanism of the optical properties upon fibril binding
CR has a torsional angle of 20° between the two central phenyl rings in solution [132], whereas an increased planarity has been suggested upon binding to amyloid fibrils [127]. Most studies suggest that CR bound to amyloid fibrils have a planar conformation [116, 127, 132, 139], although recently a twisted conformation has also been suggested [101, 133]. The latter is based on the chirality of the CR-binding site revealed by the induced CD bands [130, 140] and a proposed conformation of CR in the ribbon-like micelles [133]. Evidence for CR binding in a planar conformation is provided by a Raman study on two conformations of CR; one having a twisted biphenyl group and one having a planar conformation [127]. The spectroscopic properties of the planar CR are similar to the ones in amyloid fibrils, in contrast to its twisted conformation [127].
The characteristic red shift in absorbance maximum from about 490 nm to about 540 nm upon binding to amyloid fibrils has been suggested to occur from an expansion of the conjugated π-electron system of CR accompanied by a conformational change [127]. Such a conformation will be provided in a planar CR conformation but may also be obtained in one of the supramolecular forms of CR.
A combination of birefringence and absorption has been shown to explain the occurrence of “apple-green” birefringence when examining a specimen between crossed polarizer and analyzer [32, 119]. Instead of the traditional “apple-green” color, a more accurate term is suggested to be anomalous colors [32, 119]. The birefringence of CR has been discussed thoroughly in the recent review by Howie et al. [32] and will not be covered in more detail here.
Plausible binding modes of Congo Red in amyloid fibrils
A planar CR form is likely to somehow bind to the predominating cross β-pleated sheets in the amyloid fibrils with the long axis parallel to the long axis of the fibril. The nature of the molecular form of CR upon binding to amyloid fibrils needs further examination to clarify whether it is the unimolecular or the supramolecular form that binds. Plausible binding locations are regular grooves formed by the β-strands along the wet side of the β-sheet extension direction, for example, at the surface of the fibril. The binding mode is likely to be a combination of hydrophobic interactions and specific interactions, such as salt bridges. The absorbance is likely to occur from formation of a more conjugated system, while a combination of absorbance and birefringence explains the occurrence of anomalous colors between crossed polarizer and analyzer.
Do Congo Red and Thioflavin T have similar binding modes?
Knowledge of the binding mode of different types of molecular probes to amyloid fibrils is important for development of new probes towards amyloid fibrils. Different distinct types of binding sites are likely to be present in amyloid fibrils [73], although both CR and ThT have been suggested to bind with the long axis parallel to the long axis of the fibril [53, 93, 116, 123].
Several studies suggest different types of binding sites. For example, three distinct types of binding sites have been identified in Aβ(1–40) fibrils: those for CR, ThT, and 2-(1-{6-[(2-[F-18]fluoroethyl) (methyl)amino]-2-naphthyl} ethylidene) malononitrile (FDDNP), respectively [73, 141, 142]. The differentiation between ThT and CR-type binding sites is suggested since these dyes do not displace each other when bound to Aβ fibrils, presumably due to each dye binding to distinct non-overlapping sites on the Aβ aggregates [142]. The opposite charges of CR and ThT have also led to the proposal that they bind at different binding sites [108, 121]. Furthermore, the large difference in binding stoichiometry indicates different binding sites, at least for ThT inducing the characteristic fluorescence and CR. Different binding sites have also been revealed for CR-type ligands, where a second binding site exists which has significantly reduced ionic interactions but increased affinity [73].
Are Thioflavin T and Congo Red amyloid-specific dyes?
Molecular probes for detection of amyloid fibrils should be specific towards amyloid due to them undergoing a spectral shift that does not occur in the presence of other forms of the proteins. Unfortunately, the specificity of the two most commonly applied dyes, ThT and CR, can be questioned, and care should be taken when interpreting the signal. For both dyes, confirmation of the presence of fibrils using complementary methods, such as electron microscopy, X-ray fiber diffraction, Fourier transform infrared spectroscopy, etc. are needed.
The binding specificity of CR to amyloid fibrils, and thus the specificity of the resulting apple-green birefringence, is generally considered to be specific for amyloid fibrils and this pattern generally distinguishes them from other fibrillary proteins, including collagen, elastin, and non-amyloid deposits [25]. This specificity, however, has recently been questioned [130, 143]. CR has been observed to bind to native proteins with various secondary structures [130, 134], as well as partially unfolded proteins [130], protein oligomers [144], and lipids [145]. Notably, the sensitivity of CR staining can depend on the method used and may make for instance collagen fibers and cytoskeletal proteins produce a false-positive birefringence [130, 146]. Finally, in a few cases, fibrils with a typical appearance of amyloid fibrils lack the ability to induce apple-green birefringence upon CR staining [147, 148].
