Abstract
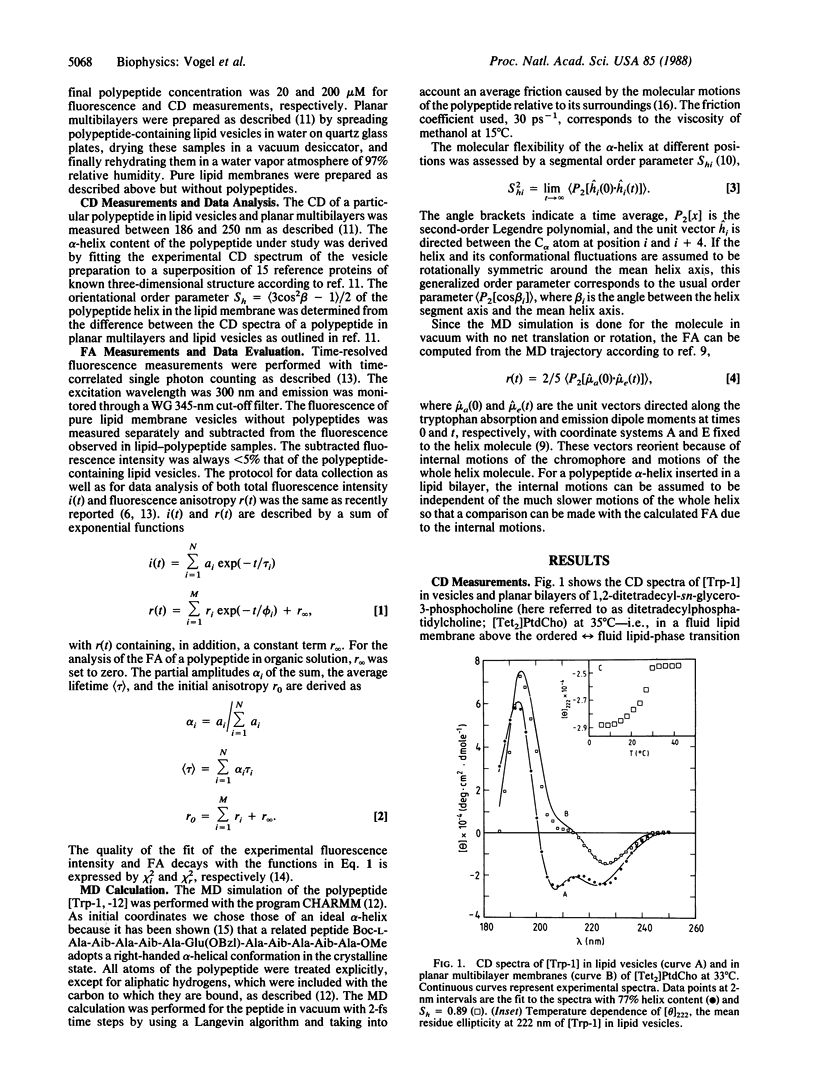
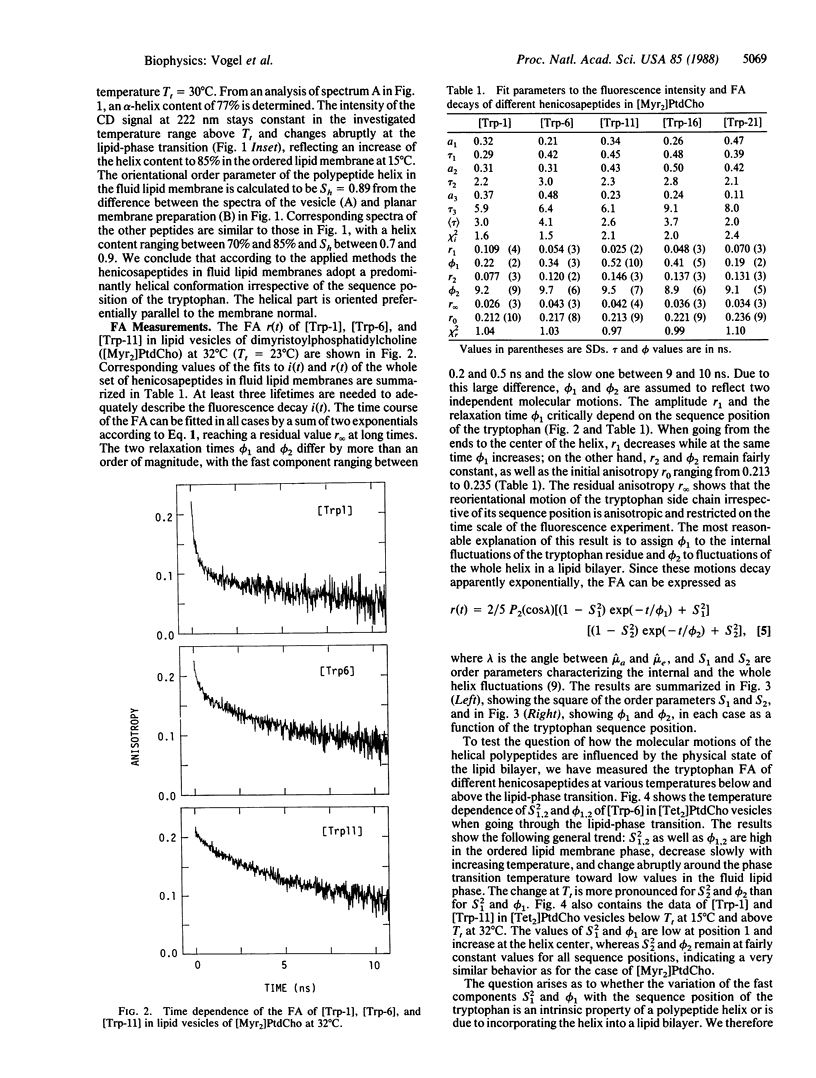
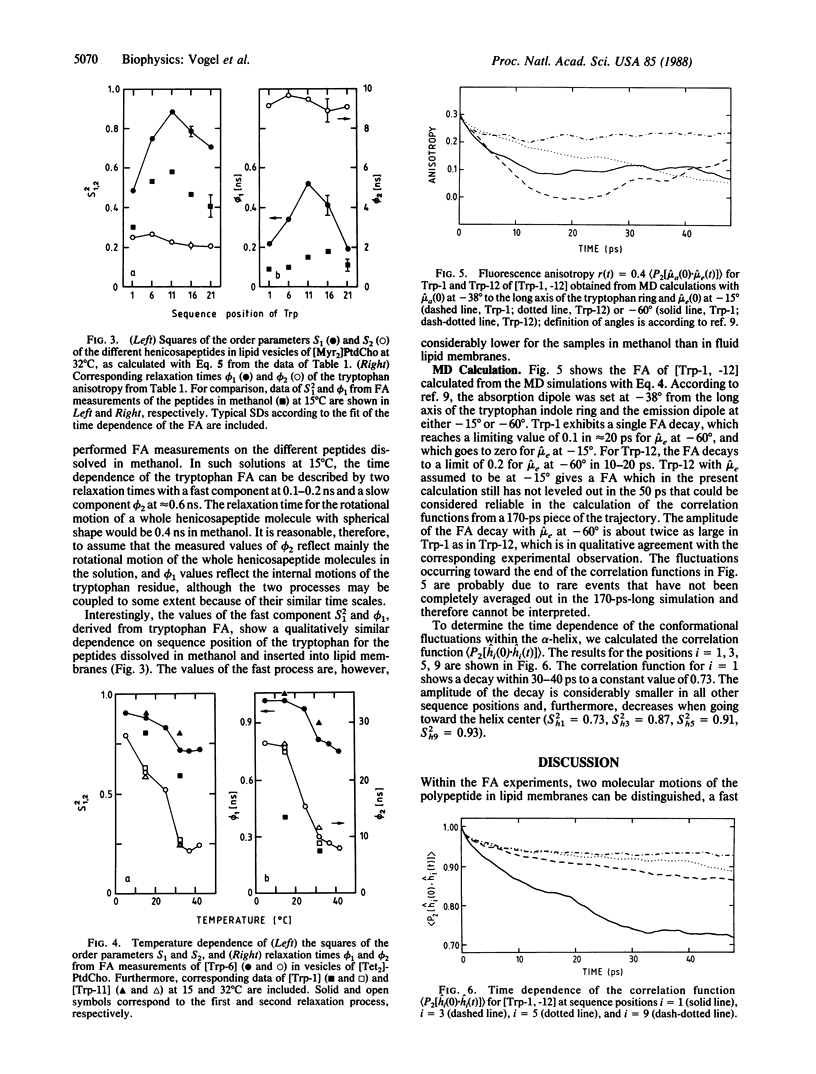
Time-resolved fluorescence anisotropy (FA) measurements are reported for five helical bilayer-spanning henicosapeptides, each containing one tryptophan at sequence position 1, 6, 11, 16, or 21. The FA decay reflects two molecular processes in all cases: local internal fluctuations of the tryptophan side chain with a relaxation time of 200-500 ps, and motions of the whole polypeptide molecule with a relaxation time of 9-10 ns. The amplitudes of the fast fluctuation are largest at the helix ends and decrease toward the center of the helix. A similar observation was made for the helical polypeptides dissolved in organic solvents showing that the mobility gradient along the polypeptide sequence is an inherent property of the polypeptide helix and not due to the lipid bilayer. However, the amplitudes of the fast fluctuations can be modulated by the physical state of the lipid bilayer. With decreasing temperature, the internal mobility of the tryptophan in all positions decreases with an abrupt change at the lipid-phase transition. Concomitant molecular dynamics calculations indicate a correlation between the fast FA decay and conformational fluctuations within the helix. According to the simulation, the conformation of the polypeptide is on average predominantly helical, but actually the molecule can fluctuate between a variety of different substructures. The conformational fluctuations are largest at the helix ends and provide the free space required for rotation of the indole ring around the tryptophan side chain bonds.