ThT is generally considered to be more specific towards amyloid fibrils than CR, and the characteristic fluorescence of ThT does not generally occur upon binding to the precursor proteins or amorphous aggregates of proteins [40, 46, 60, 108]. There are, however, several exceptions to this general rule, e.g., the fluorescent binding of ThT to native AChE [85, 87]. In a few other cases, dimers, trimers, and larger aggregates of, for example, β-lactoglobulin cause fluorescence, indicating that ThT binding is not restricted to complete amyloid fibrils [149]. Similarly, oligomers formed early on the fibrillation pathway of TTR [21, 60], Aβ oligomers [144], and distinct β-sheet-rich insulin oligomers [150] have been shown to be ThT positive. Apart from proteins, ThT has also been shown to yield its characteristic fluorescence upon binding to DNA [118], γ-CD [87, 105, 109], and sodium dodecyl sulfate micelles [151]. Furthermore, sulfated polysaccharides, polyols, and polycationic polymers have been shown to induce the fluorescence [45]. Applying ThT to detect amyloid in tissue sections should also be interpreted with care, since it may also bind to other tissues, such as cartilage matrix, elastic fibers, and mucopolysaccharides [44, 54, 152].
Future perspectives and concluding remarks
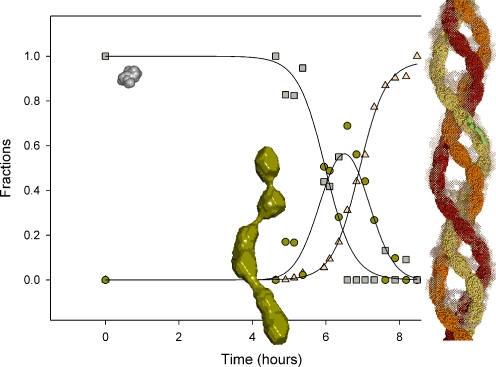
Specific dyes for amyloid fibril detection used as diagnostics tools or as assays for high-throughput screening are of great importance for increased understanding of the process of amyloidogenesis in vitro and in vivo. Since the kinetics of interconversion of all species on the reaction pathway is essential, molecular probes for detection of early events in the fibrillation process as well as different fibril morphologies are needed. Interestingly, oligomers appear to be yet a common denominator in the early stages of the amyloid fibrillation pathway, as recently shown for insulin fibrillation above the supercritical concentration [20] (Fig. 8). The cytotoxicity of the soluble, oligomeric forms is dependent on the type of oligomeric species [153–159], and existence of both on- and off-pathway oligomers has been proposed [20, 60, 150, 153, 160]. Hydrophobic probes such as 8-anilino-1-naphthalenesulfonic acid and the molecular rotor 9-dicyanovinyl-julolidine have been shown to respond to earlier species such as TTR oligomeric species [60]. Furthermore, the LCPs may be promising for oligomer detection [82], as they are already able to differentiate between different fibril morphologies. Finally, ThT and PiB, etc., could be used as lead compounds for development of imaging agents with higher affinity towards oligomers [144].
Fig. 8.
Insulin fibrillation process above supercritical concentration shown as the relative volume fractions as a function of time of the three components (monomer (squares), distinct oligomer (circles), and fibril (triangles)) in the fibrillation process of 5 mg/ml insulin in 20% acetic acid with 0.5 M NaCl (pH 2.0) at 45 °C determined using small-angle X-ray scattering (SAXS) [20]. The sizes of monomer and oligomer are on scale, the scaling to the fibril is visualized by the superposition of an oligomer onto the protofibril. The monomer is a surface representation of the isolated monomer from the hexameric structure (1MSO.pdb). The oligomer and fibril structures are ab initio models based on SAXS solution data [20]
The most often used dye for amyloid fibril detection in vitro, ThT, is not always optimal for the detection, which stresses the need for development of other complimentary probes. Some reasons for weak ThT fluorescence are: at basic pH ThT is hydroxylated [161], and some amyloid fibrils only show modest ThT fluorescence, which is dependent on morphology [55–57], pH [46, 54, 60], and protein forming the fibril [49]. Further care should be taken when applying the in situ ThT assay instead of the ex situ ThT method. For example, in several cases, polyphenolic compounds have been shown to interfere with the ThT binding and can give misleading results when examining the effect of these compounds on the possible disaggregation [162, 163]. Importantly, this finding may reflect an interconnection between the specific binding of probes and inhibition of fibrillation. Therefore, increased knowledge of the binding mode of the molecular probes, such as ThT and CR, may advance the understanding of the detection of amyloid fibrils and be useful to guide the development of new compounds in the attempt to study, monitor, and inhibit amyloid formation.
Acknowledgements
The Danish Medical Research Council, Novo Nordisk A/S, and the Drug Research Academy are thanked for financial support. The author thanks Bente Vestergaard and Per Hammarström for discussion and critical review of the manuscript.
References
- 1.Virchow R. Virchows Arch Pathol Anat Physiol Klin Med. 1854;6:416–426. [Google Scholar]
- 2.Pettersson T, Konttinen YT (2009) Semin Arthritis Rheum (in press) [DOI] [PubMed]
- 3.Sambashivan S, Eisenberg D. Bio Tech International. 2006;18(3):6–10. [Google Scholar]
- 4.Weert M, Jorgensen L, Moeller EH, et al. Expert Opin Drug Deliv. 2005;2(6):1029–1037. doi: 10.1517/17425247.2.6.1029. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen L, Frokjaer S, Carpenter JF, et al. J Pharm Sci. 2001;90(1):29–37. doi: 10.1002/1520-6017(200101)90:1<29::aid-jps4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang W. Int J Pharm. 2005;289:1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Brange J, Andersen L, Laursen ED, et al. J Pharm Sci. 1997;86(5):517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 8.Grillo AO, Edwards KT, Kashi RS, et al. Biochemistry. 2001;40:586–595. doi: 10.1021/bi001547t. [DOI] [PubMed] [Google Scholar]
- 9.Onoue S, Ohshima K, Debari K, et al. Pharm Res. 2004;21(7):1274–1283. doi: 10.1023/b:pham.0000033016.36825.2c. [DOI] [PubMed] [Google Scholar]
- 10.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 11.Maji SK, Schubert D, Rivier C, et al. PLoS Biol. 2008;6(2):e17. doi: 10.1371/journal.pbio.0060017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otzen D, Nielsen PH. Cell Mol Life Sci. 2008;65(6):910–927. doi: 10.1007/s00018-007-7404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen AS, Calkins E. Nature. 1959;183:1202–1203. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez JL, Nettleton EJ, Bouchard M, et al. Proc Natl Acad Sci U S A. 2002;99(14):9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khurana R, Ionescu-Zanetti C, Pope M, et al. Biophys J. 2003;85(2):1135–1144. doi: 10.1016/S0006-3495(03)74550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs MRH, MacPhee CE, Miller AF, et al. Proc Natl Acad Sci U S A. 2004;101(40):14420–14424. doi: 10.1073/pnas.0405933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serpell LC, Sunde M, Benson MD, et al. J Mol Biol. 2000;300(5):1033–1039. doi: 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- 18.Shirahama T, Cohen AS. J Cell Biol. 1967;33(3):679–708. doi: 10.1083/jcb.33.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westermark P, Benson MD, Buxbaum JN, et al. Amyloid. 2007;14(3):179–183. doi: 10.1080/13506120701460923. [DOI] [PubMed] [Google Scholar]
- 20.Vestergaard B, Groenning M, Roessle M, et al. PLoS Biol. 2007;5(5):e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurshman AR, White JT, Powers ET, et al. Biochemistry. 2004;43(23):7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 22.Naiki H, Gejyo F. Methods Enzymol. 1999;20:305–318. doi: 10.1016/s0076-6879(99)09022-9. [DOI] [PubMed] [Google Scholar]
- 23.Jansen R, Dzwolak W, Winter R. Biophys J. 2005;88:1344–1353. doi: 10.1529/biophysj.104.048843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fodera V, Librizzi F, Groenning M, et al. J Phys Chem B. 2008;112(12):3853–3858. doi: 10.1021/jp710131u. [DOI] [PubMed] [Google Scholar]
- 25.Giorgadze TA, Shiina N, Baloch ZW, et al. Diagn Cytopathol. 2004;31(5):300–306. doi: 10.1002/dc.20131. [DOI] [PubMed] [Google Scholar]