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brooks C. L., 3rd, Brünger A., Karplus M. Active site dynamics in protein molecules: a stochastic boundary molecular-dynamics approach. Biopolymers. 1985 May;24(5):843–865. doi: 10.1002/bip.360240509. [DOI] [PubMed] [Google Scholar]
- Cherry R. J., Godfrey R. E. Anisotropic rotation of bacteriorhodopsin in lipid membranes. Comparison of theory with experiment. Biophys J. 1981 Oct;36(1):257–276. doi: 10.1016/S0006-3495(81)84727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H., Petsko G. A., Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979 Aug 16;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- Ichiye T., Karplus M. Fluorescence depolarization of tryptophan residues in proteins: a molecular dynamics study. Biochemistry. 1983 Jun 7;22(12):2884–2893. doi: 10.1021/bi00281a017. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Kinosita K., Jr, Kawato S., Ikegami A. Dynamic structure of biological and model membranes: analysis by optical anisotropy decay measurement. Adv Biophys. 1984;17:147–203. doi: 10.1016/0065-227x(84)90027-3. [DOI] [PubMed] [Google Scholar]
- Levy R. M., Perahia D., Karplus M. Molecular dynamics of an alpha-helical polypeptide: Temperature dependence and deviation from harmonic behavior. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1346–1350. doi: 10.1073/pnas.79.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A. D., Jr, Rigler R., Nilsson L., Hahn U., Saenger W. Protein dynamics. A time-resolved fluorescence, energetic and molecular dynamics study of ribonuclease T1. Biophys Chem. 1987 May 9;26(2-3):247–261. doi: 10.1016/0301-4622(87)80027-3. [DOI] [PubMed] [Google Scholar]