- 26.Nelson R, Sawaya MR, Balbirnie M, et al. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaya MR, Sambashivan S, Nelson R, et al. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 28.Mishra R, Sellin D, Radovan D, et al. ChemBioChem. 2009;10(3):445–449. doi: 10.1002/cbic.200800762. [DOI] [PubMed] [Google Scholar]
- 29.Porat Y, Abramowitz A, Gazit E. Chem Biol Drug Des. 2006;67(1):27–37. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 30.Cohen T, Frydman-Marom A, Rechter M, et al. Biochemistry. 2006;45(15):4727–4735. doi: 10.1021/bi051525c. [DOI] [PubMed] [Google Scholar]
- 31.Frid P, Anisimov SV, Popovic N. Brain Res Rev. 2007;53:135–160. doi: 10.1016/j.brainresrev.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Howie AJ, Brewer DB. Micron. 2009;40(3):285–301. doi: 10.1016/j.micron.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Ban T, Hamada D, Hasegawa K, et al. J Biol Chem. 2003;278(19):16462–16465. doi: 10.1074/jbc.C300049200. [DOI] [PubMed] [Google Scholar]
- 34.McParland VJ, Kad NM, Kalverda AP, et al. Biochemistry. 2000;39(30):8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 35.Turnell WG, Finch JT. J Mol Biol. 1992;227(4):1205–1223. doi: 10.1016/0022-2836(92)90532-o. [DOI] [PubMed] [Google Scholar]
- 36.Kim YS, Randolph TW, Manning MC, et al. J Biol Chem. 2003;278(12):10842–10850. doi: 10.1074/jbc.M212540200. [DOI] [PubMed] [Google Scholar]
- 37.Sen S, Basdemir G. Pathol Int. 2003;53(8):534–538. doi: 10.1046/j.1440-1827.2003.01513.x. [DOI] [PubMed] [Google Scholar]
- 38.Klunk WE, Pettegrew JW, Abraham DJ. J Histochem Cytochem. 1989;37(8):1293–1297. doi: 10.1177/37.8.2666512. [DOI] [PubMed] [Google Scholar]
- 39.Klunk WE, Jacob RF, Mason RP. Anal Biochem. 1999;266(1):66–76. doi: 10.1006/abio.1998.2933. [DOI] [PubMed] [Google Scholar]
- 40.LeVine H., III Arch Biochem Biophys. 1997;342(2):306–316. doi: 10.1006/abbi.1997.0137. [DOI] [PubMed] [Google Scholar]
- 41.Caughey B, Ernst D, Race RE. J Virol. 1993;67:6270–6272. doi: 10.1128/jvi.67.10.6270-6272.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorenzo A, Yankner BA. Proc Natl Acad Sci U S A. 1994;91:12243–12247. doi: 10.1073/pnas.91.25.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chander H, Chauhan A, Chauhan V. J Alzheimers Dis. 2007;12(3):261–269. doi: 10.3233/jad-2007-12308. [DOI] [PubMed] [Google Scholar]
- 44.Vassar PS, Culling CFA. Arch Pathol. 1959;68(4):487–494. [PubMed] [Google Scholar]
- 45.LeVine H., III Protein Sci. 1993;2(3):404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naiki H, Higuchi K, Hosokawa M, et al. Anal Biochem. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 47.Saeed SM, Fine G. Am J Clin Pathol. 1967;47(5):588–593. doi: 10.1093/ajcp/47.5.588. [DOI] [PubMed] [Google Scholar]
- 48.Andersen CB, Yagi H, Manno M, et al. Biophys J. 2009;96(4):1529–1536. doi: 10.1016/j.bpj.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabate R, Saupe SJ. Biochem Biophys Res Commun. 2007;360(1):135–138. doi: 10.1016/j.bbrc.2007.06.063. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen L, Khurana R, Coats A, et al. Biochemistry. 2001;40(20):6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 51.Mauro M, Craparo EF, Podesta A, et al. J Mol Biol. 2007;366(1):258–274. doi: 10.1016/j.jmb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Naiki H, Higuchi K, Nakakuki K, et al. Lab Invest. 1991;65(1):104–110. [PubMed] [Google Scholar]
- 53.Groenning M, Norrman M, Flink JM, et al. J Struct Biol. 2007;159(3):483–497. doi: 10.1016/j.jsb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 54.LeVine H., III Int J Exp Clin Invest. 1995;2:1–6. [Google Scholar]