- Maliwal B. P., Hermetter A., Lakowicz J. R. A study of protein dynamics from anisotropy decays obtained by variable frequency phase-modulation fluorometry: internal motions of N-methylanthraniloyl melittin. Biochim Biophys Acta. 1986 Sep 26;873(2):173–181. doi: 10.1016/0167-4838(86)90043-9. [DOI] [PubMed] [Google Scholar]
- Munro I., Pecht I., Stryer L. Subnanosecond motions of tryptophan residues in proteins. Proc Natl Acad Sci U S A. 1979 Jan;76(1):56–60. doi: 10.1073/pnas.76.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigler R., Ehrenberg M. Fluorescence relaxation spectroscopy in the analysis of macromolecular structure and motion. Q Rev Biophys. 1976 Feb;9(1):1–19. doi: 10.1017/s0033583500002122. [DOI] [PubMed] [Google Scholar]
- Seelig J., Seelig A. Lipid conformation in model membranes and biological membranes. Q Rev Biophys. 1980 Feb;13(1):19–61. doi: 10.1017/s0033583500000305. [DOI] [PubMed] [Google Scholar]
- Vogel H. Comparison of the conformation and orientation of alamethicin and melittin in lipid membranes. Biochemistry. 1987 Jul 14;26(14):4562–4572. doi: 10.1021/bi00388a060. [DOI] [PubMed] [Google Scholar]
- Voges K. P., Jung G., Sawyer W. H. Depth-dependent fluorescent quenching of a tryptophan residue located at defined positions on a rigid 21-peptide helix in liposomes. Biochim Biophys Acta. 1987 Jan 9;896(1):64–76. doi: 10.1016/0005-2736(87)90357-9. [DOI] [PubMed] [Google Scholar]