- 55.Pedersen JS, Dikov D, Flink JL, et al. J Mol Biol. 2006;355(3):501–523. doi: 10.1016/j.jmb.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 56.Wood SJ, Maleeff B, Hart T, et al. J Mol Biol. 1996;256(5):870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 57.Kardos J, Okuno D, Kawai T, et al. Biochim Biophys Acta, Proteins Proteomics. 2005;1753(1):108–120. doi: 10.1016/j.bbapap.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 58.Wall J, Murphy CL, Solomon A. Methods Enzymol. 1999;309:204–217. doi: 10.1016/s0076-6879(99)09016-3. [DOI] [PubMed] [Google Scholar]
- 59.Ahn JS, Lee JH, Kim JH, et al. Anal Biochem. 2007;367(2):259–265. doi: 10.1016/j.ab.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 60.Lindgren M, Sörgjerd K, Hammarström P. Biophys J. 2005;88:4200–4212. doi: 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kowa H, Sakakura T, Matsuura Y, et al. Am J Pathol. 2004;165(1):273–281. doi: 10.1016/s0002-9440(10)63295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt ML, Robinson KA, Lee VMY, et al. Am J Pathol. 1995;147(2):503–515. [PMC free article] [PubMed] [Google Scholar]
- 63.Crystal AS, Giasson BI, Crowe A, et al. J Neurochem. 2003;86(6):1359–1368. doi: 10.1046/j.1471-4159.2003.01949.x. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt ML, Schuck T, Sheridan S, et al. Am J Pathol. 2001;159(3):937–943. doi: 10.1016/s0002-9440(10)61769-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Styren SD, Hamilton RL, Styren GC, et al. J Histochem Cytochem. 2000;48(9):1223–1232. doi: 10.1177/002215540004800906. [DOI] [PubMed] [Google Scholar]
- 66.Volkova KD, Kovalska VB, Balanda AO, et al. Bioorg Med Chem. 2008;16(3):1452–1459. doi: 10.1016/j.bmc.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 67.Volkova KD, Kovalska VB, Balanda AO, et al. J Biochem Biophys Methods. 2007;70(5):727–733. doi: 10.1016/j.jbbm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Luna-Munoz J, Peralta-Ramirez J, Chavez-Macias L, et al. Acta Neuropathol. 2008;116(5):507–515. doi: 10.1007/s00401-008-0431-x. [DOI] [PubMed] [Google Scholar]
- 69.Klunk WE, Debnath ML, Pettegrew JW. Neurobiol Aging. 1995;16(4):541–548. doi: 10.1016/0197-4580(95)00058-m. [DOI] [PubMed] [Google Scholar]
- 70.Klunk WE, Engler H, Nordberg A, et al. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 71.Klunk WE, Wang Y, Huang GF, et al. Life Sci. 2001;69(13):1471–1484. doi: 10.1016/s0024-3205(01)01232-2. [DOI] [PubMed] [Google Scholar]
- 72.Mathis CA, Bacskai BJ, Kajdasz ST, et al. Bioorg Med Chem Lett. 2002;12(3):295–298. doi: 10.1016/s0960-894x(01)00734-x. [DOI] [PubMed] [Google Scholar]
- 73.Cai L, Innis RB, Pike VW. Curr Med Chem. 2007;14(1):19–52. doi: 10.2174/092986707779313471. [DOI] [PubMed] [Google Scholar]
- 74.McNamee RL, Yee SH, Price JC, et al. J Nucl Med. 2009;50(3):348–355. doi: 10.2967/jnumed.108.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiley CA, Lopresti BJ, Venneti S, et al. Arch Neurol. 2009;66(1):60–67. doi: 10.1001/archneurol.2008.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sigurdson CJ, Nilsson KP, Hornemann S, et al. Nat Methods. 2007;4(12):1023–1030. doi: 10.1038/nmeth1131. [DOI] [PubMed] [Google Scholar]
- 77.Nilsson KP, Hammarstrom P, Ahlgren F, et al. ChemBioChem. 2006;7(7):1096–1104. doi: 10.1002/cbic.200500550. [DOI] [PubMed] [Google Scholar]
- 78.Nilsson KP, Aslund A, Berg I, et al. ACS Chem Biol. 2007;2(8):553–560. doi: 10.1021/cb700116u. [DOI] [PubMed] [Google Scholar]
- 79.Sigurdson CJ, Nilsson KP, Hornemann S, et al. Proc Natl Acad Sci U S A. 2009;106(1):304–309. doi: 10.1073/pnas.0810680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nilsson KPR (2009) FEBS Lett In press
- 81.Åslund A, Nilsson KPR, Konradsson P (2009) J Chem Biol (in press). doi:10.1007/s12154-009-0024-8 [DOI] [PMC free article] [PubMed]
- 82.Nilsson KPR, Hammarstrom P. Adv Mater. 2008;20:2639–2645. [Google Scholar]
- 83.Nilsson KPR, Herland A, Hammarstrom P, et al. Biochemistry. 2005;44(10):3718–3724. doi: 10.1021/bi047402u. [DOI] [PubMed] [Google Scholar]
- 84.Groenning M (2007) Studies on insulin amyloid fibrillation; formation, structure, and detection. Faculty of Pharmaceutical Sciences, University of Copenhagen
- 85.Ferrari GV, Mallender WD, Inestrosa NC. J Biol Chem. 2001;276(26):23282–23287. doi: 10.1074/jbc.M009596200. [DOI] [PubMed] [Google Scholar]
- 86.Johnson JL, Cusack B, Davies MP, et al. Biochemistry. 2003;42(18):5438–5452. doi: 10.1021/bi027065u. [DOI] [PubMed] [Google Scholar]
- 87.Groenning M, Olsen L, Weert M, et al. J Struct Biol. 2007;158:358–369. doi: 10.1016/j.jsb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 88.Cundall RB, Davies AK, Morris PG, et al. J Photochem. 1981;17:369–376. [Google Scholar]
- 89.Khurana R, Coleman C, Ionescu-Zanetti C, et al. J Struct Biol. 2005;151(3):229–238. doi: 10.1016/j.jsb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 90.Sabate R, Lascu I, Saupe SJ. J Struct Biol. 2008;162(3):387–396. doi: 10.1016/j.jsb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 91.Lockhart A, Ye L, Judd DB, et al. J Biol Chem. 2004;280(9):7677–7684. doi: 10.1074/jbc.M412056200. [DOI] [PubMed] [Google Scholar]
- 92.Bourhim M, Kruzel M, Srikrishnan T, et al. J Neurosci Methods. 2007;160(2):264–268. doi: 10.1016/j.jneumeth.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 93.Krebs MRH, Bromley EHC, Donald AM. J Struct Biol. 2005;149(1):30–37. doi: 10.1016/j.jsb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Biancalana M, Makabe K, Koide A, et al. J Mol Biol. 2009;385(4):1052–1063. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu C, Wang Z, Lei H, et al. J Mol Biol. 2008;384(3):718–729. doi: 10.1016/j.jmb.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eanes ED, Glenner GG. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 97.Serpell LC, Smith JM. J Mol Biol. 2000;299(1):225–231. doi: 10.1006/jmbi.2000.3650. [DOI] [PubMed] [Google Scholar]
- 98.Stsiapura VI, Maskevich AA, Kuzmitsky VA, et al. J Phys Chem A. 2007;111(22):4829–4835. doi: 10.1021/jp070590o. [DOI] [PubMed] [Google Scholar]
- 99.Voropai ES, Samtsov MP, Kaplevskii KN, et al. J Appl Spectrosc. 2003;70(6):868–874. [Google Scholar]
- 100.Harel M, Sonoda LK, Silman I, et al. J Am Chem Soc. 2008;130(25):7856–7861. doi: 10.1021/ja7109822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dzwolak W, Pecul M. FEBS L. 2005;579(29):6601–6603. doi: 10.1016/j.febslet.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 102.Loksztejn A, Dzwolak W. J Mol Biol. 2008;379(1):9–16. doi: 10.1016/j.jmb.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 103.Stsiapura VI, Maskevich AA, Kuzmitsky VA et al (2008) J Phys Chem B 112(49):15893–15902 [DOI] [PubMed]
- 104.Friedhoff P, Schneider A, Mandelkow E-M, et al. Biochemistry. 1998;37(28):10223–10230. doi: 10.1021/bi980537d. [DOI] [PubMed] [Google Scholar]
- 105.Retna Raj C, Ramaraj R. J Photochem Photobiol A. 1999;122(1):39–46. [Google Scholar]
- 106.Maskevich AA, Stsiapura VI, Kuzmitsky VA, et al. J Proteome Res. 2007;6(4):1392–1401. doi: 10.1021/pr0605567. [DOI] [PubMed] [Google Scholar]
- 107.Pedersen JS (2006) In vitro studies of amyloid-like protein fibrils using glucagon as model system. Aalborg University.
- 108.LeVine H., III Methods Enzymol. 1999;309(18):274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 109.Retna Raj C, Ramaraj R. Chem Phys Lett. 1997;273(3–4):285–290. [Google Scholar]
- 110.Blake C, Serpell L. Structure. 1996;4(8):989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 111.Perutz MF, Finch JT, Berriman J, et al. Proc Natl Acad Sci U S A. 2002;99(8):5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jimenez JL, Guijarro JI, Orlova E, et al. EMBO J. 1999;18(4):815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Serpell LC, Sunde M, Fraser PE, et al. J Mol Biol. 1995;254(2):113–118. doi: 10.1006/jmbi.1995.0604. [DOI] [PubMed] [Google Scholar]
- 114.Elam JS, Taylor AB, Strange R, et al. Nat Struct Biol. 2003;10(6):461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 115.Malinchik SB, Inouye H, Szumowski KE, et al. Biophys J. 1998;74(1):537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu C, Wang Z, Lei H, et al. J Am Chem Soc. 2007;129(5):1225–1232. doi: 10.1021/ja0662772. [DOI] [PubMed] [Google Scholar]
- 117.Lakowicz JR. Principles of fluorescence spectroscopy. 2. New York: Kluwer; 1999. [Google Scholar]
- 118.Ilanchelian M, Ramaraj R. J Photochem Photobiol A. 2004;162(1):129–137. [Google Scholar]
- 119.Howie AJ, Brewer DB, Howell D, et al. Lab Invest. 2008;88(3):232–242. doi: 10.1038/labinvest.3700714. [DOI] [PubMed] [Google Scholar]
- 120.Cooper JH. Lab Invest. 1974;31(3):232–238. [PubMed] [Google Scholar]
- 121.Klunk WE, Pettegrew JW, Abraham DJ. J Histochem Cytochem. 1989;37(8):1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 122.Glenner GG. New Engl J Med. 1980;302(23):1283–1292. doi: 10.1056/NEJM198006053022305. [DOI] [PubMed] [Google Scholar]
- 123.Jin LW, Claborn KA, Kurimoto M, et al. Proc Natl Acad Sci U S A. 2003;100(26):15294–15298. doi: 10.1073/pnas.2534647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Romhanyi G. Virchows Arch A Pathol Pathol Anat. 1971;354(3):209–222. doi: 10.1007/BF00544254. [DOI] [PubMed] [Google Scholar]
- 125.Carter DB, Chou KC. Neurobiol Aging. 1998;19(1):37–40. doi: 10.1016/s0197-4580(97)00164-4. [DOI] [PubMed] [Google Scholar]
- 126.Li L, Darden TA, Bartolotti L, et al. Biophys J. 1999;76(6):2871–2878. doi: 10.1016/S0006-3495(99)77442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miura T, Yamamiya C, Sasaki M, et al. J Raman Spectrosc. 2002;33:530–535. [Google Scholar]
- 128.Nilsson MR. Methods. 2004;34(1):151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 129.Zhen W, Han H, Anguiano M, et al. J Med Chem. 1999;42(15):2805–2815. doi: 10.1021/jm990103w. [DOI] [PubMed] [Google Scholar]
- 130.Khurana R, Uversky VN, Nielsen L, et al. J Biol Chem. 2001;276(25):22715–22721. doi: 10.1074/jbc.M011499200. [DOI] [PubMed] [Google Scholar]
- 131.Klunk WE, Debnath ML, Pettegrew JW. Neurobiol Aging. 1994;15(6):691–698. doi: 10.1016/0197-4580(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 132.Cavillon F, Elhaddaoui A, Alix AJP, et al. J Mol Struct. 1997;408/409:185–189. [Google Scholar]
- 133.skowronek M, Stopa B, Konieczny L, et al. Biopolymers. 1998;46:267–281. [Google Scholar]
- 134.Stopa B, Piekarska B, Konieczny L, et al. Acta Biochim Pol. 2003;50(4):1213–1227. [PubMed] [Google Scholar]
- 135.Roterman I, Król M, Nowak M, et al. Med Sci Monit. 2001;7(4):771–784. [PubMed] [Google Scholar]
- 136.Mourtzis N, Cordoyiannis G, Nounesis G, et al. Supramol Chem. 2003;15(7–8):639–649. [Google Scholar]
- 137.Sereikaite J, Bumelis VA. Acta Biochim Pol. 2006;53(1):87–92. [PubMed] [Google Scholar]
- 138.stopa B, Gorny M, Konieczny L, et al. Biochimie. 1998;80(12):963–968. doi: 10.1016/s0300-9084(99)80001-7. [DOI] [PubMed] [Google Scholar]
- 139.Demaimay R, Harper J, Gordon H, et al. J Neurochem. 1998;71(6):2534–2541. doi: 10.1046/j.1471-4159.1998.71062534.x. [DOI] [PubMed] [Google Scholar]
- 140.Benditt EP, Eriksen N, Berglund C. Proc Natl Acad Sci U S A. 1970;66(4):1044–1051. doi: 10.1073/pnas.66.4.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Agdeppa ED, Kepe V, Liu J, et al. J Neurosci. 2001;21:189–193. doi: 10.1523/JNEUROSCI.21-24-j0004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhuang ZP, Kung MP, Hou C, et al. J Med Chem. 2001;44(12):1905–1914. doi: 10.1021/jm010045q. [DOI] [PubMed] [Google Scholar]
- 143.Bousset L, Redeker V, Decottignies P, et al. Biochemistry. 2004;43:5022–5032. doi: 10.1021/bi049828e. [DOI] [PubMed] [Google Scholar]
- 144.Maezawa I, Hong HS, Liu R, et al. J Neurochem. 2008;104(2):457–468. doi: 10.1111/j.1471-4159.2007.04972.x. [DOI] [PubMed] [Google Scholar]
- 145.Hahn Ch, Kaiser S, Wokaun A. Tenside Surf Det. 1996;33(3):209–213. [Google Scholar]
- 146.Bely M, Makovitzky J. Acta Histochem. 2006;108(3):175–180. doi: 10.1016/j.acthis.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 147.Booth DR, Sunde M, Bellotti V, et al. Nature. 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 148.Chaiban JT, Kalache SM, bu Alfa AK, et al. Am J Med Sci. 2008;336(3):293–296. doi: 10.1097/MAJ.0b013e31815b9d51. [DOI] [PubMed] [Google Scholar]
- 149.Carrotta R, Bauer R, Waninge R, et al. Protein Sci. 2001;10(7):1312–1318. doi: 10.1110/ps.42501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Grudzielanek S, Smirnovas V, Winter R. J Mol Biol. 2006;356(2):497–509. doi: 10.1016/j.jmb.2005.11.075. [DOI] [PubMed] [Google Scholar]
- 151.Kumar S, Singh AK, Krishnamoorthy G, et al. J Fluoresc. 2008;18(6):1199–1205. doi: 10.1007/s10895-008-0378-2. [DOI] [PubMed] [Google Scholar]
- 152.Kelenyi G. J Histochem Cytochem. 1967;15(3):172–180. doi: 10.1177/15.3.172. [DOI] [PubMed] [Google Scholar]
- 153.Hoshi M, Sato M, Matsumoto S, et al. Proc Natl Acad Sci U S A. 2003;100(11):6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bucciantini M, Giannoni E, Chiti F, et al. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 155.Conway KA, Lee SJ, Rochet JC, et al. Proc Natl Acad Sci U S A. 2000;97(2):571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lambert MP, Barlow AK, Chromy BA, et al. Proc Natl Acad Sci U S A. 1998;95(11):6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nilsberth C, Westlind-Danielsson A, Eckman CB, et al. Nat Neurosci. 2001;4(9):887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 158.Sousa MM, Cardoso I, Fernandes R, et al. Am J Pathol. 2001;159(6):1993–2000. doi: 10.1016/s0002-9440(10)63050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stefani M, Dobson CM. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 160.Morozova-Roche LA, Zamotin V, Malisauskas M, et al. Biochemistry. 2004;43(30):9610–9619. doi: 10.1021/bi0494121. [DOI] [PubMed] [Google Scholar]
- 161.Fodera V, Groenning M, Vetri V, et al. J Phys Chem B. 2008;112(47):15174–15181. doi: 10.1021/jp805560c. [DOI] [PubMed] [Google Scholar]
- 162.Meng F, Marek P, Potter KJ, et al. Biochemistry. 2008;47(22):6016–6024. doi: 10.1021/bi702518m. [DOI] [PubMed] [Google Scholar]
- 163.Kroes-Nijboer A, Lubbersen YS, Venema P, et al. J Struct Biol. 2009;165(3):140–145. doi: 10.1016/j.jsb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 164.Petkova AT, Buntkowsky G, Dyda F, et al. J Mol Biol. 2004;335(1):247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 165.Sikorski P, Atkins E. Biomacromolecules. 2005;6(1):425–432. doi: 10.1021/bm0494388. [DOI] [PubMed] [Google Scholar]
- 166.Losic D, Martin LL, Mechler A, et al. J Struct Biol. 2006;155(1):104–110. doi: 10.1016/j.jsb.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 167.Balbach JJ, Petkova AT, Oyler NA, et al. Biophys J. 2002;83(2):1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Benzinger TL, Gregory DM, Burkoth TS, et al. Proc Natl Acad Sci U S A. 1998;95(23):13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dobson CM. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]